Identification of a 2′-O-Methyluridine Nucleoside Hydrolase Using the Metagenomic Libraries
Abstract
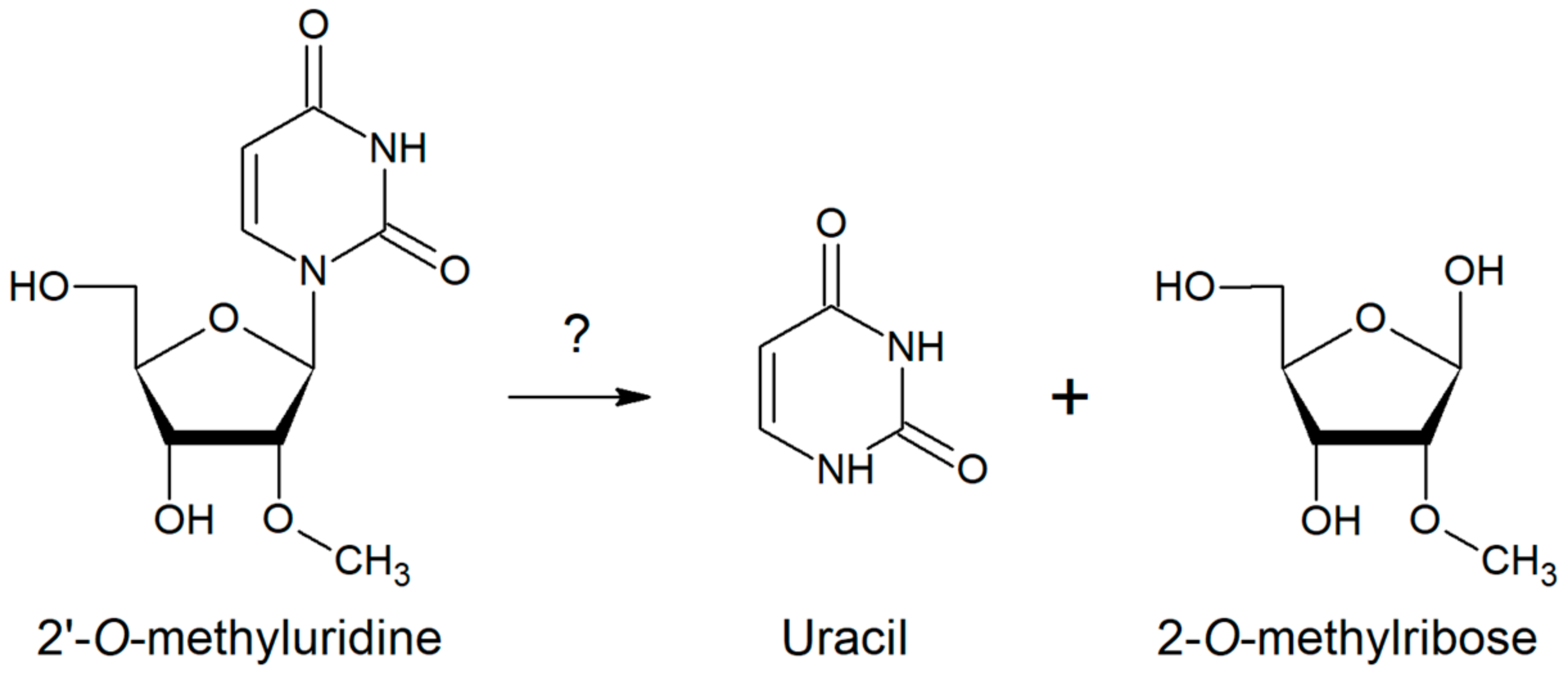
1. Introduction
2. Results
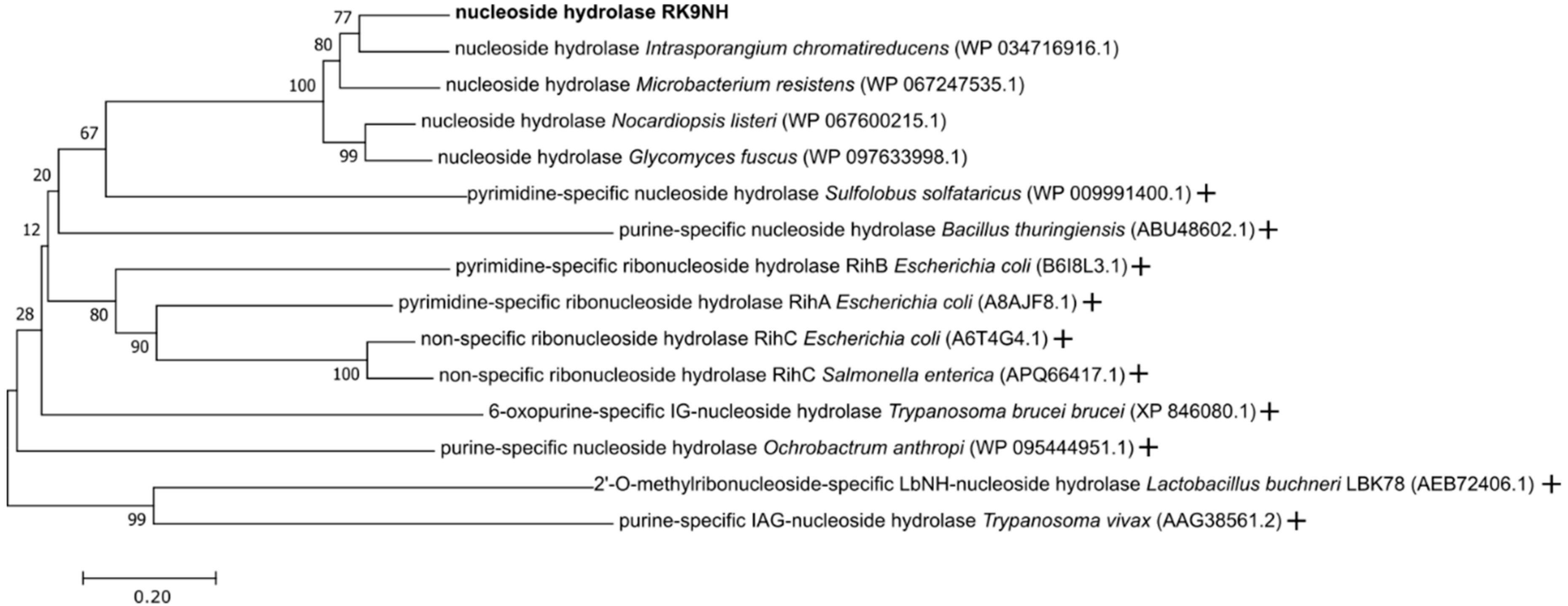
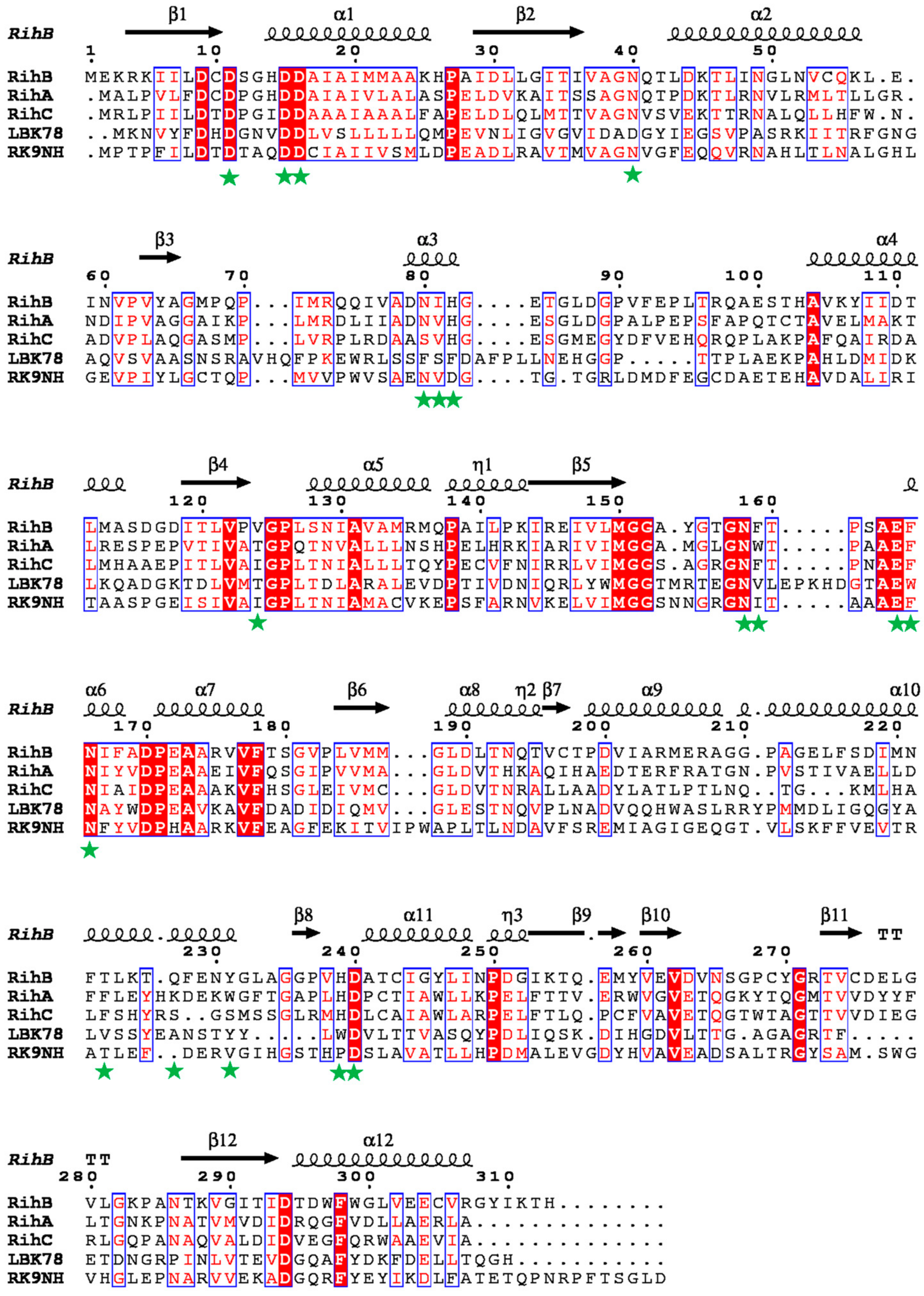
2.1. Discovery of RK9 Nucleoside Hydrolase and Deoxyribose-Phosphate Aldolase
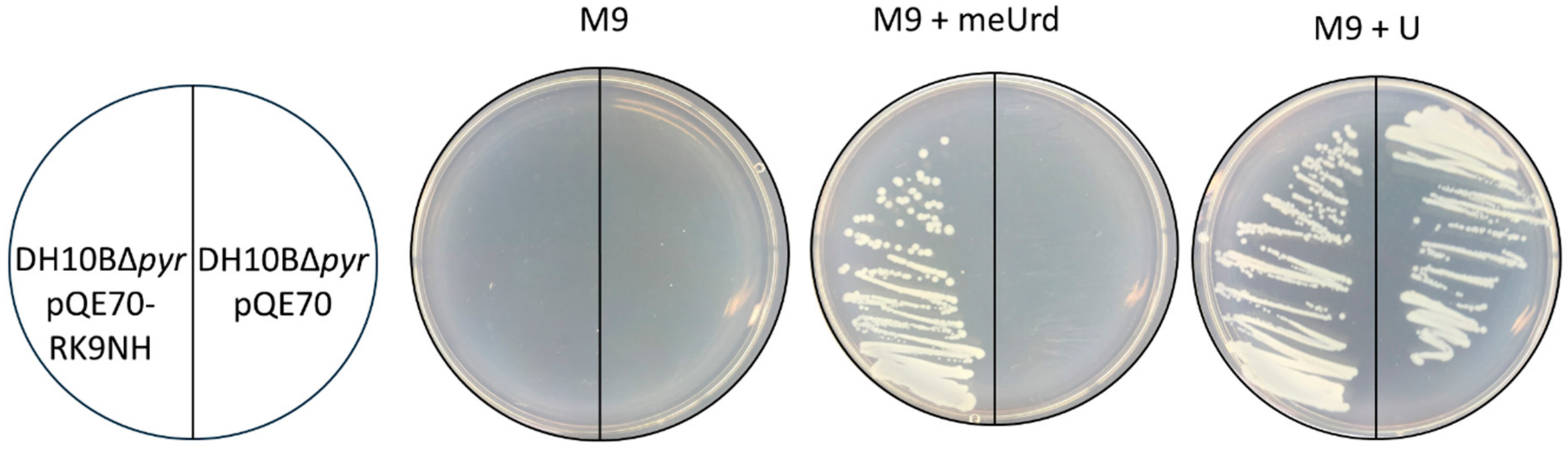
2.2. Activity of RK9NH and RK9DPA Proteins in E. coli Cells
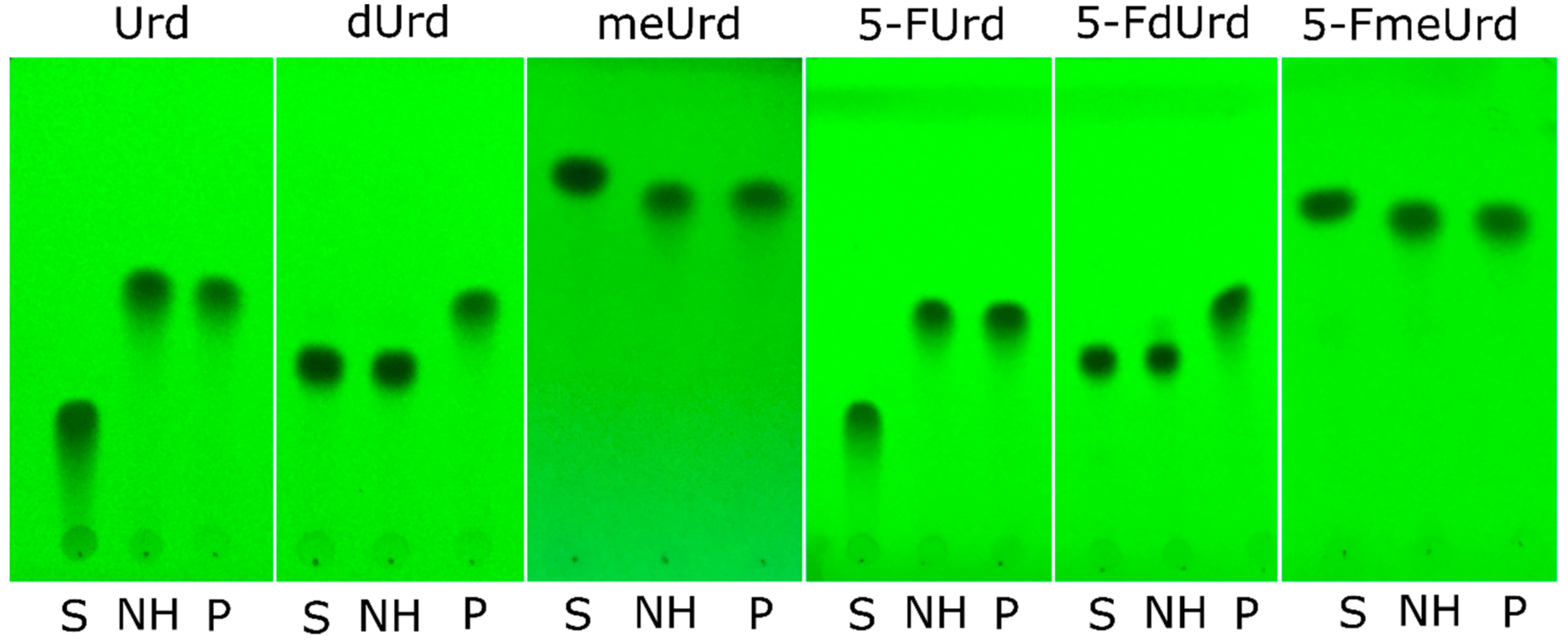
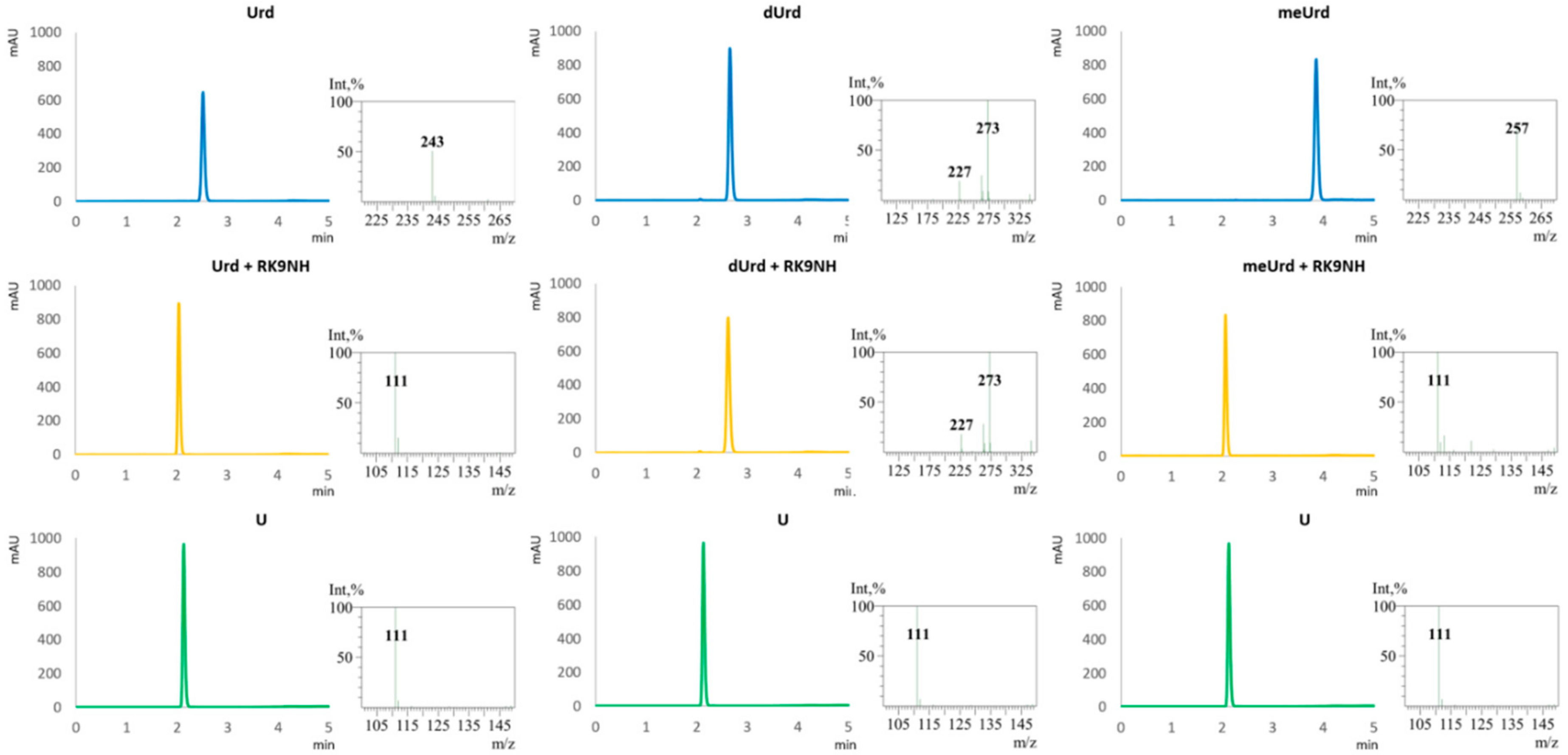
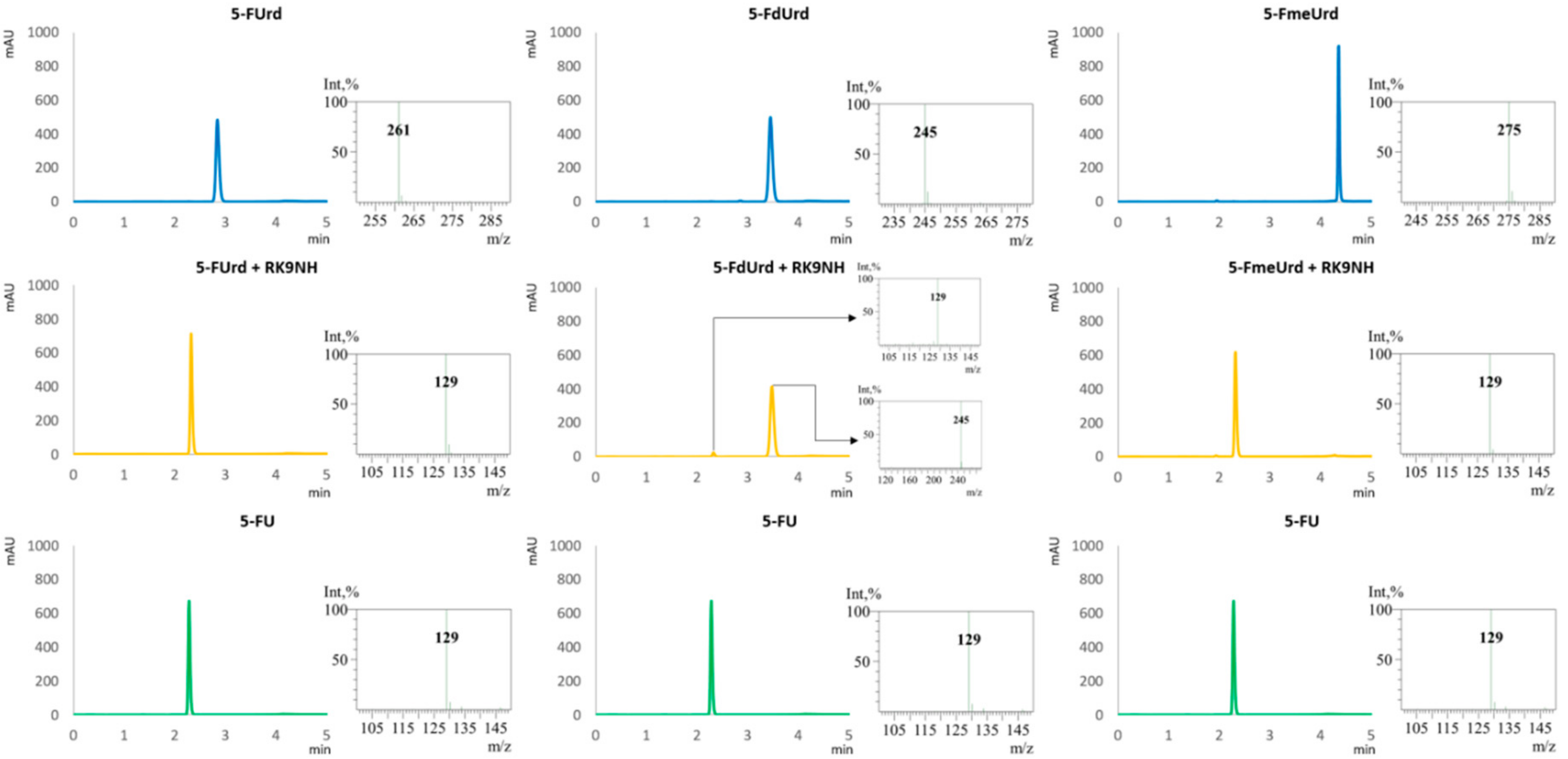
2.3. Substrate Specificity of RK9NH Protein
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, Primers, Media and Reagents
4.2. The E. coli Uracil Auxotroph Strain and the Metagenomic Libraries
4.3. Synthesis of 5-fluoro-2′-O-methyluridine
4.4. Over-Expression and Purification of the Recombinant RK9NH Protein
4.5. Substrate Specificity Measurements
4.6. Thin-Layer Chromatography (TLC)
4.7. HPLC-MS
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P.; Bagiński, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Dezi, V.; Ivanov, C.; Haussmann, I.U.; Soller, M. Nucleotide modifications in messenger RNA and their role in development and disease. Biochem. Soc. Trans. 2016, 44, 1385–1393. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.S.; Sarin, L.P. Transfer RNA modification and infection—Implications for pathogenicity and host responses. Biochim. Biophys. Acta 2018, 1861, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cozen, A.E.; Liu, Y.; Chen, Q.; Lowe, T.M. Small RNA modifications: Integral to function and disease. Trends Mol. Med. 2016, 22, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Sergiev, P.V.; Aleksashin, N.A.; Chugunova, A.A.; Polikanov, Y.S.; Dontsova, O.A. Structural and evolutionary insights into ribosomal RNA methylation. Nat. Chem. Biol. 2018, 14, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Jöckel, S.; Nees, G.; Sommer, R.; Zhao, Y.; Cherkasov, D.; Hori, H.; Ehm, G.; Schnare, M.; Nain, M.; Kaufmann, A.; et al. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 2012, 209, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.-H.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef] [PubMed]
- Züst, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Helm, M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006, 34, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.J.; Kim, H.-W.; Yoon, H.-J.; Lee, B.I.; Suh, S.W.; Yang, J.K. Crystal structure of tRNA(m1G37)methyltransferase: Insights into tRNA recognition. EMBO J. 2003, 22, 2593–2603. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Páez, A.; Villarroya, M.; Armengod, M.-E. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA 2012, 18, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
- Björk, G.R.; Hagervall, T.G. Transfer RNA modification: Presence, synthesis, and function. EcoSal Plus 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bailly, M.; de Crécy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- De Crécy-Lagard, V.; Brochier-Armanet, C.; Urbonavicius, J.; Fernandez, B.; Phillips, G.; Lyons, B.; Noma, A.; Alvarez, S.; Droogmans, L.; Armengaud, J.; et al. Biosynthesis of wyosine derivatives in tRNA: An ancient and highly diverse pathway in Archaea. Mol. Biol. Evol. 2010, 27, 2062–2077. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Lai, S.; Jia, R.; Xu, A.; Zhang, L.; Lu, J.; Ye, K. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature 2011, 469, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Somme, J.; Van Laer, B.; Roovers, M.; Steyaert, J.; Versées, W.; Droogmans, L. Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA 2014, 20, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.S.; Fedorov, A.A.; Marti-Arbona, R.; Fedorov, E.V.; Kolb, P.; Sauder, J.M.; Burley, S.K.; Shoichet, B.K.; Almo, S.C.; Raushel, F.M. The hunt for 8-oxoguanine deaminase. J. Am. Chem. Soc. 2010, 132, 1762–1763. [Google Scholar] [CrossRef] [PubMed]
- Preumont, A.; Snoussi, K.; Stroobant, V.; Collet, J.-F.; Van Schaftingen, E. Molecular identification of pseudouridine-metabolizing enzymes. J. Biol. Chem. 2008, 283, 25238–25246. [Google Scholar] [CrossRef] [PubMed]
- Seley-Radtke, K.L.; Yates, M.K. The evolution of nucleoside analogue antivirals: A review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antiviral Res. 2018, 154, 66–86. [Google Scholar] [CrossRef] [PubMed]
- Makita, S.; Maeshima, A.M.; Maruyama, D.; Izutsu, K.; Tobinai, K. Forodesine in the treatment of relapsed/refractory peripheral T-cell lymphoma: An evidence-based review. OncoTargets Ther. 2018, 11, 2287–2293. [Google Scholar] [CrossRef] [PubMed]
- Maio, G.; Enweronye, O.; Zumrut, H.E.; Batool, S.; Van, N.; Mallikaratchy, P. Systematic optimization and modification of a DNA aptamer with 2′-O-methyl RNA analogues. ChemistrySelect 2017, 2, 2335–2340. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Mapa, K.; Maiti, S. Understanding the effect of locked nucleic acid and 2′-O-methyl modification on the hybridization thermodynamics of a miRNA-mRNA pair in the presence and absence of AfPiwi protein. Biochemistry 2014, 53, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, I.; Kierzek, E.; Kierzek, R.; Schatz, G.C. Interplay of LNA and 2′-O-methyl RNA in the structure and thermodynamics of RNA hybrid systems: A molecular dynamics study using the revised AMBER force field and comparison with experimental results. J. Phys. Chem. B 2014, 118, 14177–14187. [Google Scholar] [CrossRef] [PubMed]
- Mitsukawa, Y.; Hibi, M.; Matsutani, N.; Horinouchi, N.; Takahashi, S.; Ogawa, J. A novel nucleoside hydrolase from Lactobacillus buchneri LBK78 catalyzing hydrolysis of 2′-O-methylribonucleosides. Biosci. Biotechnol. Biochem. 2016, 80, 1568–1576. [Google Scholar] [CrossRef] [PubMed]
- Mitsukawa, Y.; Hibi, M.; Matsutani, N.; Horinouchi, N.; Takahashi, S.; Ogawa, J. Enzymatic synthesis of 2′-O-methylribonucleosides with a nucleoside hydrolase family enzyme from Lactobacillus buchneri LBK78. J. Biosci. Bioeng. 2017, 123, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Mitsukawa, Y.; Hibi, M.; Matsutani, N.; Horinouchi, N.; Takahashi, S.; Ogawa, J. New nucleoside hydrolase with transribosylation activity from Agromyces sp. MM-1 and its application for enzymatic synthesis of 2′-O-methylribonucleosides. J. Biosci. Bioeng. 2018, 125, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Aučynaitė, A.; Rutkienė, R.; Gasparavičiūtė, R.; Meškys, R.; Urbonavičius, J. A gene encoding a DUF523 domain protein is involved in the conversion of 2-thiouracil into uracil. Environ. Microbiol. Rep. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.; Møller, L.B. The RihA, RihB, and RihC ribonucleoside hydrolases of Escherichia coli. Substrate specificity, gene expression, and regulation. J. Biol. Chem. 2001, 276, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Heine, A.; DeSantis, G.; Luz, J.G.; Mitchell, M.; Wong, C.H.; Wilson, I.A. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science 2001, 294, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence, in Proteins; Elsevier: Amsterdam, The Netherlands, 1965. [Google Scholar]
- Giabbai, B.; Degano, M. Crystal structure to 1.7 Å of the Escherichia coli pyrimidine nucleoside hydrolase YeiK, a novel candidate for cancer gene therapy. Structure 2004, 12, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Han, T.K.; Zhu, Z.; Dao, M.L. Identification, molecular cloning, and sequence analysis of a deoxyribose aldolase in Streptococcus mutans GS-5. Curr. Microbiol. 2004, 48, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Popovic, A.; Tchigvintsev, A.; Tran, H.; Chernikova, T.N.; Golyshina, O.V.; Yakimov, M.M.; Golyshin, P.N.; Yakunin, A.F. Metagenomics as a tool for enzyme discovery: Hydrolytic enzymes from marine-related metagenomes. Adv. Exp. Med. Biol. 2015, 883, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Mikhailopulo, I.A.; Miroshnikov, A.I. Biologically important nucleosides: Modern trends in biotechnology and application. Mendeleev Commun. 2011, 21, 57–68. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Malekshah, O.M.; Chen, X.; Nomani, A.; Sarkar, S.; Hatefi, A. Enzyme/prodrug systems for cancer gene therapy. Curr. Pharmacol. Rep. 2016, 2, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Aučynaitė, A.; Rutkienė, R.; Tauraitė, D.; Meškys, R.; Urbonavičius, J. Discovery of bacterial deaminases that convert 5-fluoroisocytosine into 5-fluorouracil. Front. Microbiol. 2018, 9, 2375. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Lab Press: New York, NY, USA, 2001. [Google Scholar]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K.; Wiley, C.J.; Allison, R.D.; Bittner, M.; et al. Current Protocols in Molecular Biology; Wiley: Hoboken, NJ, USA, 2003; ISBN 047150338X. [Google Scholar]
- Urbelienė, N.; Kutanovas, S.; Meškienė, R.; Gasparavičiūtė, R.; Tauraitė, D.; Koplūnaitė, M.; Meškys, R. Application of the uridine auxotrophic host and synthetic nucleosides for a rapid selection of hydrolases from metagenomic libraries. Microb. Biotechnol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
Sample Availability: Samples of the plasmids created in this work are available from the authors on request. |
| Strain | Carbon Source | ||
|---|---|---|---|
| None | Glucose | Thymidine | |
| E. coli BW25113 | - | ++ | + |
| E. coli BW25113 deoC::kan | - | ++ | - |
| E. coli BW25113 deoC::kan pQE70-RK9DPA | - | ++ | - |
| Substrate | Conversion (%) |
|---|---|
| Uridine | 100 |
| 2′-Deoxyuridine | 0 |
| 2′-O-methyluridine | 100 |
| 3′-O-methyluridine | 0 |
| 2′-O-allyluridine | 0 |
| 3′-O-allyluridine | 0 |
| 5-Fluorouridine | 100 |
| 5-Fluoro-2′-deoxyuridine | 4 ± 0.1 |
| 5-Fluoro-2′-O-methyluridine | 100 |
| 2′-Amino-2′-deoxyuridine | 0 |
| Cytidine | 65 ± 1 |
| 2′-Deoxycytidine | 0 |
| 2′-O-methylcytidine | 70 ± 1 |
| 5-Methyluridine | 100 |
| Thymidine | 0 |
| Adenosine | 10 ± 0.2 |
| 2′-Deoxyadenosine | 0 |
| 2′-O-methyladenosine | 5 ± 0.1 |
| Guanosine | 100 |
| 2′-Deoxyguanosine | 3 ± 0.1 |
| 2′-O-methylguanosine | 5 ± 0.1 |
| Inosine | 22 ± 0.5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aučynaitė, A.; Rutkienė, R.; Tauraitė, D.; Meškys, R.; Urbonavičius, J. Identification of a 2′-O-Methyluridine Nucleoside Hydrolase Using the Metagenomic Libraries. Molecules 2018, 23, 2904. https://doi.org/10.3390/molecules23112904
Aučynaitė A, Rutkienė R, Tauraitė D, Meškys R, Urbonavičius J. Identification of a 2′-O-Methyluridine Nucleoside Hydrolase Using the Metagenomic Libraries. Molecules. 2018; 23(11):2904. https://doi.org/10.3390/molecules23112904
Chicago/Turabian StyleAučynaitė, Agota, Rasa Rutkienė, Daiva Tauraitė, Rolandas Meškys, and Jaunius Urbonavičius. 2018. "Identification of a 2′-O-Methyluridine Nucleoside Hydrolase Using the Metagenomic Libraries" Molecules 23, no. 11: 2904. https://doi.org/10.3390/molecules23112904
APA StyleAučynaitė, A., Rutkienė, R., Tauraitė, D., Meškys, R., & Urbonavičius, J. (2018). Identification of a 2′-O-Methyluridine Nucleoside Hydrolase Using the Metagenomic Libraries. Molecules, 23(11), 2904. https://doi.org/10.3390/molecules23112904





