A Turn-On Detection of DNA Sequences by Means of Fluorescence of DNA-Templated Silver Nanoclusters via Unique Interactions of a Hydrated Ionic Liquid
Abstract
1. Introduction
2. Results
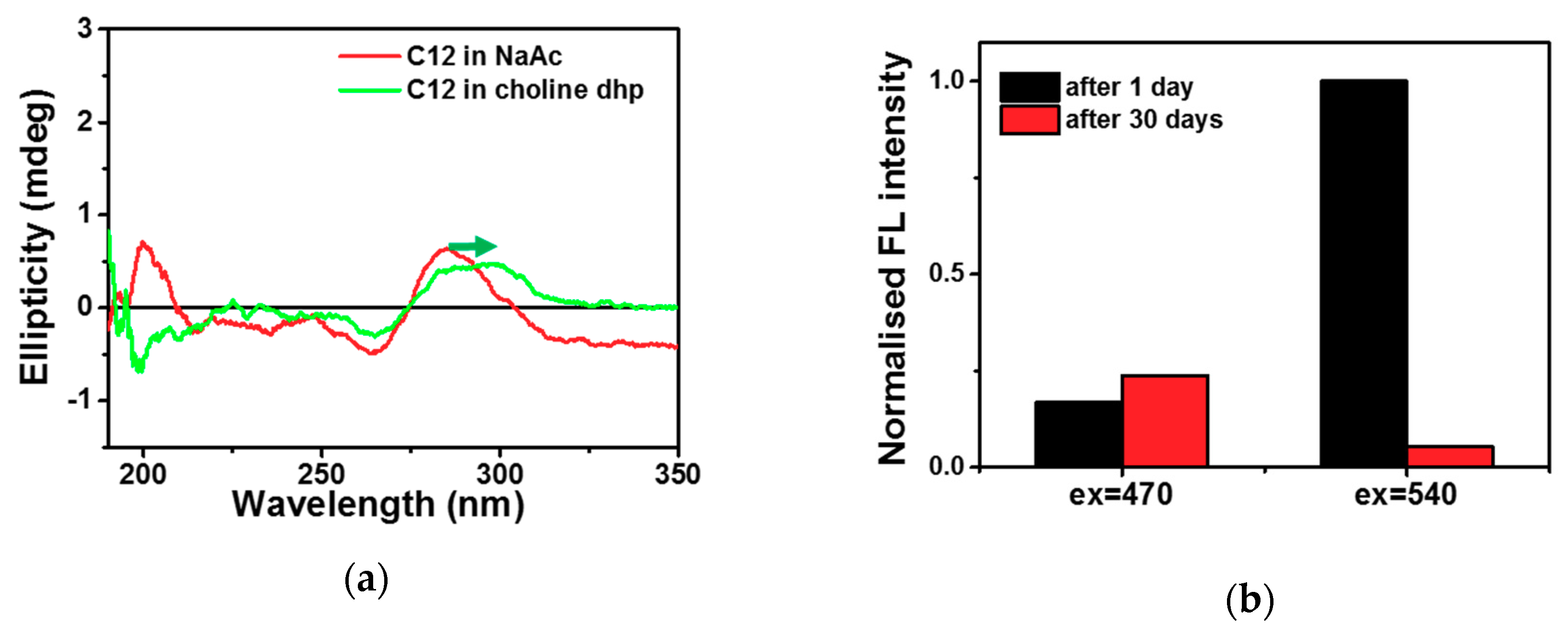
2.1. C-Ag+-C Interaction in Choline Dhp
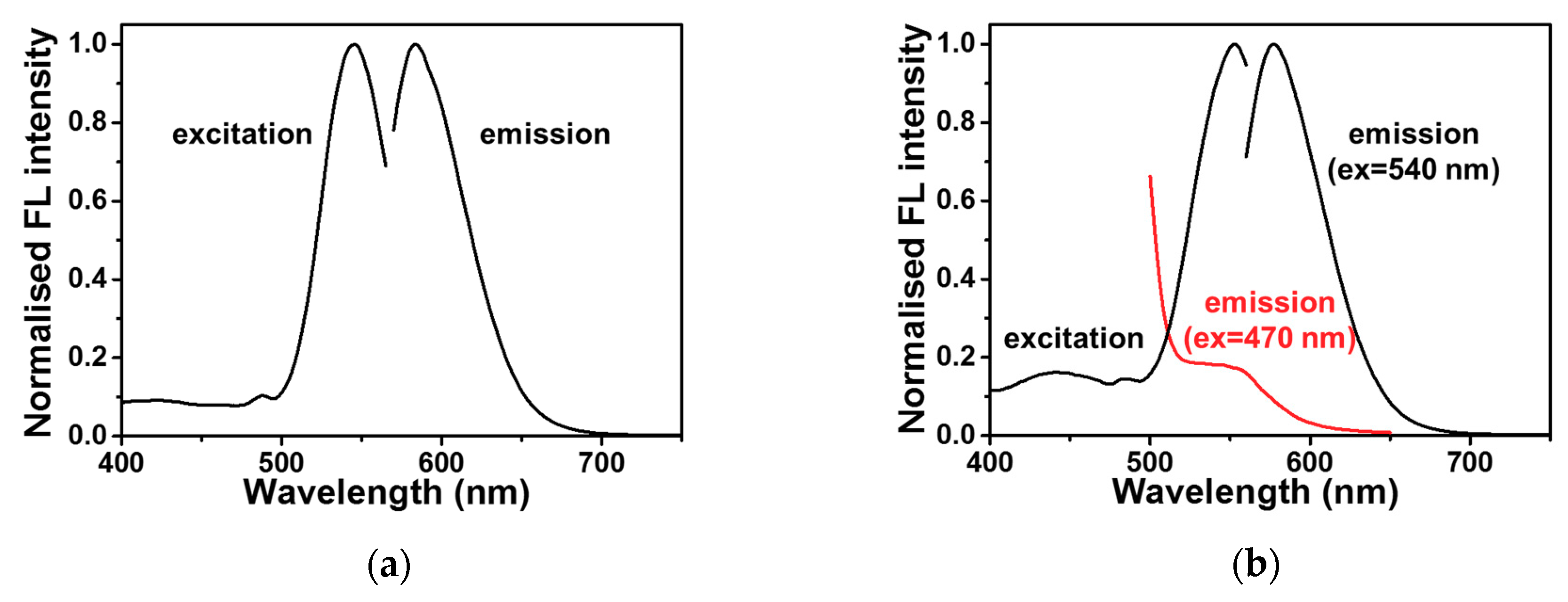
2.2. DNA-Ag NCs in Choline Dhp
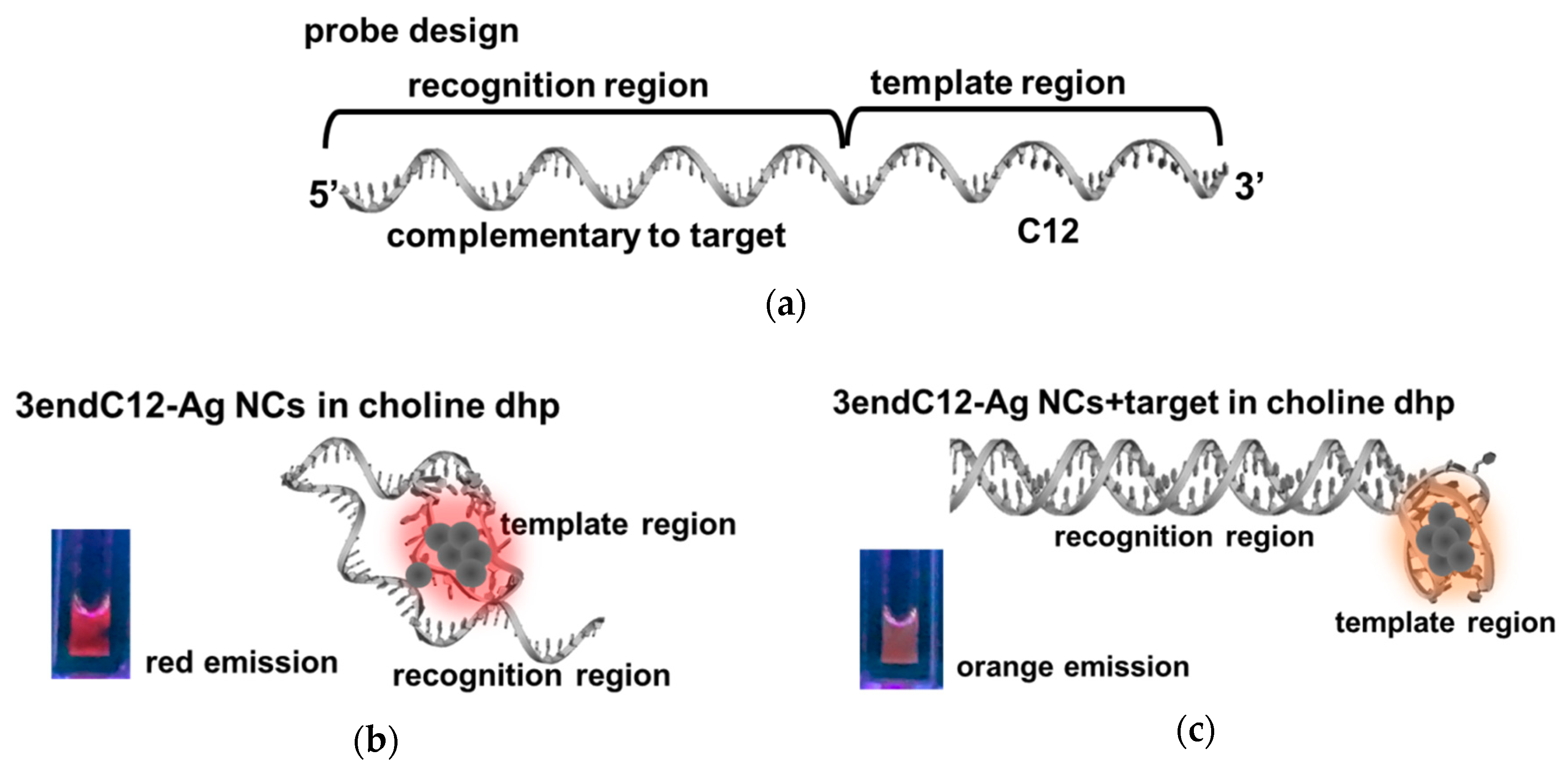
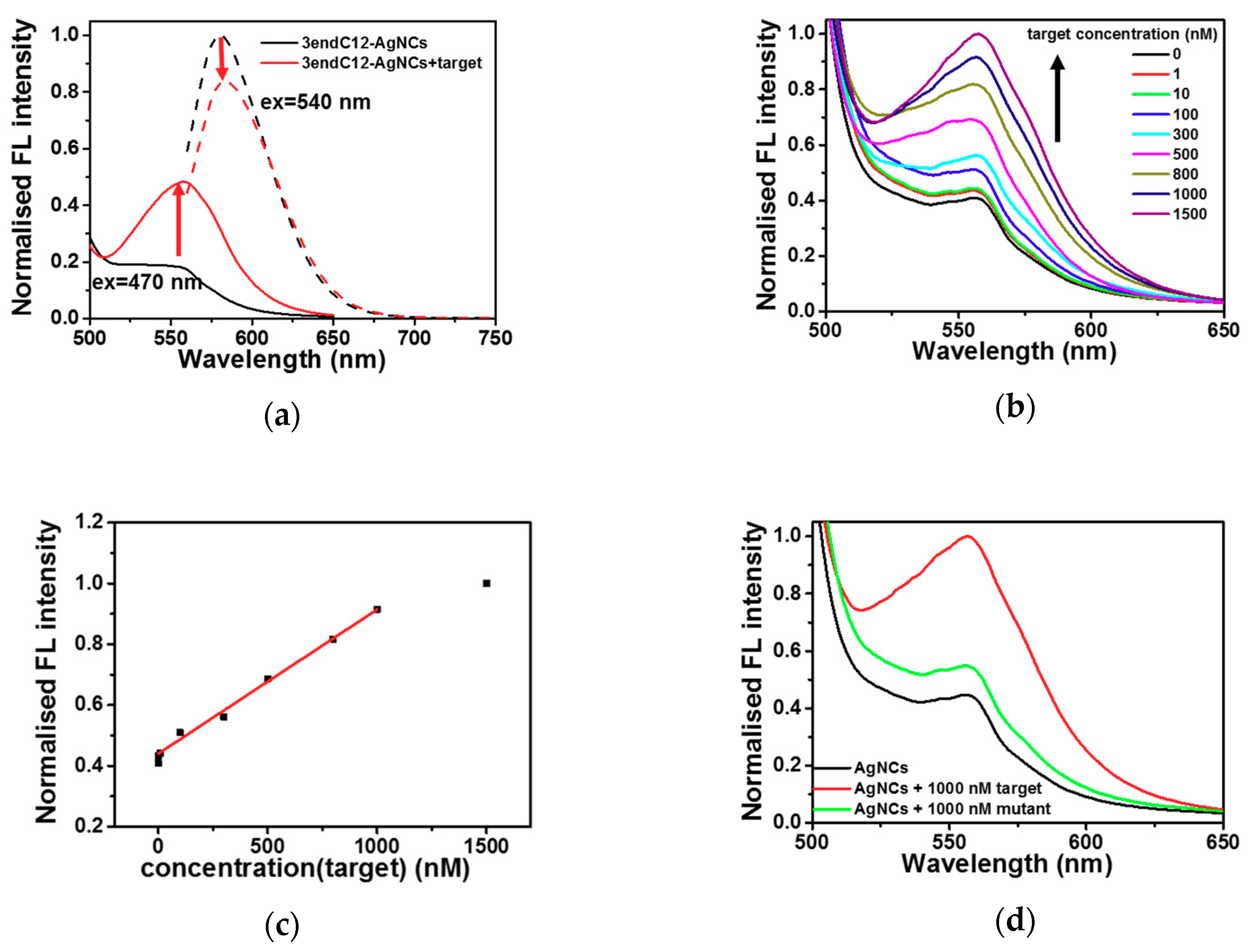
2.3. Detection of DNA by Means of DNA-Ag NCs in Choline Dhp
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. The UV Melting Assay
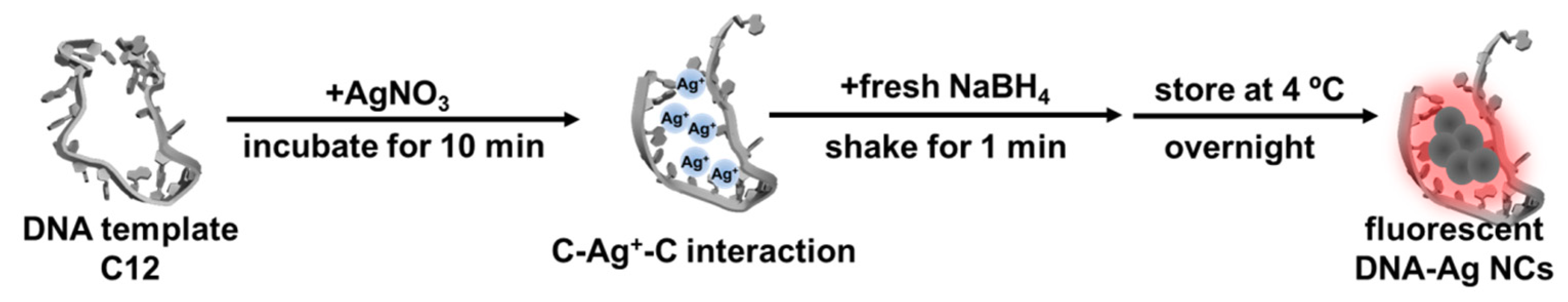
4.3. Synthesis of DNA-Ag NCs
4.4. Detection Methods in Choline Dhp
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Earle, M.J.; Seddon, K.R. Ionic liquids. Green solvents for the future. Pure Appl. Chem. 2000, 72, 1391–1398. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Sugimoto, N. Biological and nanotechnological applications using interactions between ionic liquids and nucleic acids. Biophys. Rev. 2018, 10, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Chandran, A.; Ghoshdastidar, D.; Senapati, S. Groove Binding Mechanism of Ionic Liquids: A Key Factor in Long-Term Stability of DNA in Hydrated Ionic Liquids? J. Am. Chem. Soc. 2012, 134, 20330–20339. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Mukherjee, A. Effect of water and ionic liquids on biomolecules. Biophys. Rev. 2018, 10, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraghavan, R.; Izgorodin, A.; Ganesh, V.; Surianarayanan, M.; MacFarlane, D.R. Long-term structural and chemical stability of DNA in hydrated ionic liquids. Angew. Chem. Int. Ed. Engl. 2010, 49, 1631–1633. [Google Scholar] [CrossRef] [PubMed]
- Mukesh, C.; Mondal, D.; Sharma, M.; Prasad, K. Rapid dissolution of DNA in a novel bio-based ionic liquid with long-term structural and chemical stability: Successful recycling of the ionic liquid for reuse in the process. Chem. Commun. 2013, 49, 6849–6851. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; MacFarlane, D.R.; Forsyth, M.; Yoshizawa-Fujita, M.; Murata, K.; Nakamura, N.; Ohno, H. Solubility and Stability of Cytochrome c in Hydrated Ionic Liquids: Effect of Oxo Acid Residues and Kosmotropicity. Biomacromolecules 2007, 8, 2080–2086. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Tateishi-Karimata, H.; Tanaka, S.; Sugimoto, N. Choline Ion Interactions with DNA Atoms Explain Unique Stabilization of A-T Base Pairs in DNA Duplexes: A Microscopic View. J. Phys. Chem. B 2014, 118, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Tateishi-Karimata, H.; Sugimoto, N. A-T Base Pairs are More Stable Than G-C Base Pairs in a Hydrated Ionic Liquid. Angew. Chem. Int. Ed. 2012, 51, 1416–1419. [Google Scholar] [CrossRef] [PubMed]
- Tateishi-Karimata, H.; Nakano, M.; Pramanik, S.; Tanaka, S.; Sugimoto, N. i-Motifs are more stable than G-quadruplexes in a hydrated ionic liquid. Chem. Commun. 2015, 51, 6909–6912. [Google Scholar] [CrossRef] [PubMed]
- Tateishi-Karimata, H.; Nakano, M.; Sugimoto, N. Comparable Stability of Hoogsteen and Watson-Crick Base Pairs in Ionic Liquid Choline Dihydrogen Phosphate. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Mazid, R.R.; Cooper, A.; Zhang, Y.; Vijayaraghavan, R.; MacFarlane, D.R.; Cortez-Jugo, C.; Cheng, W.L. Enhanced enzymatic degradation resistance of plasmid DNA in ionic liquids. RSC Adv. 2015, 5, 43839–43844. [Google Scholar] [CrossRef]
- Behera, K.; Pandey, S.; Kadyan, A.; Pandey, S. Ionic Liquid-Based Optical and Electrochemical Carbon Dioxide Sensors. Sensors 2015, 15, 30487–30503. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.D.; Trujillo-Rodriguez, M.J.; Anderson, J.L. Advances in the analysis of biological samples using ionic liquids. Anal. Bioanal. Chem. 2018, 410, 4567–4573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Cao, Y.H.; Chen, D.; Cui, P.L.; Yang, J. Ionic liquid assisted synthesis of palladium nanoclusters for highly efficient formaldehyde oxidation. Electrochim. Acta 2018, 269, 38–44. [Google Scholar] [CrossRef]
- Jagannath, B.; Muthukumar, S.; Prasad, S. Electrical double layer modulation of hybrid room temperature ionic liquid/aqueous buffer interface for enhanced sweat based biosensing. Anal. Chim. Acta 2018, 1016, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Pandya, A.; Lad, A.N.; Singh, S.P.; Shanker, R. DNA assembled metal nanoclusters: Synthesis to novel applications. RSC Adv. 2016, 6, 113095–113114. [Google Scholar] [CrossRef]
- Petty, J.T.; Zheng, J.; Hud, N.V.; Dickson, R.M. DNA-templated Ag nanocluster formation. J. Am. Chem. Soc. 2004, 126, 5207–5212. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.I.; Choi, S.; Hsiang, J.C.; Antoku, Y.; Vosch, T.; Bongiorno, A.; Tzeng, Y.L.; Dickson, R.M. Oligonucleotide-stabilized Ag nanocluster fluorophores. J. Am. Chem. Soc. 2008, 130, 5038–5039. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.S.; Sharma, J.; Yeh, H.C.; Yoo, H.; Werner, J.H. A complementary palette of fluorescent silver nanoclusters. Chem. Commun. 2010, 46, 3280–3282. [Google Scholar]
- New, S.Y.; Lee, S.T.; Su, X.D. DNA-templated silver nanoclusters: Structural correlation and fluorescence modulation. Nanoscale 2016, 8, 17729–17746. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.W.; Yuan, J.P.; Wang, E.K. Oligonucleotide-stabilized Ag nanoclusters as novel fluorescence probes for the highly selective and sensitive detection of the Hg2+ ion. Chem. Commun. 2009, 3395–3397. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Zhou, Z.; Li, J.; Li, T.; Dong, S. Fluorescent silver nanoclusters in hybridized DNA duplexes for the turn-on detection of Hg2+ ions. Chem. Commun. 2011, 47, 11065–11067. [Google Scholar] [CrossRef] [PubMed]
- Lan, G.Y.; Huang, C.C.; Chang, H.T. Silver nanoclusters as fluorescent probes for selective and sensitive detection of copper ions. Chem. Commun. 2010, 46, 1257–1259. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.T.; Su, Y.T.; Lan, G.Y.; Chen, W.Y. Detection of Copper Ions Through Recovery of the Fluorescence of DNA-Templated Copper/Silver Nanoclusters in the Presence of Mercaptopropionic Acid. Anal. Chem. 2010, 82, 8566–8572. [Google Scholar]
- Wang, E.K.; Han, B.Y. Oligonucleotide-stabilized fluorescent silver nanoclusters for sensitive detection of biothiols in biological fluids. Biosens. Bioelectron. 2011, 26, 2585–2589. [Google Scholar]
- Zhou, Z.; Du, Y.; Dong, S. DNA-Ag nanoclusters as fluorescence probe for turn-on aptamer sensor of small molecules. Biosens. Bioelectron. 2011, 28, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Lan, G.Y.; Chen, W.Y.; Chang, H.T. One-pot synthesis of fluorescent oligonucleotide Ag nanoclusters for specific and sensitive detection of DNA. Biosens. Bioelectron. 2011, 26, 2431–2435. [Google Scholar] [CrossRef] [PubMed]
- Petty, J.T.; Sengupta, B.; Story, S.P.; Degtyareva, N.N. DNA Sensing by Amplifying the Number of Near-Infrared Emitting, Oligonucleotide-Encapsulated Silver Clusters. Anal. Chem. 2011, 83, 5957–5964. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.W.; Vosch, T. Rapid Detection of MicroRNA by a Silver Nanocluster DNA Probe. Anal. Chem. 2011, 83, 6935–6939. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.S.; Sharma, J.; Yeh, H.C.; Yoo, H.; Werner, J.H. Silver nanocluster aptamers: In situ generation of intrinsically fluorescent recognition ligands for protein detection. Chem. Commun. 2011, 47, 2294–2296. [Google Scholar]
- Lan, G.Y.; Chen, W.Y.; Chang, H.T. Characterization and application to the detection of single-stranded DNA binding protein of fluorescent DNA-templated copper/silver nanoclusters. Analyst 2011, 136, 3623–3628. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.M.; Yu, J.H.; Patel, S.A.; Tzeng, Y.L.; Dickson, R.M. Tailoring silver nanodots for intracellular staining. Photoch. Photobiol. Sci. 2011, 10, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhong, X.; Cheng, F.; Zhang, J.-R.; Jiang, L.-P.; Zhu, J.-J. One-Pot Synthesis of Aptamer-Functionalized Silver Nanoclusters for Cell-Type-Specific Imaging. Anal. Chem. 2012, 84, 4140–4146. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; He, X.; Wang, K.; Qing, Z.; Wu, X.; Shi, H.; Yang, X. One-step engineering of silver nanoclusters-aptamer assemblies as luminescent labels to target tumor cells. Nanoscale 2012, 4, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Guo, W.; Li, B.; Li, T.; Li, D.; Wang, E. DNA G-Quadruplex-Templated Formation of the Fluorescent Silver Nanocluster and Its Application to Bioimaging. Talanta 2012, 88, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wang, Y.; Wei, Y.; Liu, R.; Zhu, H.; Cui, Y.; Zhao, Y.; Gao, X. Ag cluster-aptamer hybrid: Specifically marking the nucleus of live cells. Chem. Commun. 2011, 47, 11960–11962. [Google Scholar] [CrossRef] [PubMed]
- Morishita, K.; MacLean, J.L.; Liu, B.; Jiang, H.; Liu, J. Correlation of photobleaching, oxidation and metal induced fluorescence quenching of DNA-templated silver nanoclusters. Nanoscale 2013, 5, 2840–2849. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, J.; Huang, Y.; Zhao, H.; Tian, L. Highly Stable and Multiemissive Silver Nanoclusters Synthesized in Situ in a DNA Hydrogel and Their Application for Hydroxyl Radical Sensing. ACS Appl. Mater. Interfaces 2018, 10, 26075–26083. [Google Scholar] [CrossRef] [PubMed]
- Tateishi-Karimata, H.; Pramanik, S.; Sugimoto, N. DNA sensor's selectivity enhancement and protection from contaminating nucleases due to a hydrated ionic liquid. Analyst 2015, 140, 4393–4398. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Cao, S.; Togashi, H.; Tashiro, M.; Fujimoto, T.; Machinami, T.; Oda, S.; Miyake, Y.; Okamoto, I.; Tanaka, Y. Specific interactions between silver(i) ions and cytosine-cytosine pairs in DNA duplexes. Chem. Commun. 2008, 4825–4827. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Yang, X.; Han, L.; Wang, E. The Relationship between DNA Sequences and Oligonucleotide-Templated Silver Nanoclusters and Their Fluorescence Properties. Chem.-Eur. J. 2014, 20, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Day, H.A.; Pavlou, P.; Waller, Z.A.E. i-Motif DNA: Structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014, 22, 4407–4418. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, B.; Springer, K.; Buckman, J.G.; Story, S.P.; Abe, O.H.; Hasan, Z.W.; Prudowsky, Z.D.; Rudisill, S.E.; Degtyareva, N.N.; Petty, J.T. DNA Templates for Fluorescent Silver Clusters and I-Motif Folding. J. Phys. Chem. C 2009, 113, 19518–19524. [Google Scholar] [CrossRef]
- Sharma, J.; Rocha, R.C.; Phipps, M.L.; Yeh, H.-C.; Balatsky, K.A.; Vu, D.M.; Shreve, A.P.; Werner, J.H.; Martinez, J.S. A DNA-templated fluorescent silver nanocluster with enhanced stability. Nanoscale 2012, 4, 4107–4110. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kawde, A.N. Pencil-based renewable biosensor for label-free electrochemical detection of DNA hybridization. Anal. Chim. Acta 2001, 431, 219–224. [Google Scholar] [CrossRef]
- Tong, D.; Stimpfl, M.; Reinthaller, A.; Vavra, N.; Mullauer-Ertl, S.; Leodolter, S.; Zeillinger, R. BRCAI gene mutations in sporadic ovarian carcinomas: Detection by PCR and reverse allele-specific oligonucleotide hybridization. Clin. Chem. 1999, 45, 976–981. [Google Scholar] [PubMed]
- Li, T.T.; He, N.Y.; Wang, J.H.; Li, S.; Deng, Y.; Wang, Z.L. Effects of the i-motif DNA loop on the fluorescence of silver nanoclusters. RSC Adv. 2016, 6, 22839–22844. [Google Scholar] [CrossRef]
- Nordén, B.; Matsuoka, Y.; Kurucsev, T. Nucleic acid-metal interactions. IV. Complexes of Ag(I) with thymine and cytosine from studies of UV and IR dichroic spectra. J. Crystallogr. Spectrosc. Res. 1986, 16, 217–226. [Google Scholar] [CrossRef]
Sample Availability: Not available. |


| Name | Sequences (5′-3′) |
|---|---|
| target | GAACAAAAGGAAGAAAATC |
| mutant 1 | GAACAAAAGGAATAAAATC |
| 3endC12 2 | GATTTTCTTCCTTTTGTTCCCCCCCCCCCCC |
| Excitation (nm)/Emission (nm) | In Aqueous Solution | In Choline Dhp | ||||
|---|---|---|---|---|---|---|
| Fluorescence | Stability | Sensing | Fluorescence | Stability | Sensing | |
| 470/557 | - | - | - | weak | good | suitable |
| 540/577 | strong | poor | unsuitable | strong | poor | unsuitable |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, Y.; Tateishi-Karimata, H.; Tsuruoka, T.; Sugimoto, N. A Turn-On Detection of DNA Sequences by Means of Fluorescence of DNA-Templated Silver Nanoclusters via Unique Interactions of a Hydrated Ionic Liquid. Molecules 2018, 23, 2889. https://doi.org/10.3390/molecules23112889
Teng Y, Tateishi-Karimata H, Tsuruoka T, Sugimoto N. A Turn-On Detection of DNA Sequences by Means of Fluorescence of DNA-Templated Silver Nanoclusters via Unique Interactions of a Hydrated Ionic Liquid. Molecules. 2018; 23(11):2889. https://doi.org/10.3390/molecules23112889
Chicago/Turabian StyleTeng, Ye, Hisae Tateishi-Karimata, Takaaki Tsuruoka, and Naoki Sugimoto. 2018. "A Turn-On Detection of DNA Sequences by Means of Fluorescence of DNA-Templated Silver Nanoclusters via Unique Interactions of a Hydrated Ionic Liquid" Molecules 23, no. 11: 2889. https://doi.org/10.3390/molecules23112889
APA StyleTeng, Y., Tateishi-Karimata, H., Tsuruoka, T., & Sugimoto, N. (2018). A Turn-On Detection of DNA Sequences by Means of Fluorescence of DNA-Templated Silver Nanoclusters via Unique Interactions of a Hydrated Ionic Liquid. Molecules, 23(11), 2889. https://doi.org/10.3390/molecules23112889






