Immobilization of Enzymes on a Phospholipid Bionically Modified Polysulfone Gradient-Pore Membrane for the Enhanced Performance of Enzymatic Membrane Bioreactors
Abstract
:1. Introduction
2. Results and Discussion
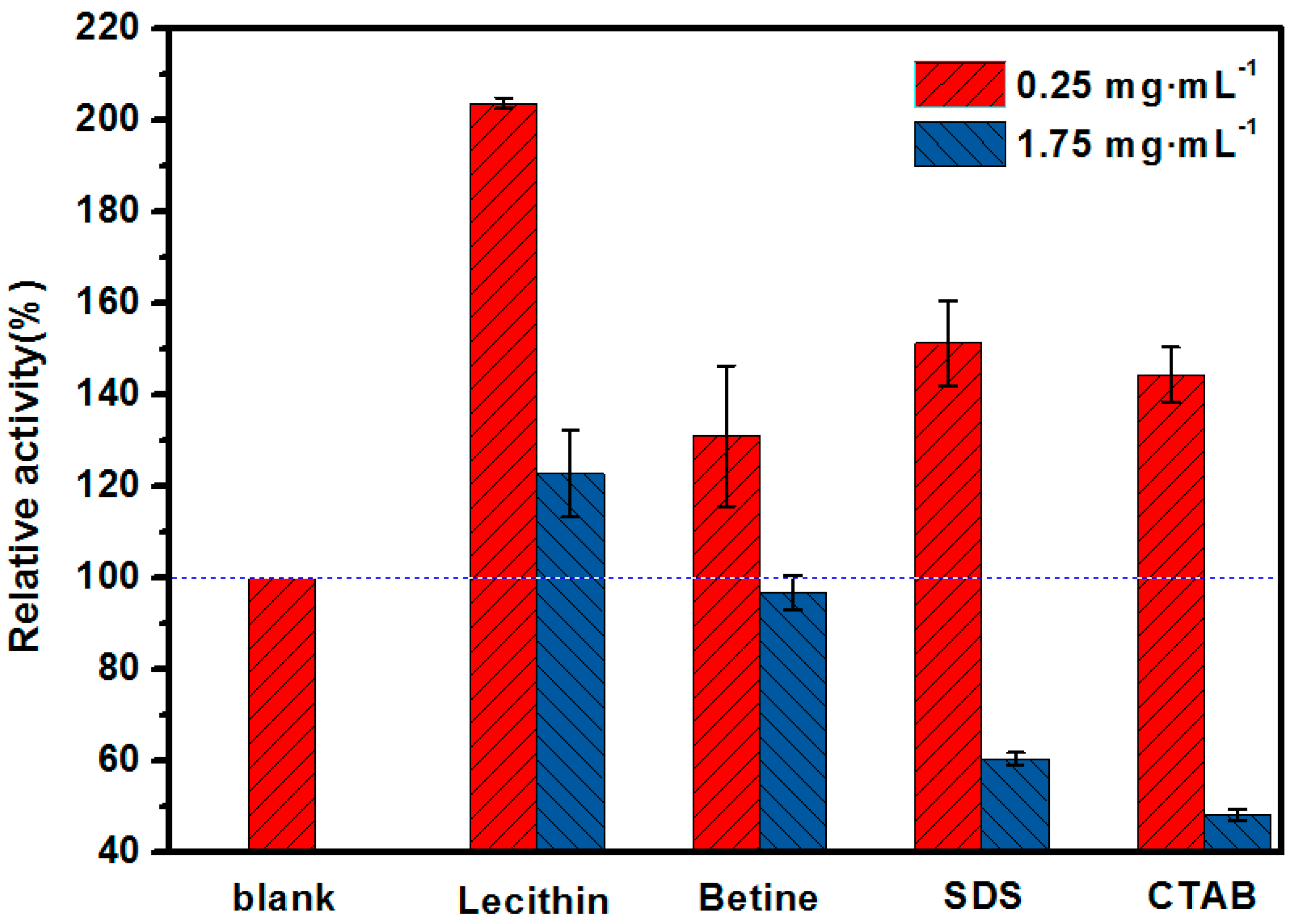
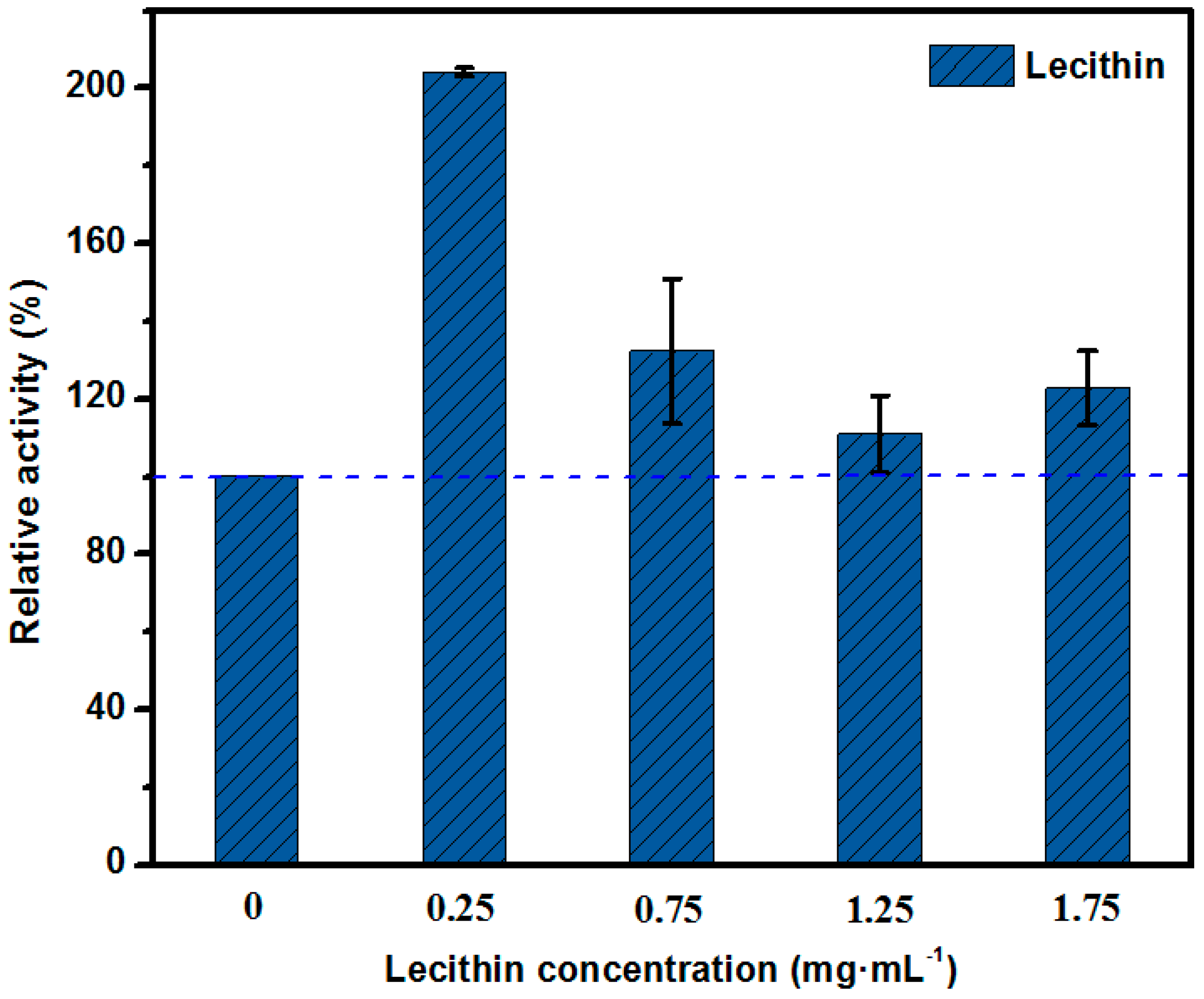
2.1. Mimetic Biological Microenvironment Based on Enzyme Surfactant Micelles
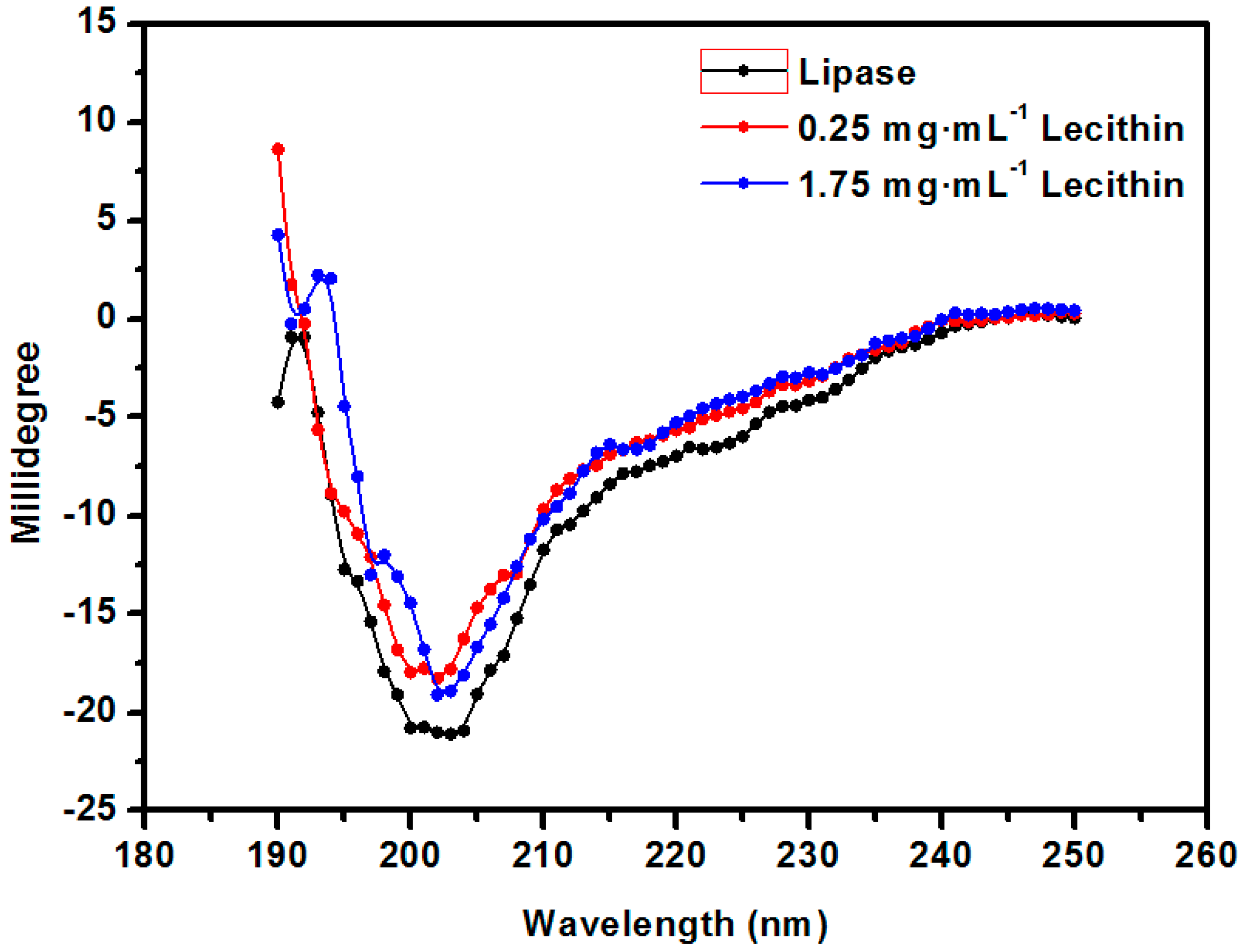
2.2. The Optimization of the Structure of the Enzyme Lecithin Complex
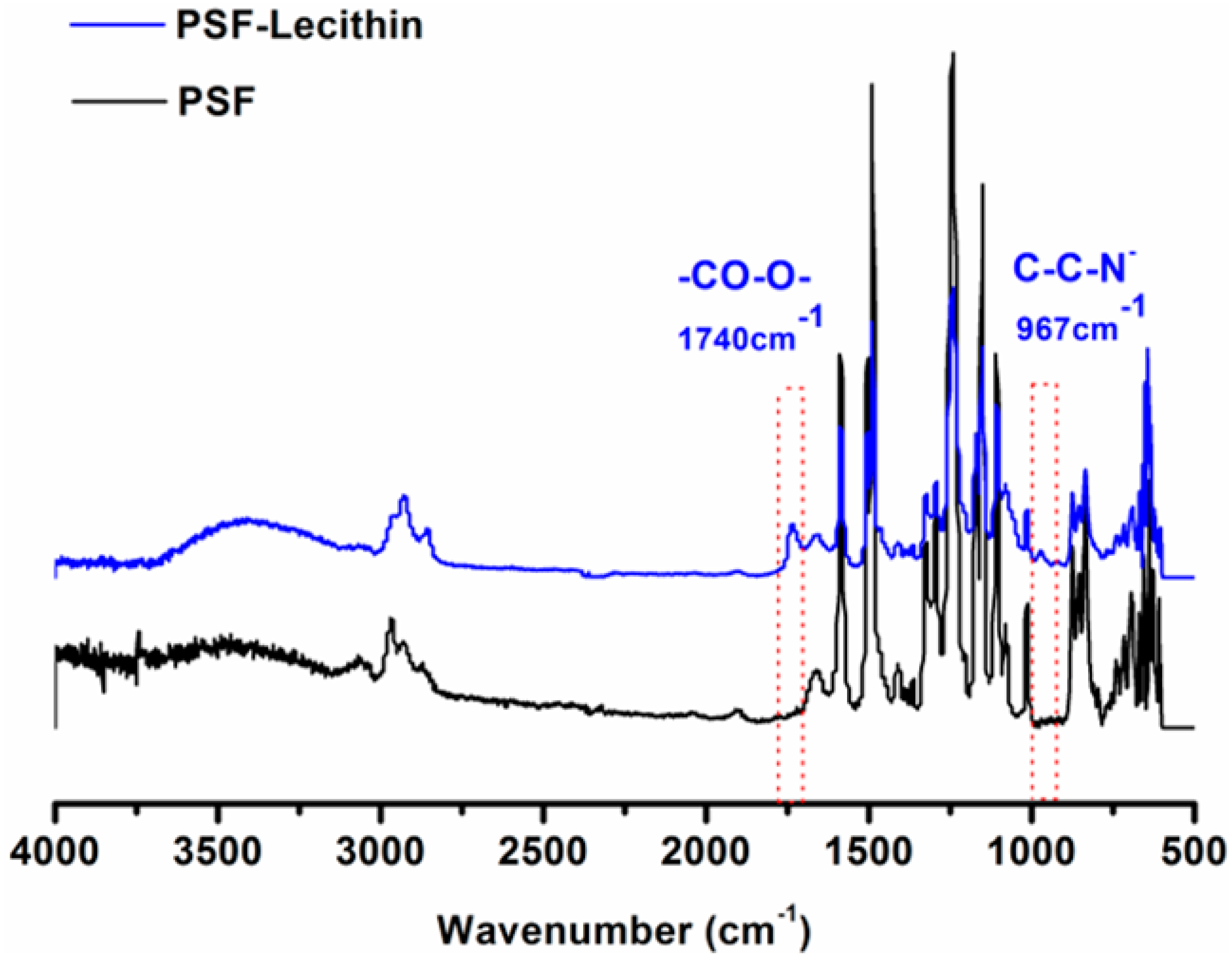
2.3. Characterization of the PSF Membrane with a Lecithin Biomimetic Interface
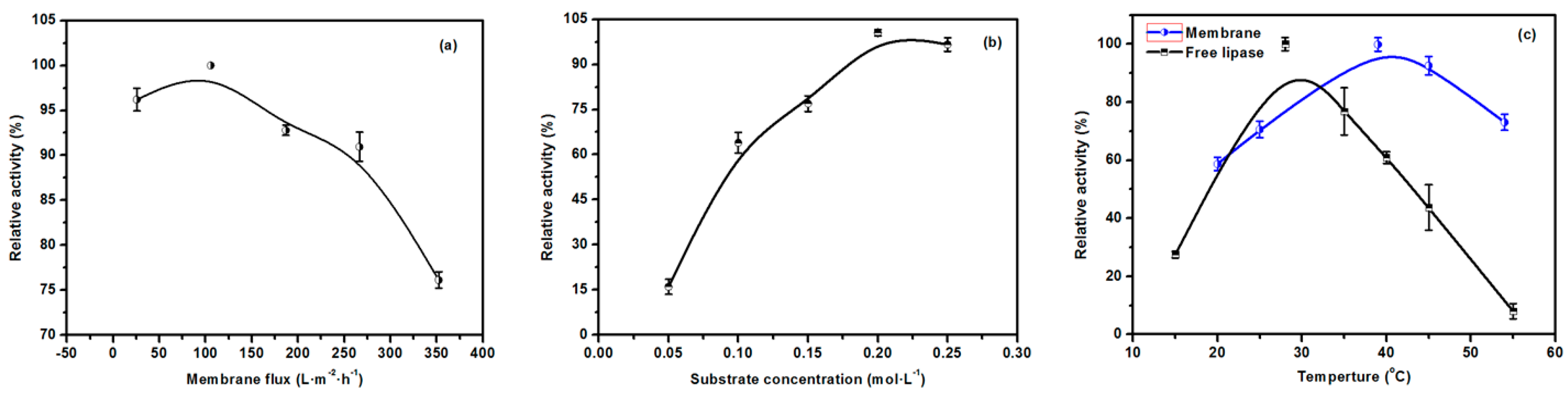
2.4. The Effect of the Lecithin Biomimetic Interface on the Performance of the EMBR
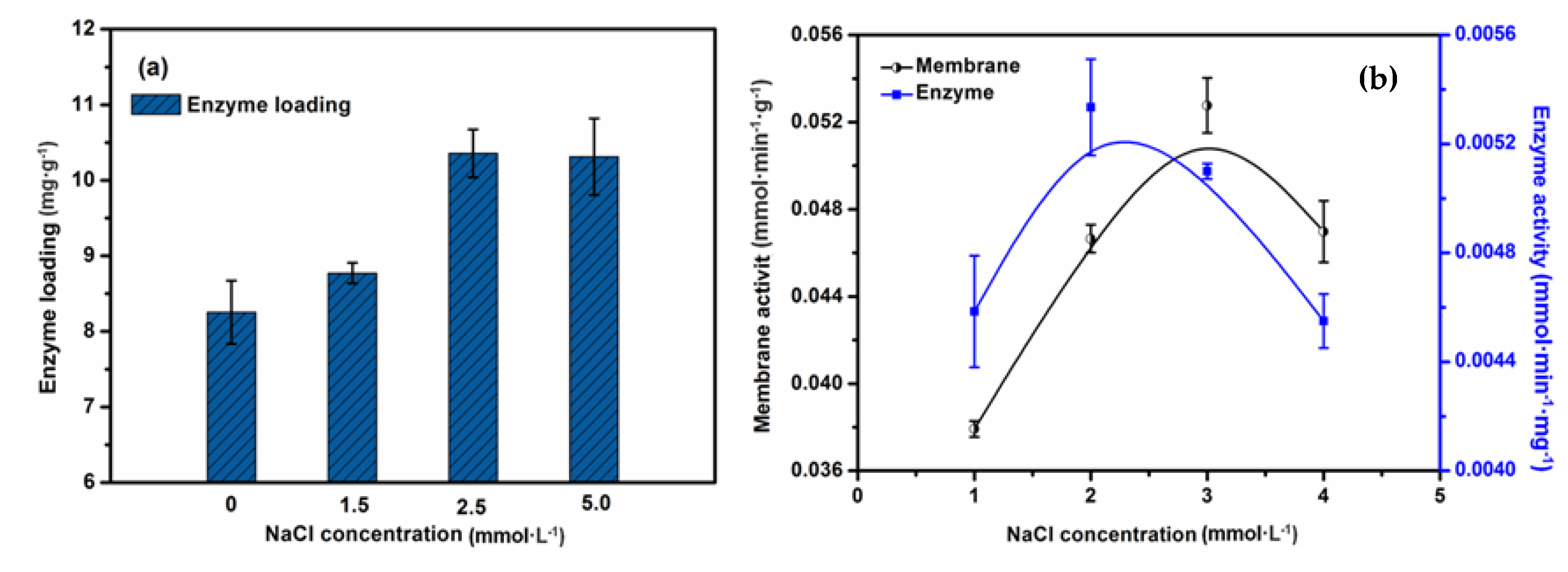
2.5. The Effect of Ion Strength on the Performance of the EMBR
2.6. The Effect of the Concentration of Lipase on Enzyme Loading and Activity of the EMBR
2.7. Optimization of the Properties of the EMBR
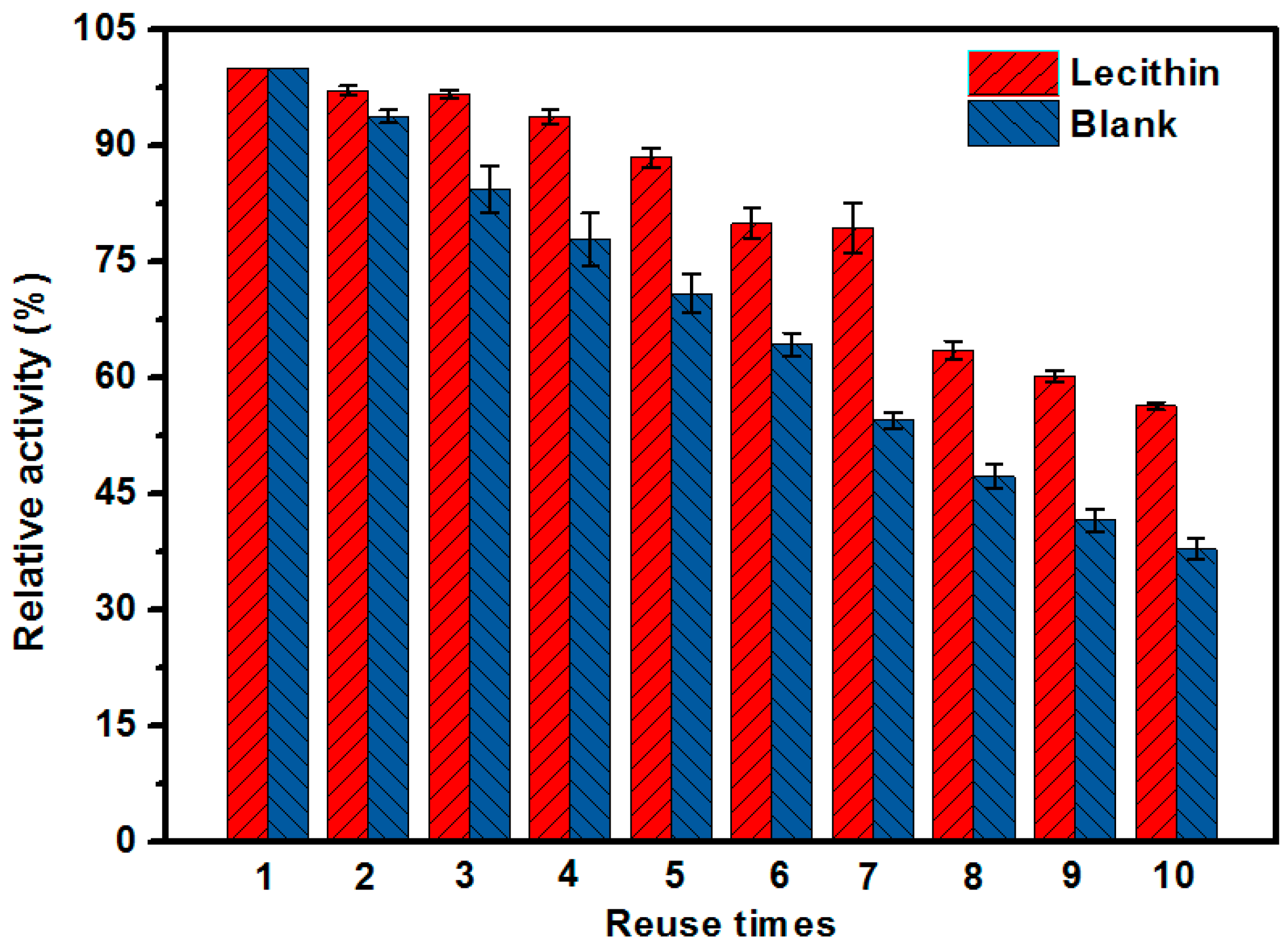
2.8. Reusability of the EMBR
3. Materials and Methods
3.1. Materials
3.2. The Determination of Optimal Surfactant to Enhance the Activity of Lipase
3.3. Preparation of an EMBR with Lecithin Biomimetic Interfaces
3.4. Activity Assay for the EMBR
3.5. Reusability of the EMBR
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Dhake, K.P.; Karoyo, A.H.; Mohamed, M.H.; Wilson, L.D.; Bhanage, B.M. Enzymatic activity studies of pseudomonas cepacia lipase adsorbed onto copolymer supports containing β-cyclodextrin. J. Mol. Catal. B Enzym. 2013, 87, 105–112. [Google Scholar] [CrossRef]
- Feng, Q.; Zhao, Y.; Wei, A.; Li, C.; Wei, Q.; Fong, H. Immobilization of catalase on electrospun PVA/PA6-Cu(II) nanofibrous membrane for the development of efficient and reusable enzyme membrane reactor. Environ. Sci. Technol. 2014, 48, 10390–10397. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-C.; Huang, X.-J.; Xu, Z.-K. Kinetics-bolstered catalytic study of a high performance lipase-immobilized nanofiber membrane bioreactor. RSC Adv. 2014, 4, 6151–6158. [Google Scholar] [CrossRef]
- Zhou, F.; Jia, X.; Yang, Y.; Yang, Q.; Gao, C.; Zhao, Y.; Fan, Y.; Yuan, X. Peptide-modified pelcl electrospun membranes for regulation of vascular endothelial cells. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 68, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Mechrez, G.; Krepker, M.A.; Harel, Y.; Lellouche, J.-P.; Segal, E. Biocatalytic carbon nanotube paper: A ‘one-pot’ route for fabrication of enzyme-immobilized membranes for organophosphate bioremediation. J. Mater. Chem. B 2014, 2, 915–922. [Google Scholar] [CrossRef]
- Sandu, T.; Sarbu, A.; Damian, C.M.; Patroi, D.; Iordache, T.V.; Budinova, T.; Tsyntsarski, B.; Ferhat Yardim, M.; Sirkecioglu, A. Functionalized bicomponent polymer membranes as supports for covalent immobilization of enzymes. React. Funct. Polym. 2015, 96, 5–13. [Google Scholar] [CrossRef]
- Nagy, E.; Lepossa, A.; Prettl, Z. Mass transfer through a biocatalytic membrane reactor. Ind. Eng. Chem. Res. 2011, 51, 1635–1646. [Google Scholar] [CrossRef]
- Chen, P.-C.; Qian, Y.-C.; Fang, F.; Zhu, X.-Y.; Huang, X.-J. Adsorption and activity of lipase on polyphosphazene-modified polypropylene membrane surface. Catalysts 2016, 6, 174. [Google Scholar] [CrossRef]
- Lee, H.R.; Chung, M.; Kim, M.I.; Ha, S.H. Preparation of glutaraldehyde-treated lipase-inorganic hybrid nanoflowers and their catalytic performance as immobilized enzymes. Enzym. Microb. Technol. 2017, 105, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Sarma, R.; Islam, M.S.; Miller, A.F.; Bhattacharyya, D. Layer-by-layer-assembled laccase enzyme on stimuli-responsive membranes for chloro-organics degradation. ACS Appl. Mater. Interfaces 2017, 9, 14858–14867. [Google Scholar] [CrossRef] [PubMed]
- Wicklein, B.; Darder, M.; Aranda, P.; Ruiz-Hitzky, E. Phospholipid-sepiolite biomimetic interfaces for the immobilization of enzymes. ACS Appl. Mater. Interfaces 2011, 3, 4339–4348. [Google Scholar] [CrossRef] [PubMed]
- Jochems, P.; Satyawali, Y.; Diels, L.; Dejonghe, W. Enzyme immobilization on/in polymeric membranes: Status, challenges and perspectives in biocatalytic membrane reactors (BMRs). Green Chem. 2011, 13, 1609–1623. [Google Scholar] [CrossRef]
- Wu, S.-C.; Wu, S.-M.; Su, F.-M. Novel process for immobilizing an enzyme on a bacterial cellulose membrane through repeated absorption. J. Chem. Technol. Biotechnol. 2017, 92, 109–114. [Google Scholar] [CrossRef]
- Khaldi, K.; Sam, S.; Lounas, A.; Yaddaden, C.; Gabouze, N.-E. Comparative investigation of two methods for acetylcholinesterase enzyme immobilization on modified porous silicon. Appl. Surf. Sci. 2017, 421, 148–154. [Google Scholar] [CrossRef]
- Li, C.; Jiang, S.; Zhao, X.; Liang, H. Co-immobilization of enzymes and magnetic nanoparticles by metal-nucleotide hydrogelnanofibers for improving stability and recycling. Molecules 2017, 22, 179. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.P.; Lau, K.H.; Ulijn, R.V. Biocatalytic self-assembly using reversible and irreversible enzyme immobilization. ACS Appl. Mater. Interfaces 2017, 9, 3266–3271. [Google Scholar] [CrossRef] [PubMed]
- Bolivar, J.M.; Rocha-Martín, J.; Mateo, C.; Guisan, J.M. Stabilization of a highly active but unstable alcohol dehydrogenase from yeast using immobilization and post-immobilization techniques. Process Biochem. 2012, 47, 679–686. [Google Scholar] [CrossRef]
- Yabuki, S. How to lengthen the long-term stability of enzyme membranes: Trends and strategies. Catalysts 2017, 7, 36. [Google Scholar] [CrossRef]
- Li, M.; Luo, M.; Li, F.; Wang, W.; Liu, K.; Liu, Q.; Wang, Y.; Lu, Z.; Wang, D. Biomimetic copper-based inorganic–protein nanoflower assembly constructed on the nanoscale fibrous membrane with enhanced stability and durability. J. Phys. Chem. C 2016, 120, 17348–17356. [Google Scholar] [CrossRef]
- Altinkaynak, C.; Tavlasoglu, S.; Ozdemir, N.; Ocsoy, I. A new generation approach in enzyme immobilization: Organic-inorganic hybrid nanoflowers with enhanced catalytic activity and stability. Enzym. Microb. Technol. 2016, 93–94, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-J.; Xu, Z.-K.; Wan, L.-S.; Innocent, C.; Seta, P. Electrospun nanofibers modified with phospholipid moieties for enzyme immobilization. Macromol. Rapid Commun. 2006, 27, 1341–1345. [Google Scholar] [CrossRef]
- Fu, C.; Yi, D.; Deng, C.; Wang, X.; Zhang, W.; Tang, Y.; Caruso, F.; Wang, Y. A partially graphitic mesoporous carbon membrane with three-dimensionally networked nanotunnels for ultrasensitive electrochemical detection. Chem. Mater. 2017, 29, 5286–5293. [Google Scholar] [CrossRef]
- Kumari, A.; Datta, S. Phospholipid bilayer functionalized membrane pores for enhanced efficiency of immobilized glucose oxidase enzyme. J. Membr. Sci. 2017, 539, 43–51. [Google Scholar] [CrossRef]
- Bali, N.; Petsi, A.J.; Skouras, E.D.; Burganos, V.N. Three-dimensional reconstruction of bioactive membranes and pore-scale simulation of enzymatic reactions: The case of lactose hydrolysis. J. Membr. Sci. 2017, 524, 225–234. [Google Scholar] [CrossRef]
- Okobira, T.; Matsuo, A.; Matsumoto, H.; Tanaka, T.; Kai, K.; Minari, C.; Goto, M.; Kawakita, H.; Uezu, K. Enhancement of immobilized lipase activity by design of polymer brushes on a hollow fiber membrane. J. Biosci. Bioeng. 2015, 120, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-Y.; Chen, C.; Chen, P.-C.; Gao, Q.-L.; Fang, F.; Li, J.; Huang, X.-J. High-performance enzymatic membrane bioreactor based on a radial gradient of pores in a psf membrane via facile enzyme immobilization. RSC Adv. 2016, 6, 30804–30812. [Google Scholar] [CrossRef]
- Ding, L.; Li, Y.; Jiang, Y.; Cao, Z.; Huang, J. New supports for enzyme immobilization based on copolymers of vinylene carbonate and ?-hydroxyethylene acrylate. J. Appl. Polym. Sci. 2002, 83, 94–102. [Google Scholar] [CrossRef]
- Patra, A.; Samanta, N.; Das, D.K.; Mitra, R.K. Enhanced catalytic activity of alpha-chymotrypsin in cationic surfactant solutions: The component specificity revisited. J. Phys. Chem. B 2017, 121, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Souza, L.T.A.; Moreno-Perez, S.; Fernandez Lorente, G.; Cipolatti, E.P.; de Oliveira, D.; Resende, R.R.; Pessela, B.C. Immobilization of moniliella spathulata r25l270 lipase on ionic, hydrophobic and covalent supports: Functional properties and hydrolysis of sardine oil. Molecules 2017, 22, 1508. [Google Scholar] [CrossRef] [PubMed]
- Jurado, E.; Bravo, V.; Luzón, G.; Fernández-Serrano, M.; García-Román, M.; Altmajer-Vaz, D.; Vicaria, J.M. Hard-surface cleaning using lipases: Enzyme–surfactant interactions and washing tests. J. Surfactants Deterg. 2007, 10, 61–70. [Google Scholar] [CrossRef]
- van de Ven, W.J.C.; van’t Sant, K.; Pünt, I.G.M.; Zwijnenburg, A.; Kemperman, A.J.B.; van der Meer, W.G.J.; Wessling, M. Hollow fiber dead-end ultrafiltration: Influence of ionic environment on filtration of alginates. J. Membr. Sci. 2008, 308, 218–229. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available. |
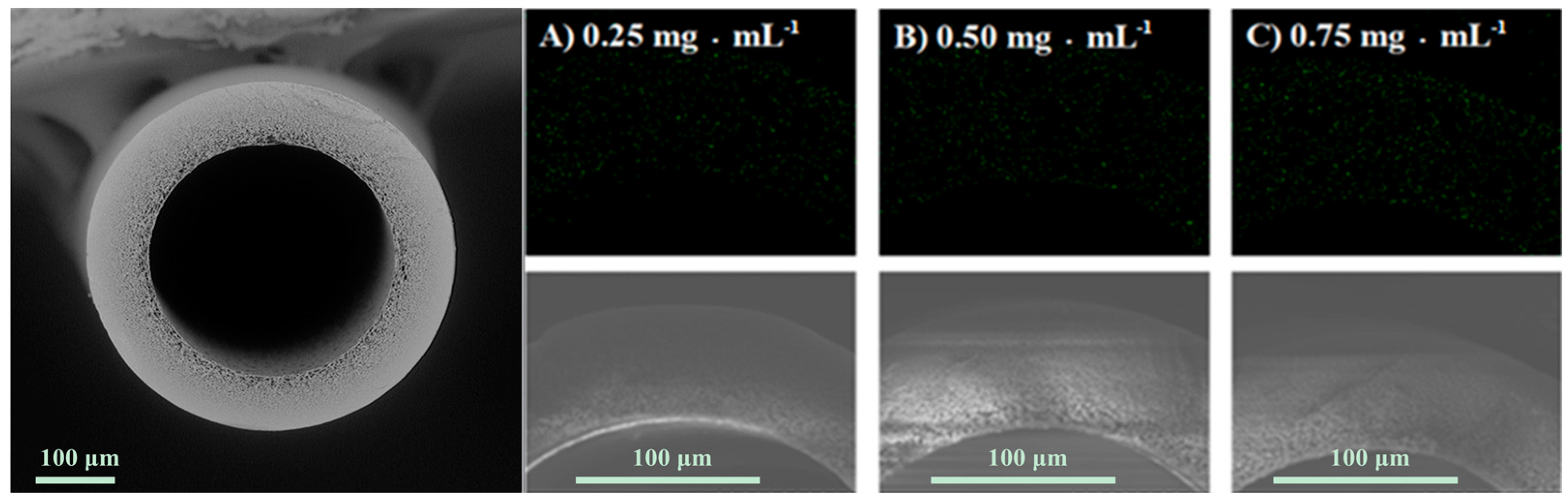



| CLecithin (mg·mL−1) Elements | C | N | O | S |
|---|---|---|---|---|
| 0.00 | 82.90% | 0.00% | 14.29% | 2.82% |
| 0.25 | 77.49% | 4.19% | 14.77% | 3.55% |
| 0.50 | 75.78% | 4.26% | 15.76% | 4.20% |
| 0.75 | 77.05% | 4.86% | 14.39% | 3.70% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Zhu, X.; Fang, F.; Hong, X.; Wu, H.; Chen, D.; Huang, X. Immobilization of Enzymes on a Phospholipid Bionically Modified Polysulfone Gradient-Pore Membrane for the Enhanced Performance of Enzymatic Membrane Bioreactors. Molecules 2018, 23, 144. https://doi.org/10.3390/molecules23010144
Guo Y, Zhu X, Fang F, Hong X, Wu H, Chen D, Huang X. Immobilization of Enzymes on a Phospholipid Bionically Modified Polysulfone Gradient-Pore Membrane for the Enhanced Performance of Enzymatic Membrane Bioreactors. Molecules. 2018; 23(1):144. https://doi.org/10.3390/molecules23010144
Chicago/Turabian StyleGuo, Yizong, Xueyan Zhu, Fei Fang, Xiao Hong, Huimin Wu, Dajing Chen, and Xiaojun Huang. 2018. "Immobilization of Enzymes on a Phospholipid Bionically Modified Polysulfone Gradient-Pore Membrane for the Enhanced Performance of Enzymatic Membrane Bioreactors" Molecules 23, no. 1: 144. https://doi.org/10.3390/molecules23010144
APA StyleGuo, Y., Zhu, X., Fang, F., Hong, X., Wu, H., Chen, D., & Huang, X. (2018). Immobilization of Enzymes on a Phospholipid Bionically Modified Polysulfone Gradient-Pore Membrane for the Enhanced Performance of Enzymatic Membrane Bioreactors. Molecules, 23(1), 144. https://doi.org/10.3390/molecules23010144





