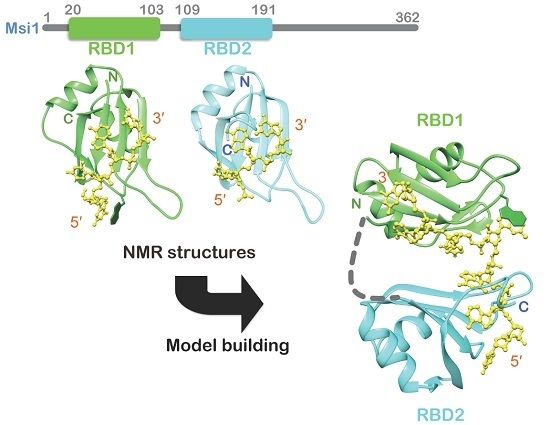
Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Abstract
1. Introduction
2. Results
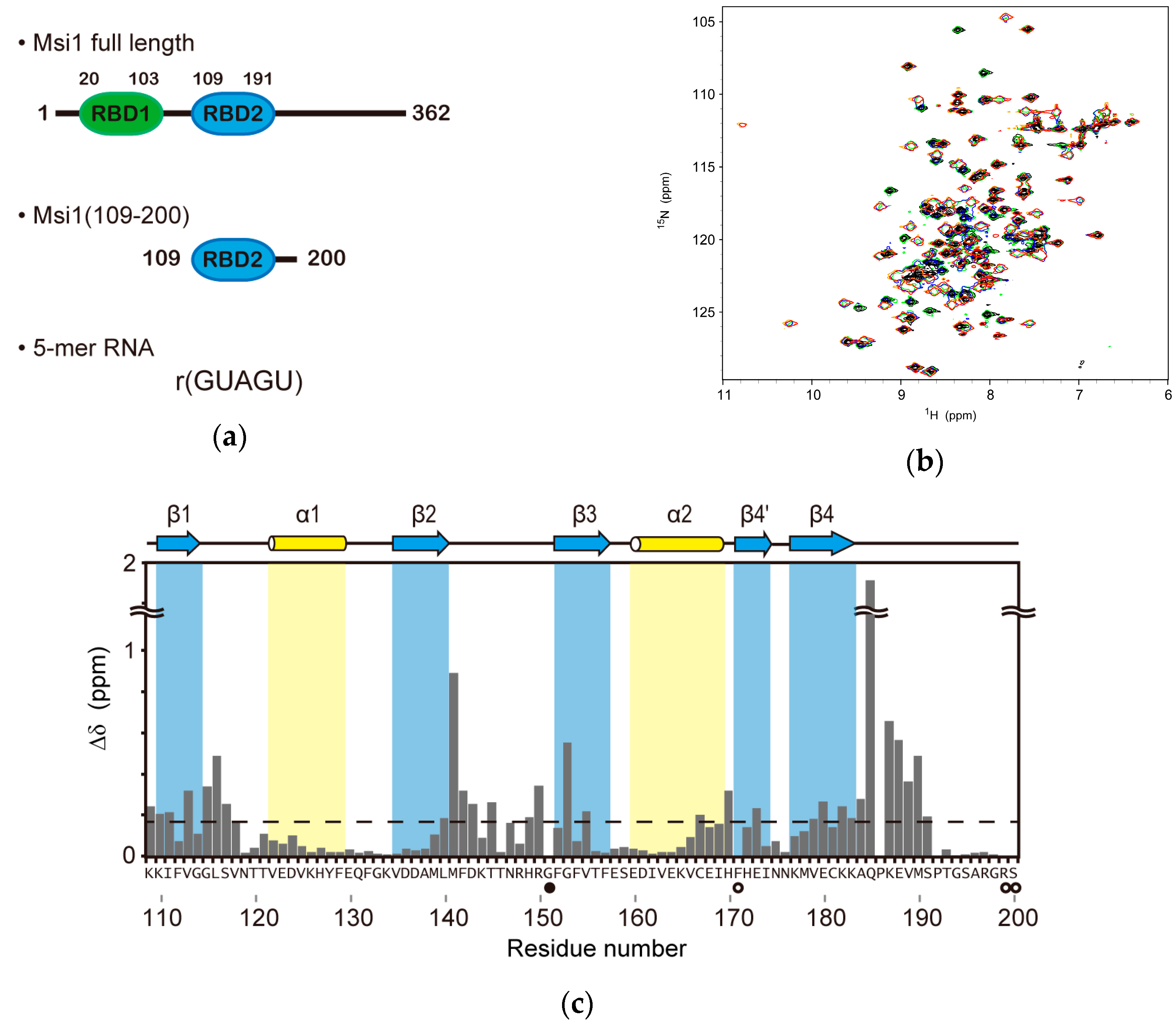
2.1. Structure Determination of Msi1 RBD2(109–191) in the Free Form
2.2. NMR Titration Experiments on Msi1(109–200) with r(GUAGU)
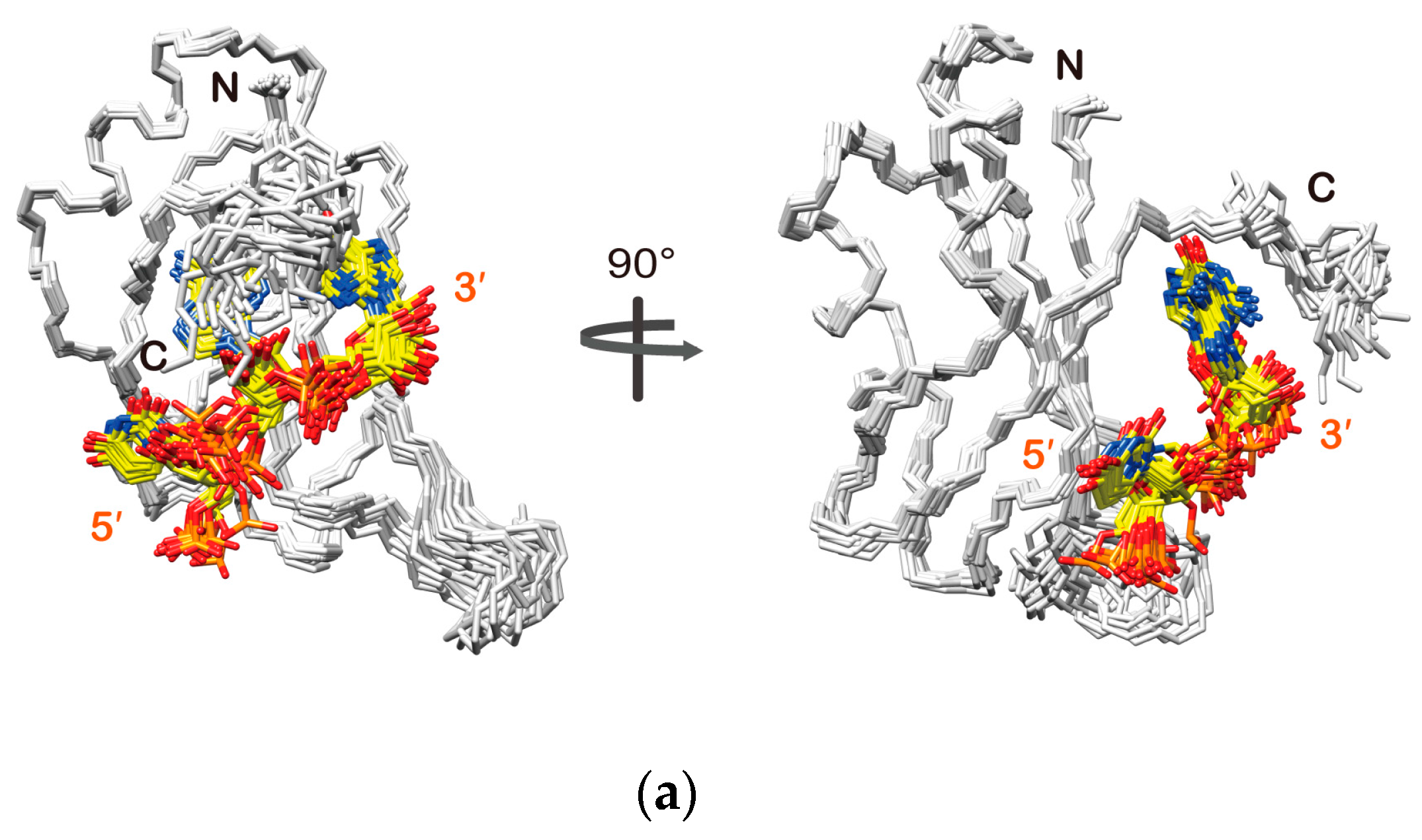
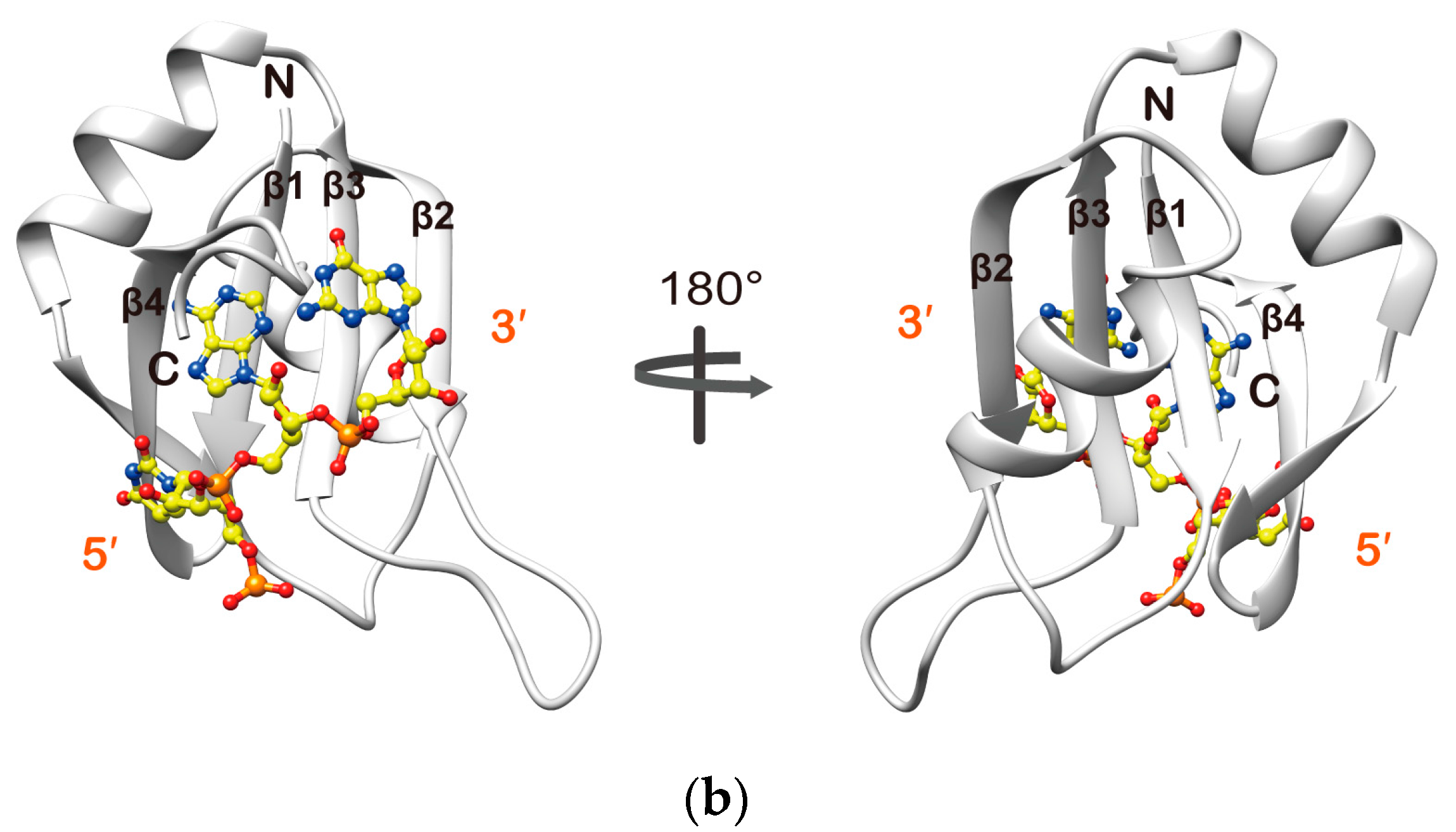
2.3. Structure Determination of the Msi1 RBD2(109–191):r(GUAGU) Complex
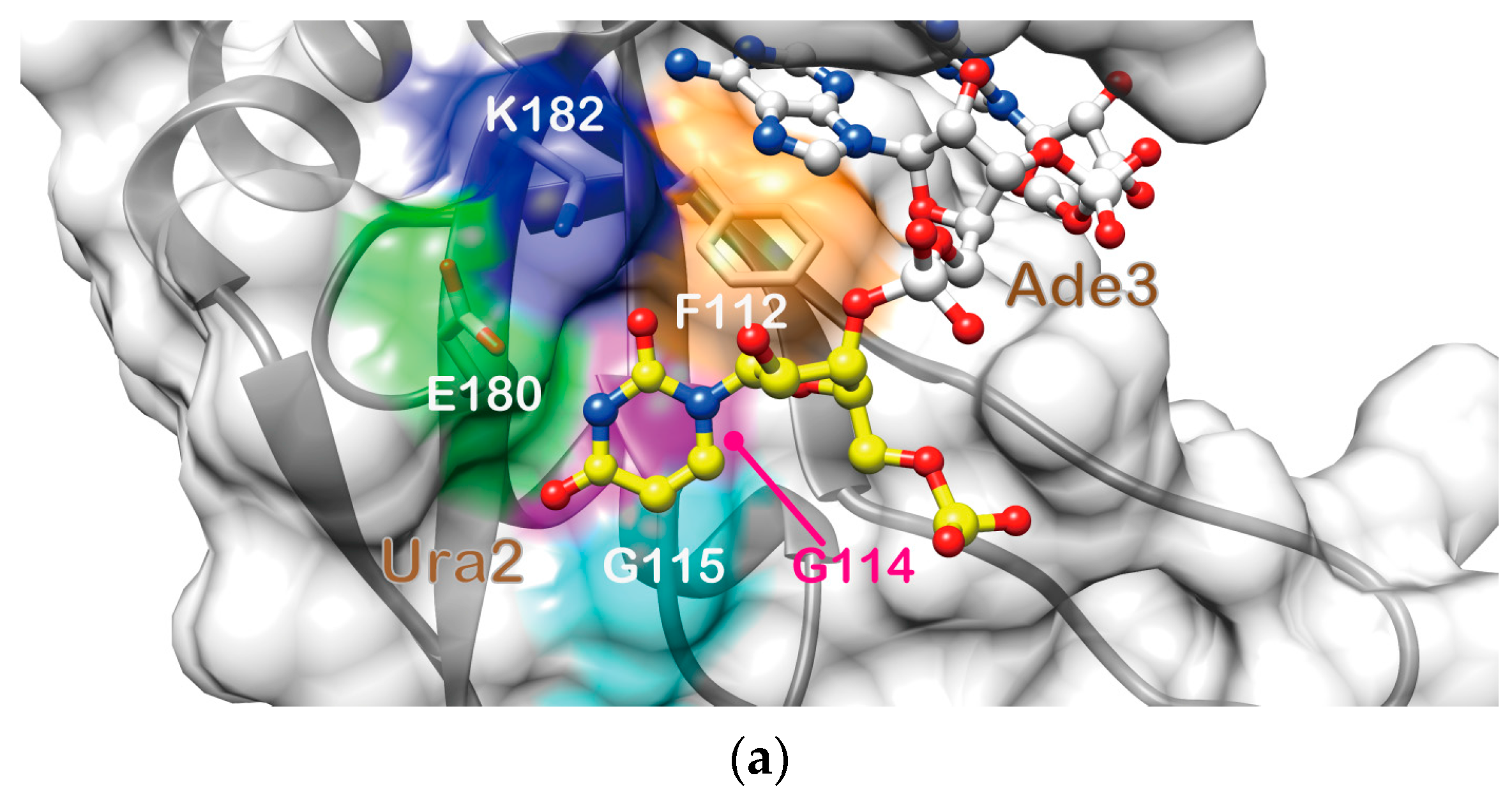
2.4. Structure Description of the Base-Specific RNA Recognition by Msi1 RBD2(109–191)
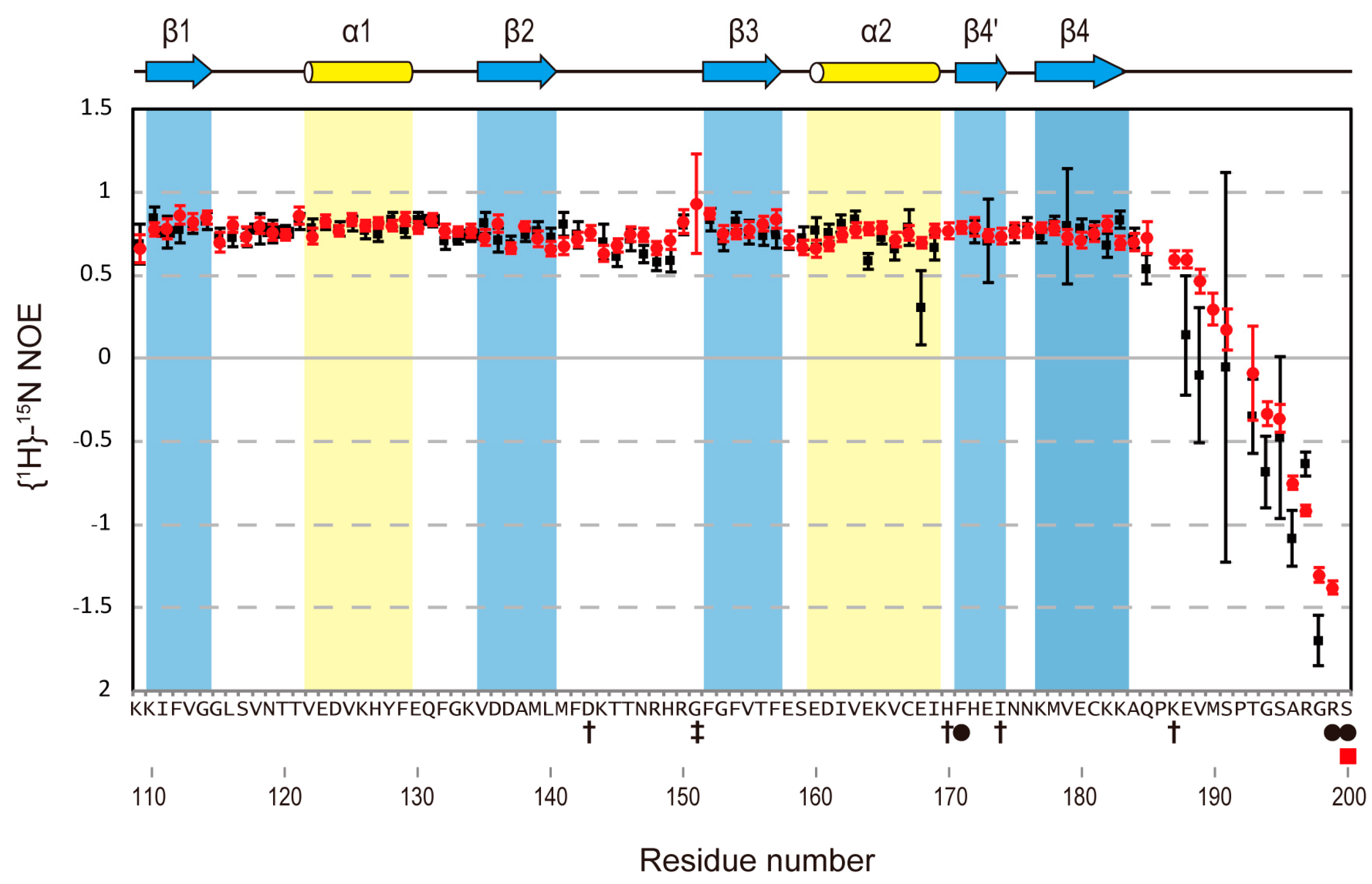
2.5. Heteronuclear Steady-State {1H}-15N Nuclear Overhauser Effect Measurement
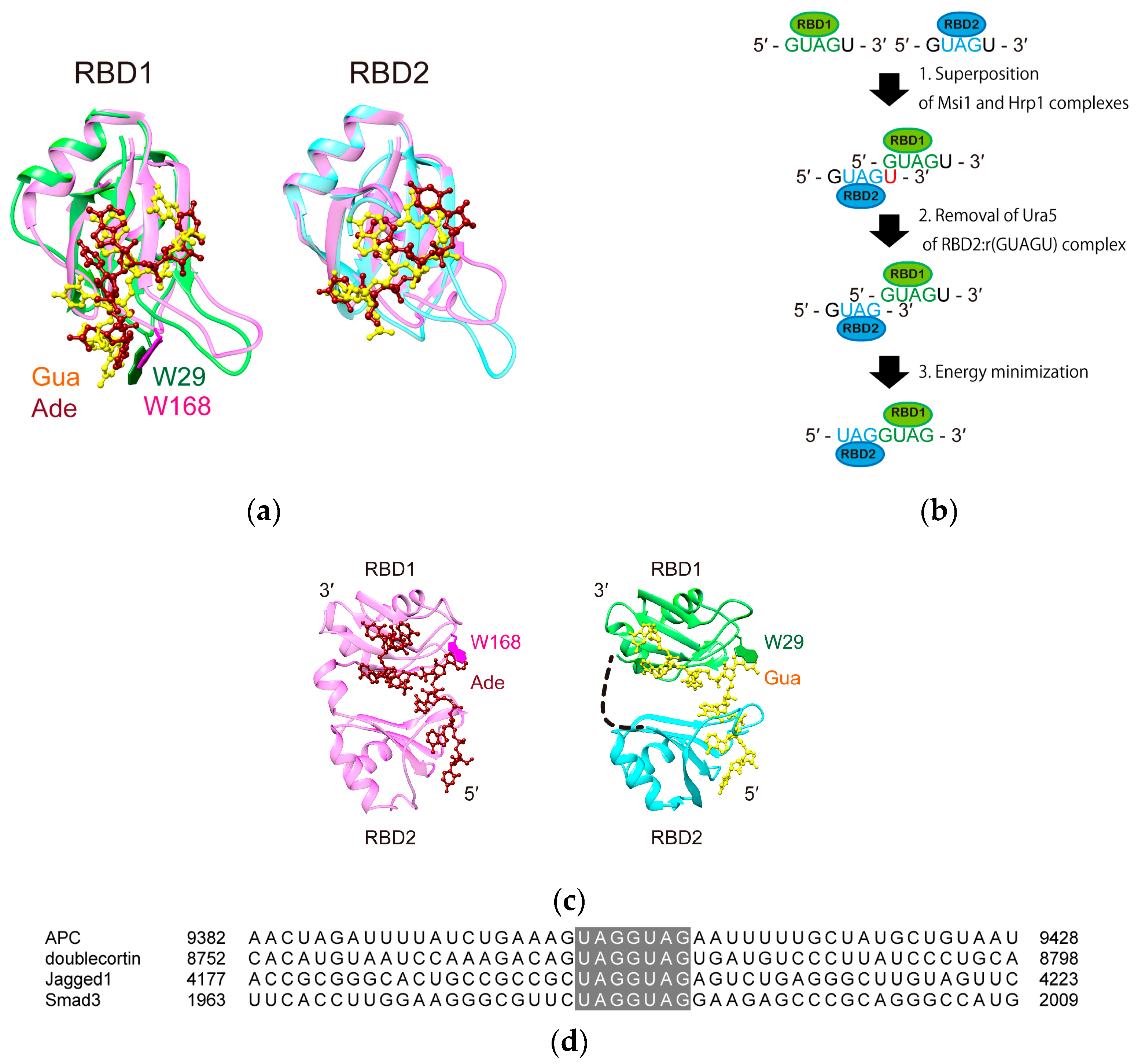
3. Discussion
4. Materials and Methods
4.1. Protein and RNA Preparation
4.2. NMR Spectroscopy
4.3. Structure Calculations
4.4. NMR Titration Experiments
4.5. Heteronuclear Steady-State {1H}-15N Nuclear Overhauser Effect (NOE) Measurements
4.6. Model Structure Generation
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sugiyama-Nakagiri, Y.; Akiyama, M.; Shibata, S.; Okano, H.; Shimizu, H. Expression of RNA-binding protein Musashi in hair follicle development and hair cycle progression. Am. J. Pathol. 2006, 168, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.M.; Fraser, B.A.; Sobinoff, A.P.; Pye, V.J.; Davidson, T.-L.; Siddall, N.A.; Koopman, P.; Hime, G.R.; McLaughlin, E.A. Developmental expression of Musashi-1 and Musashi-2 RNA-binding proteins during spermatogenesis: Analysis of the deleterious effects of dysregulated Expression1. Biol. Reprod. 2014, 90, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, S.-I.; Nakamura, Y.; Yoshida, T.; Shibata, S.; Koike, M.; Takano, H.; Ueda, S.; Uchiyama, Y.; Noda, T.; Okano, H. RNA-binding protein Musashi family: Roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl. Acad. Sci. USA 2002, 99, 15194–15199. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Tokunaga, A.; Yoshida, T.; Hashimoto, M.; Mikoshiba, K.; Weinmaster, G.; Nakafuku, M.; Okano, H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001, 21, 3888–3900. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Han, C.; Yu, Q.; Guan, Y.; Li, N.; Zhou, J.; Tian, Y.; Zhang, Y. Downregulation of Msi1 suppresses the growth of human colon cancer by targeting p21cip1. Int. J. Oncol. 2015, 46, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Cui, J.; Quan, M.; Xie, D.; Jia, Z.; Wei, D.; Wang, L.; Gao, Y.; Ma, Q.; Xie, K. The novel KLF4/MSI2 signaling pathway regulates growth and metastasis of pancreatic cancer. Clin. Cancer Res. 2017, 23, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Kanemura, Y.; Yamasaki, M.; Kanemura, Y.; Mori, K.; Fujikawa, H.; Hayashi, H.; Nakano, A.; Matsumoto, T.; Tamura, K.; Arita, N.; et al. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation 2001, 68, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-H.; Jeong, K.J.; Kim, W.Y.; Lee, H.J.; Gong, G.; Suh, N.; Győrffy, B.; Kim, S.; Jeong, S.-Y.; Mills, G.B.; et al. Musashi RNA-binding protein 2 regulates estrogen receptor 1 function in breast cancer. Oncogene 2017, 36, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Kudinov, A.E.; Deneka, A.; Nikonova, A.S.; Beck, T.N.; Ahn, Y.-H.; Liu, X.; Martinez, C.F.; Schultz, F.A.; Reynolds, S.; Yang, D.-H.; et al. Musashi-2 (MSI2) supports TGF-β signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, 6955–6960. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; An, S.; Choi, Y.M.; Lee, J.; Ahn, K.J.; Lee, J.H.; Kim, T.J.; An, I.-S.; Bae, S. Musashi-2 is a novel regulator of paclitaxel sensitivity in ovarian cancer cells. Int. J. Oncol. 2016, 49, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Penalva, L.O.; Yuan, H.; Linnoila, R.I.; Lu, J.; Okano, H.; Glazer, R.I. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol. Cancer 2010, 9, 221. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, W.; Wang, L.; Kazobinka, G.; Han, X.; Li, B.; Hou, T. Musashi-2 promotes migration and invasion in bladder cancer via activation of the JAK2/STAT3 pathway. Lab. Investig. 2016, 96, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Kharas, M.G.; Lengner, C.J.; Al-Shahrour, F.; Bullinger, L.; Ball, B.; Zaidi, S.; Morgan, K.; Tam, W.; Paktinat, M.; Okabe, R.; et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 2010, 16, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Kwon, H.Y.; Zimdahl, B.; Congdon, K.L.; Blum, J.; Lento, W.E.; Zhao, C.; Lagoo, A.; Gerrard, G.; Foroni, L.; et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 2010, 466, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Griner, L.N.; Reuther, G.W. Aggressive myeloid leukemia formation is directed by the musashi 2/numb pathway. Cancer Biol. Ther. 2010, 10, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ye, A.; Zhang, Y.; Cai, Z.; Wang, W.; Sun, L.; Jiang, S.; Wu, J.; Yu, K.; Zhang, S. Musashi-2 silencing exerts potent activity against acute myeloid leukemia and enhances chemosensitivity to daunorubicin. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.Y.; Bajaj, J.; Ito, T.; Blevins, A.; Konuma, T.; Weeks, J.; Lytle, N.K.; Koechlein, C.S.; Rizzieri, D.; Chuah, C.; et al. Tetraspanin 3 is required for the development and propagation of acute myelogenous leukemia. Cell Stem Cell 2015, 17, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, T.; Nagata, T.; Tsuda, K.; Kobayashi, N.; Imai, T.; Okano, H.; Yamazaki, T.; Katahira, M. Structure of Musashi1 in a complex with target RNA: The role of aromatic stacking interactions. Nucleic Acids Res. 2012, 40, 3218–3231. [Google Scholar] [CrossRef] [PubMed]
- Zearfoss, N.R.; Deveau, L.M.; Clingman, C.C.; Schmidt, E.; Johnson, E.S.; Massi, F.; Ryder, S.P. A conserved three-nucleotide core motif defines musashi RNA binding specificity. J. Biol. Chem. 2014, 289, 35530–35541. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, H.; Imai, T.; Imataka, H.; Tsujimoto, M.; Matsumoto, K.; Okano, H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 2008, 181, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, H.; Okada, Y.; Imai, T.; Iwanami, A.; Mischel, P.S.; Okano, H. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 2011, 286, 16121–16130. [Google Scholar] [CrossRef] [PubMed]
- Cragle, C.; MacNicol, A.M. Musashi protein-directed translational activation of target mRNAs is mediated by the poly(A) polymerase, germ line development defective-2. J. Biol. Chem. 2014, 289, 14239–14251. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, S.; Nakamura, Y.; Satoh, H.; Okano, H. Rna-binding protein Musashi2: Developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J. Neurosci. 2001, 21, 8091–8107. [Google Scholar] [PubMed]
- Battelli, C.; Nikopoulos, G.N.; Mitchell, J.G.; Verdi, J.M. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell. Neurosci. 2006, 31, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Vo, D.T.; Subramaniam, D.; Remke, M.; Burton, T.L.; Uren, P.J.; Gelfond, J.A.; de Sousa Abreu, R.; Burns, S.C.; Qiao, M.; Suresh, U.; et al. The RNA-binding protein musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am. J. Pathol. 2012, 181, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Katz, Y.; Li, F.; Lambert, N.J.; Sokol, E.S.; Tam, W.L.; Cheng, A.W.; Airoldi, E.M.; Lengner, C.J.; Gupta, P.B.; Yu, Z.; et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. Elife 2014, 3, e03915. [Google Scholar] [CrossRef] [PubMed]
- Afroz, T.; Cienikova, Z.; Cléry, A.; Allain, F.H.T. Chapter Nine-one, two, three, four! How multiple RRMs read the genome sequence. Methods Enzymol. 2015, 558, 235–278. [Google Scholar] [CrossRef] [PubMed]
- Nagata, T.; Kanno, R.; Kurihara, Y.; Uesugi, S.; Imai, T.; Sakakibara, S.; Okano, H.; Katahira, M. Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J. Mol. Biol. 1999, 287, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Miyanoiri, Y.; Kobayashi, H.; Imai, T.; Watanabe, M.; Nagata, T.; Uesugi, S.; Okano, H.; Katahira, M. Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, Musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics. J. Biol. Chem. 2003, 278, 41309–41315. [Google Scholar] [CrossRef] [PubMed]
- Iwaoka, R.; Nagata, T.; Tsuda, K.; Imai, T.; Okano, H.; Kobayashi, N.; Katahira, M. Backbone and side chain assignments of the second RNA-binding domain of Musashi-1 in its free form and in complex with 5-mer RNA. status (manuscript in preparation).
- Kessler, M.M.; Henry, M.F.; Shen, E.; Zhao, J.; Gross, S.; Silver, P.A.; Moore, C.L. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3’-end formation in yeast. Genes Dev. 1997, 11, 2545–2556. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cañadillas, J.M. Grabbing the message: Structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J. 2006, 25, 3167–3178. [Google Scholar] [CrossRef] [PubMed]
- Spears, E.; Neufeld, K.L. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J. Biol. Chem. 2011, 286, 4946–4950. [Google Scholar] [CrossRef] [PubMed]
- Horisawa, K.; Imai, T.; Okano, H.; Yanagawa, H. 3′-Untranslated region of doublecortin mRNA is a binding target of the Musashi1 RNA-binding protein. FEBS Lett. 2009, 583, 2429–2434. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-M.; Deering, R.P.; Lu, Y.; Tivnan, P.; Lianoglou, S.; Al-Shahrour, F.; Ebert, B.L.; Hacohen, N.; Leslie, C.; Daley, G.Q.; et al. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J. Exp. Med. 2014, 211, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Vandivier, L.; Parada, K.; Woo, D.H.; Wang, S.; Naqvi, A.S.; Rao, S.; et al. The Msi family of RNA-binding proteins function redundantly as intestinal oncoproteins. Cell Rep. 2015, 13, 2440–2455. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Parada, K.; Rao, S.; Minuesa, G.; Katz, Y.; Gregory, B.D.; et al. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat. Commun. 2015, 6, 6517. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Lin, L.-T.; Wang, M.-L.; Lee, S.-H.; Tsai, M.-L.; Tsai, C.-C.; Liu, W.-H.; Chen, T.-C.; Yang, Y.-P.; Lee, Y.-Y.; et al. Musashi-1 regulates AKT-derived IL-6 autocrinal/paracrinal malignancy and chemoresistance in glioblastoma. Oncotarget 2016, 7, 42485–42501. [Google Scholar] [CrossRef] [PubMed]
- Chiou, G.-Y.; Yang, T.-W.; Huang, C.-C.; Tang, C.-Y.; Yen, J.-Y.; Tsai, M.-C.; Chen, H.-Y.; Fadhilah, N.; Lin, C.-C.; Jong, Y.-J. Musashi-1 promotes a cancer stem cell lineage and chemoresistance in colorectal cancer cells. Sci. Rep. 2017, 7, 2172. [Google Scholar] [CrossRef] [PubMed]
- Muto, J.; Imai, T.; Ogawa, D.; Nishimoto, Y.; Okada, Y.; Mabuchi, Y.; Kawase, T.; Iwanami, A.; Mischel, P.S.; Saya, H.; et al. RNA-binding protein musashi1 modulates glioma cell growth through the post-transcriptional regulation of notch and PI 3 kinase/Akt signaling pathways. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Choi, Y.M.; Kim, K.B.; Lee, J.H.; Chun, Y.K.; An, I.S.; An, S.; Bae, S. DBC2/RhoBTB2 functions as a tumor suppressor protein via Musashi-2 ubiquitination in breast cancer. Oncogene 2017, 36, 2802–2912. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.G.; Lytle, N.K.; Jaquish, D.V.; Park, F.D.; Ito, T.; Bajaj, J.; Koechlein, C.S.; Zimdahl, B.; Yano, M.; Kopp, J.L.; et al. Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature 2016, 534, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Appelman, C.; Smith, A.R.; Yu, J.; Larsen, S.; Marquez, R.T.; Liu, H.; Wu, X.; Gao, P.; Roy, A.; et al. Natural product (−)-gossypol inhibits colon cancer cell growth by targeting RNA-binding protein Musashi-1. Mol. Oncol. 2015, 9, 1406–1420. [Google Scholar] [CrossRef] [PubMed]
- Clingman, C.C.; Deveau, L.M.; Hay, S.A.; Genga, R.M.; Shandilya, S.M.D.; Massi, F.; Ryder, S.P. Allosteric inhibition of a stem cell RNA-binding protein by an intermediary metabolite. Elife 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Iwahara, J.; Koshiba, S.; Tomizawa, T.; Tochio, N.; Güntert, P.; Kigawa, T.; Yokoyama, S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR 2007, 39, 31–52. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Harano, Y.; Tochio, N.; Nakatani, E.; Kigawa, T.; Yokoyama, S.; Mading, S.; Ulrich, E.L.; Markley, J.L.; Akutsu, H.; et al. An automated system designed for large scale NMR data deposition and annotation: Application to over 600 assigned chemical shift data entries to the BioMagResBank from the Riken Structural Genomics/Proteomics Initiative internal database. J. Biomol. NMR 2012, 53, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Blevins, R.A. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Kneller, D.G. Sparky—NMR Assignment and Integration Software. Available online: https://www.cgl.ucsf.edu/home/sparky/ (accessed on 24 June 2017).
- Güntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [CrossRef] [PubMed]
- Güntert, P.; Mumenthaler, C.; Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program Dyana. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, T.; Güntert, P.; Wüthrich, K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002, 319, 209–227. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Powers, R.; Garrett, D.S.; March, C.J.; Frieden, E.A.; Gronenborn, A.M.; Clore, G.M. The high-resolution, three-dimensional solution structure of human interleukin-4 determined by multidimensional heteronuclear magnetic resonance spectroscopy. Biochemistry 1993, 32, 6744–6762. [Google Scholar] [CrossRef] [PubMed]
- Amber Home Page. Available online: http://ambermd.org (accessed on 24 June 2017).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Farrow, N.A.; Muhandiram, R.; Singer, A.U.; Pascal, S.M.; Kay, C.M.; Gish, G.; Shoelson, S.E.; Pawson, T.; Forman-Kay, J.D.; Kay, L.E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N-NMR relaxation. Biochemistry 1994, 33, 5984–6003. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

| Free Msi1(109–200)(109–200) | Msi1(109–200):r(GUAGU) Complex | |
|---|---|---|
| NMR Restraints | ||
| Number of NOE distance restraints | 1116 | 1351 |
| Intraresidue | 315 | 341 |
| Sequential (|i − j| = 1) | 300 | 324 |
| Medium-range (1 < |i − j| < 5) | 165 | 191 |
| Long-range (|i − j| ≥ 5) | 336 | 468 |
| Proteins—RNA Intermolecular | NA | 26 |
| RNA intramolecule | NA | 1 |
| Hydrogen bond restraints a | 27 | 27 |
| Dihedral angle restraints | ||
| φ angle | 32 | 0 |
| χ1 and χ2 angles | 19 | 19 |
| Structure statistics (20 structures) | ||
| AMBER energies (kcal/mol) | ||
| Mean AMBER energy | −2780 | −3800 |
| Mean restraints violation energy | 4.698 | 4.359 |
| Ramachandran plot statistics (%) | ||
| Residues in most favored regions | 91.5 | 91.5 |
| Residues in additionally allowed regions | 7.0 | 7.0 |
| Residues in generously allowed regions | 1.3 | 0.2 |
| sidues in disallowed regions | 0.2 | 1.4 |
| Average RMSD to mean structure (Å) | ||
| Protein backbone | 0.55 ± 0.11 b | 0.74 ± 0.28 c (0.35 ± 0.07 b) |
| Protein heavy atoms | 1.45 ± 0.21 b | 1.45 ± 0.29 c (1.13 ± 0.15 b) |
| Protein heavy atoms and r(UAG) | NA | 1.42 ± 0.28 c (1.13 ± 0.15 b) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwaoka, R.; Nagata, T.; Tsuda, K.; Imai, T.; Okano, H.; Kobayashi, N.; Katahira, M. Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA. Molecules 2017, 22, 1207. https://doi.org/10.3390/molecules22071207
Iwaoka R, Nagata T, Tsuda K, Imai T, Okano H, Kobayashi N, Katahira M. Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA. Molecules. 2017; 22(7):1207. https://doi.org/10.3390/molecules22071207
Chicago/Turabian StyleIwaoka, Ryo, Takashi Nagata, Kengo Tsuda, Takao Imai, Hideyuki Okano, Naohiro Kobayashi, and Masato Katahira. 2017. "Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA" Molecules 22, no. 7: 1207. https://doi.org/10.3390/molecules22071207
APA StyleIwaoka, R., Nagata, T., Tsuda, K., Imai, T., Okano, H., Kobayashi, N., & Katahira, M. (2017). Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA. Molecules, 22(7), 1207. https://doi.org/10.3390/molecules22071207