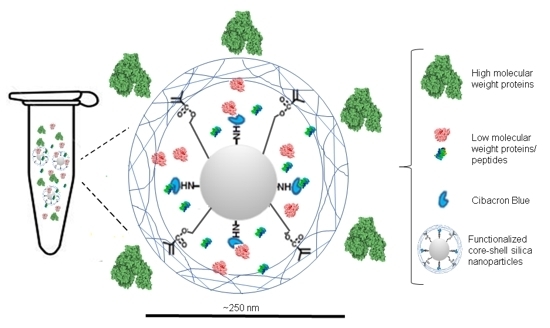
Novel Synthesis of Core-Shell Silica Nanoparticles for the Capture of Low Molecular Weight Proteins and Peptides
Abstract
:1. Introduction
2. Results and Discussion
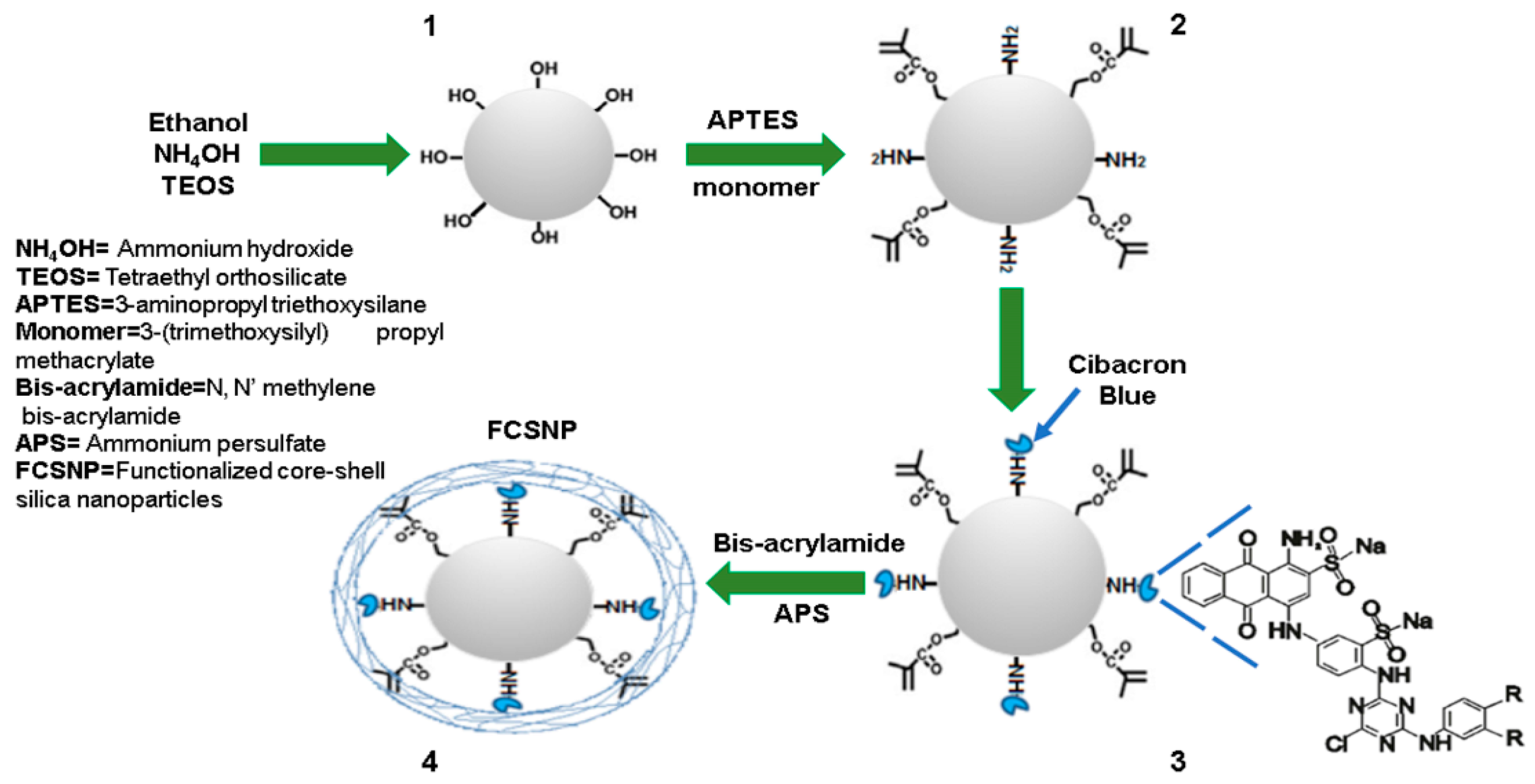
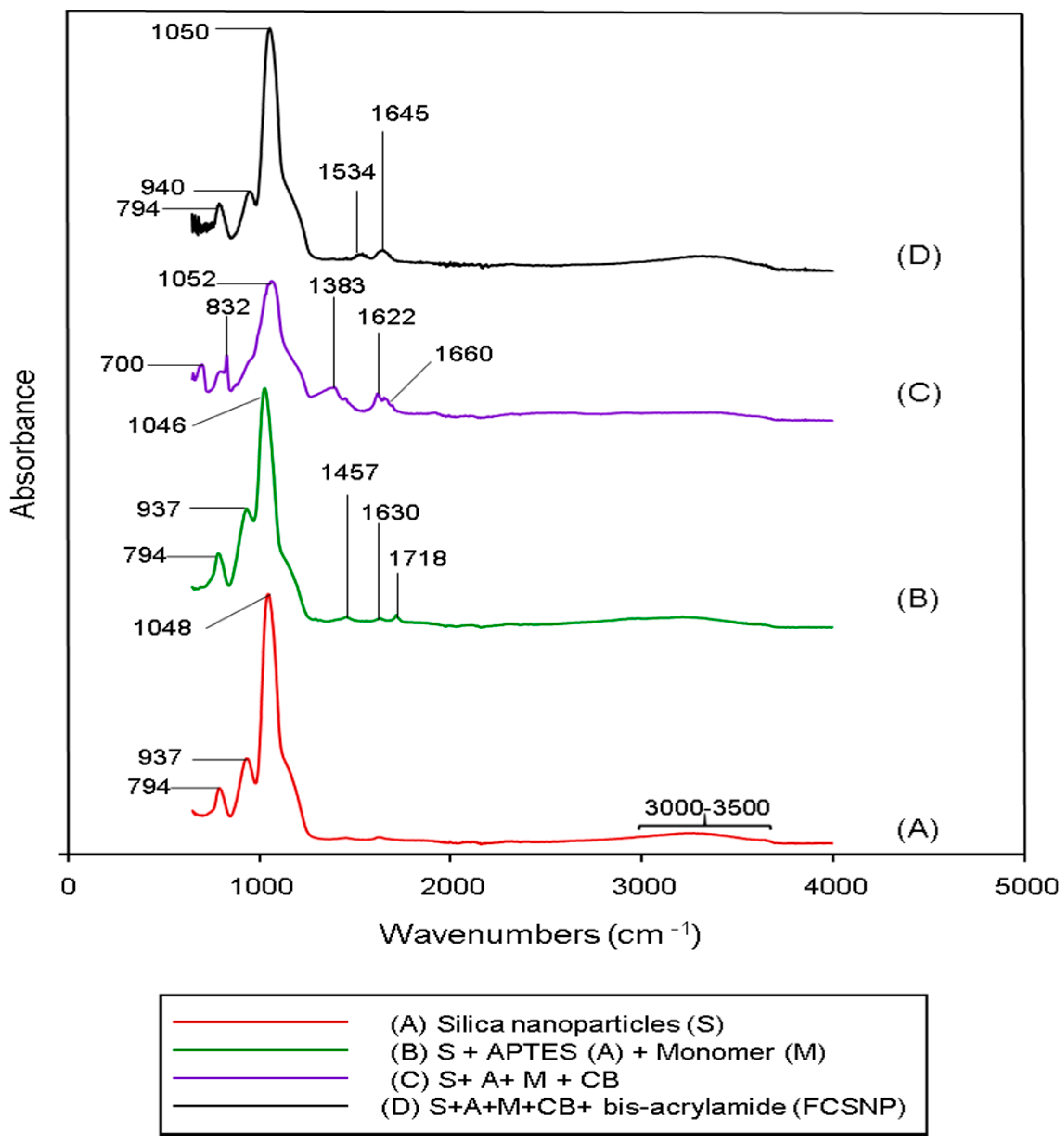
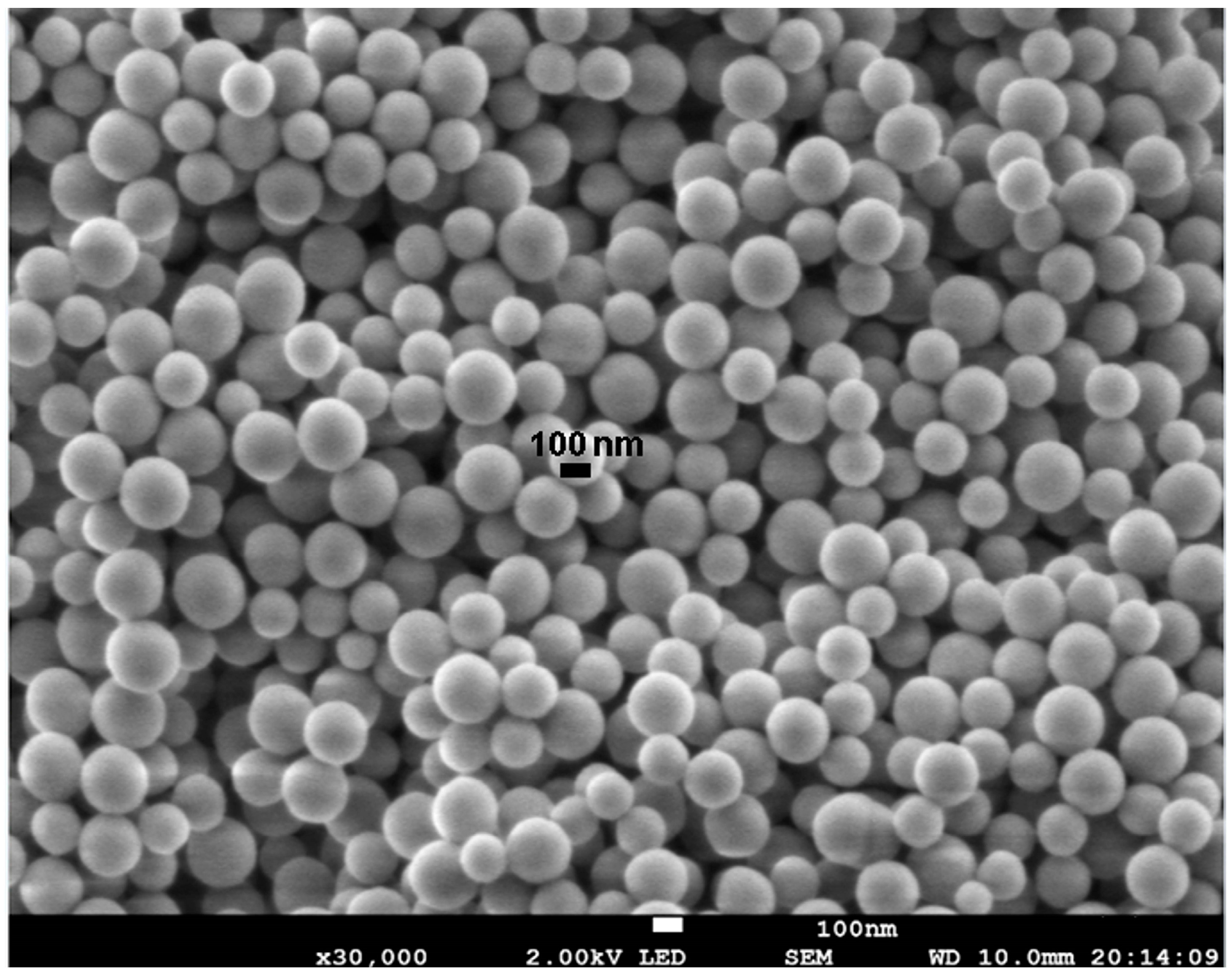
2.1. Synthesis of FCSNP and Characterization
2.2. Evaluation of the FCSNP in the Capture of Peptides
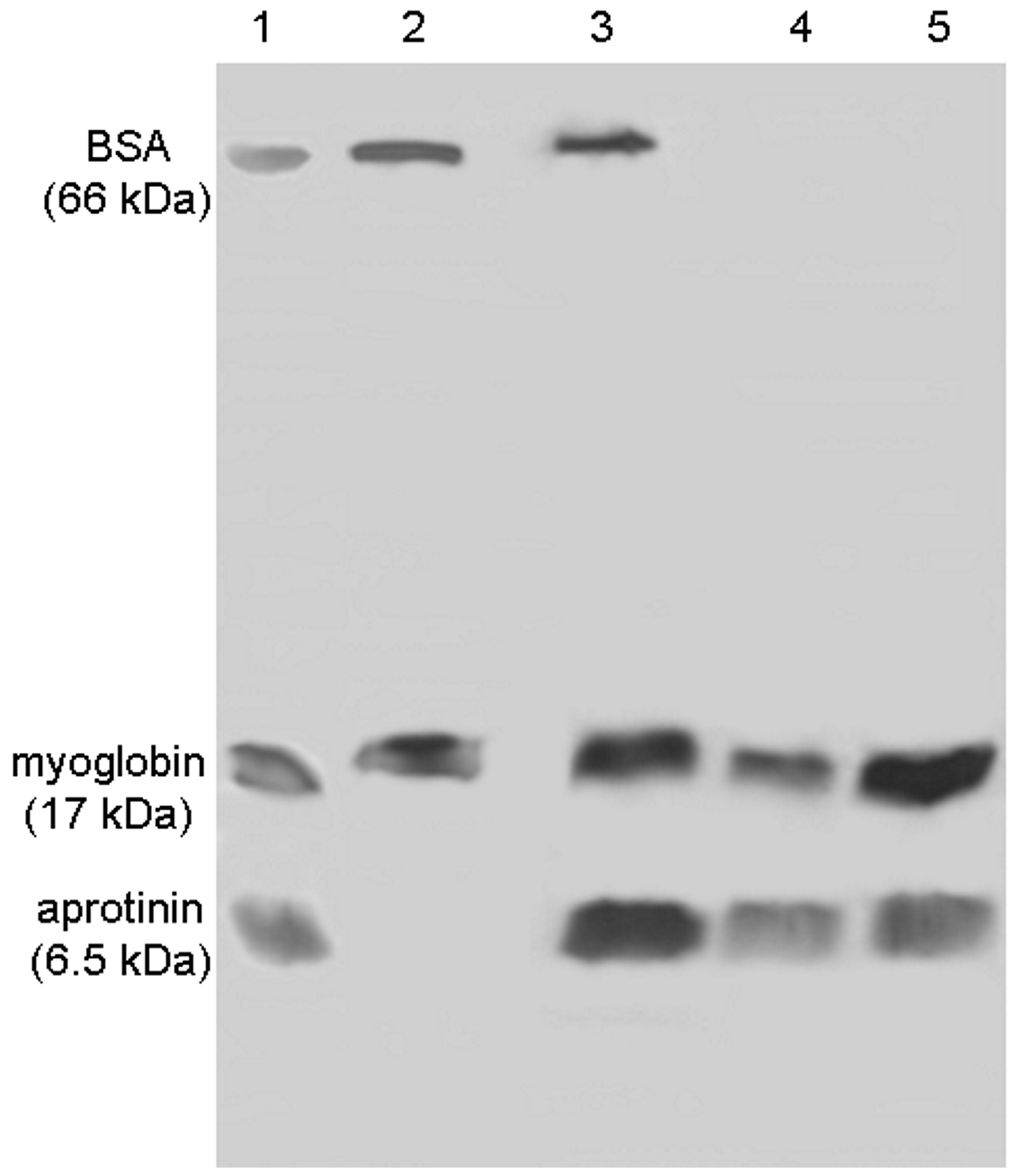
2.3. Effectiveness of Molecular Bait to Capture LMW Proteins
2.4. FCSNP in the Exclusion of HMW Proteins and Capture of LMW Proteins
2.5. Estimation of the Nanoparticles Capacity
3. Materials and Methods
3.1. General Information
3.2. Synthesis of Functionalized Core-Shell Silica Nanoparticles (FCSNP)
3.3. Size and Surface Charge Characterization of the Nanoparticles
3.4. ATR-FTIR Spectroscopic Studies
3.5. Morphological Characterization of the Nanoparticles
3.6. Evaluation of the FCSNP in the Capture of Peptides
3.7. Effectiveness of Molecular Bait in the Capture of LMW Proteins
3.8. FCSNP in the Exclusion of HMW Proteins and Capture of LMW Proteins
3.9. Estimation of the Nanoparticles Capacity
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tirumalai, R.S.; Chan, K.C.; Prieto, D.A.; Issaq, H.J.; Conrads, T.P.; Veenstra, T.D. Characterization of the Low Molecular Weight Human Serum Proteome. Mol. Cell. Proteomics 2003, 2, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Luchini, A.; Fredolini, C.; Espina, B.H.; Meani, F.; Reeder, A.; Rucker, A.; Petricoin, E.F., III; Liotta, L.A. Nanoparticle Technology: Addressing the fundamental roadblocks to protein biomarker discovery. Curr. Mol. Med. 2010, 10, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Merrell, K.; Southwick, K.; Graves, S.W.; Esplin, M.S.; Lewis, N.E.; Thulin, C.D. Analysis of low-abundance, low-molecular-weight serum proteins using mass spectrometry. J. Biomol. Tech. 2004, 15, 238–248. [Google Scholar] [PubMed]
- González-Buitrago, J.M.; Ferreira, L.; Lorenzo, I. Urinary proteomics. Clin. Chim. Acta 2007, 375, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Ni, J.; Beretov, J.; Cozzi, P.; Willcox, M.; Wasinger, V.; Walsh, B.; Graham, P.; Li, Y. Urinary biomarkers in prostate cancer detection and monitoring progression. Crit. Rev. Oncol. Hematol. 2017, 118, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Longo, C.; Patanarut, A.; George, T.; Bishop, B.; Zhou, W.; Fredolini, C.; Ross, M.M.; Espina, V.; Pellacani, G.; Petricoin, E.F.; et al. Core-shell hydrogel particles harvest, concentrate and preserve labile low abundance biomarkers. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.; Donadio-Andréi, S.; Louwagie, M.; Couté, Y.; Picard, G.; Lacoste, C.; Bruley, C.; Garin, J.; Ichai, P.; Faivre, J.; et al. Introducing plasma/serum glycodepletion for the targeted proteomics analysis of cytolysis biomarkers. Talanta 2017, 170, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Hardman, R. A toxicologic review of quantum dots: Toxicity depends on physicochemical and environmental factors. Environ. Health Perspect. 2006, 114, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Pasinszki, T.; Krebsz, M.; Tung, T.T.; Losic, D. Carbon Nanomaterial Based Biosensors for Non-Invasive Detection of Cancer and Disease Biomarkers for Clinical Diagnosis. Sensors 2017, 17, 1919. [Google Scholar] [CrossRef] [PubMed]
- Balakumaran, M.D.; Ramachandran, R.; Balashanmugam, P.; Mukeshkumar, D.J.; Kalaichelvan, P.T. Mycosynthesis of silver and gold nanoparticles: Optimization, characterization and antimicrobial activity against human pathogens. Microbiol. Res. 2016, 182, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Yuan, Z.; Han, H.; Li, T.; Li, L.; Guo, X. Chitosan cross-linked poly(acrylic acid) hydrogels: Drug release control and mechanism. Colloids Surf. B Biointerfaces 2017, 152, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Tamburro, D.; Fredolini, C.; Espina, V.; Douglas, T.A.; Ranganathan, A.; Ilag, L.; Zhou, W.; Russo, P.; Espina, B.H.; Muto, G.; et al. Multifunctional core-shell nanoparticles: Discovery of previously invisible biomarkers. J. Am. Chem. Soc. 2011, 133, 19178–19188. [Google Scholar] [CrossRef] [PubMed]
- Negin, C.; Ali, S.; Xie, Q. Application of nanotechnology for enhancing oil recovery—A review. Petroleum 2016, 2, 324–333. [Google Scholar] [CrossRef]
- Pham, T.; Jackson, J.B.; Halas, N.J.; Lee, T.R. Preparation and Characterization of Gold Nanoshells Coated with Self-Assembled Monolayers. Langmuir 2002, 18, 4915–4920. [Google Scholar] [CrossRef]
- Watermann, A.; Brieger, J. Mesoporous Silica Nanoparticles as Drug Delivery Vehicles in Cancer. Nanomaterials 2017, 7, 189. [Google Scholar] [CrossRef] [PubMed]
- Liberman, A.; Mendez, N.; Trogler, W.C.; Kummel, A.C. Synthesis and surface functionalization of silica nanoparticles for nanomedicine. Surf. Sci. Rep. 2014, 69, 132–158. [Google Scholar] [CrossRef] [PubMed]
- Stöber, W.; Fink, A.; Bohn, E. Controlled growth of monodisperse silica spheres in the micron size range. J. Colloid Interface Sci. 1968, 26, 62–69. [Google Scholar] [CrossRef]
- Ferreira, G.; Hernandez-Martinez, A.R.; Pool, H.; Molina, G.; Cruz-Soto, M.; Luna-Barcenas, G.; Estevez, M. Synthesis and functionalization of silica-based nanoparticles with fluorescent biocompounds extracted from Eysenhardtia polystachya for biological applications. Mater. Sci. Eng. C 2015, 57, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Kopperschläger, G.; Böhme, H.-J.; Hofmann, E. Cibacron blue F3G-A and related dyes as ligands in affinity chromatography. Chromatography 1982, 25, 101–138. [Google Scholar] [CrossRef]
- Pinto Reis, C.; Neufeld, R.J.; Ribeiro, A.J.; Veiga, F. Nanoencapsulation I. Methods for preparation of drug-loaded polymeric nanoparticles. Nanomedicine 2006, 2, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Achilleos, D.S.; Vamvakaki, M. End-grafted polymer chains onto inorganic nano-objects. Materials 2010, 3, 1981–2026. [Google Scholar] [CrossRef]
- Morini, M.A.; Sierra, M.B.; Pedroni, V.I.; Alarcon, L.M.; Appignanesi, G.A.; Disalvo, E.A. Influence of temperature, anions and size distribution on the zeta potential of DMPC, DPPC and DMPE lipid vesicles. Colloids Surf. B Biointerfaces 2015, 131, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, M.A.; Hassanajili, S.; Rahimpour, M.R. Synthesis of fluorinated nano-silica and its application in wettability alteration near-wellbore region in gas condensate reservoirs. Appl. Surf. Sci. 2013, 273, 205–214. [Google Scholar] [CrossRef]
- Latthe, S.S.; Imai, H.; Ganesan, V.; Rao, A.V. Superhydrophobic silica films by sol-gel co-precursor method. Appl. Surf. Sci. 2009, 256, 217–222. [Google Scholar] [CrossRef]
- Majoul, N.; Aouida, S.; Bessaïs, B. Progress of porous silicon APTES-functionalization by FTIR investigations. Appl. Surf. Sci. 2015, 331, 388–391. [Google Scholar] [CrossRef]
- Zhang, D.H.; Chen, N.; Yang, M.N.; Dou, Y.F.; Sun, J.; Liu, Y.D.; Zhi, G.Y. Effects of different spacer arms on Cibacron Blue modification and protein affinity adsorption on magnetic microspheres. J. Mol. Catal. B Enzym. 2016, 133, 136–143. [Google Scholar] [CrossRef]
- Chiem, L.T.; Huynh, L.; Ralston, J.; Beattie, D.A. An in situ ATR-FTIR study of polyacrylamide adsorption at the talc surface. J. Colloid Interface Sci. 2006, 297, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, M.; Lu, Q.; Shi, Z. Superhydrophobic polyimide films with a hierarchical topography: Combined replica molding and layer-by-layer assembly. Langmuir 2008, 24, 12651–12657. [Google Scholar] [CrossRef] [PubMed]
- Ishii, H.; Ikuno, T.; Shimojima, A.; Okubo, T. Preparation of core-shell mesoporous silica nanoparticles with bimodal pore structures by regrowth method. J. Colloid Interface Sci. 2015, 448, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yang, Q.; Jin, S.; Song, Y.; Gao, J.; Wang, Y.; Mi, H. Core-shell molecularly imprinted polymer nanoparticles with assistant recognition polymer chains for effective recognition and enrichment of natural low-abundance protein. Acta Biomater. 2014, 10, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Pyo, J.H.; Jo, E.H.; Hwang, S.I.; Kang, S.C.; Jung, J.H.; Park, E.K.; Kim, S.Y.; Choi, J.Y.; Lim, J. Establishment of a near-standard two-dimensional human urine proteomic map. Proteomics 2004, 4, 3485–3497. [Google Scholar] [CrossRef] [PubMed]
- Pieper, R.; Gatlin, C.L.; McGrath, A.M.; Makusky, A.J.; Mondal, M.; Seonarain, M.; Field, E.; Schatz, C.R.; Estock, M.A.; Ahmed, N.; et al. Characterization of the human urinary proteome: A method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics 2004, 4, 1159–1174. [Google Scholar] [CrossRef] [PubMed]
- Candiano, G.; Santucci, L.; Petretto, A.; Bruschi, M.; Dimuccio, V.; Urbani, A.; Bagnasco, S.; Ghiggeri, G.M. 2D-electrophoresis and the urine proteome map: Where do we stand? J. Proteomics 2010, 73, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
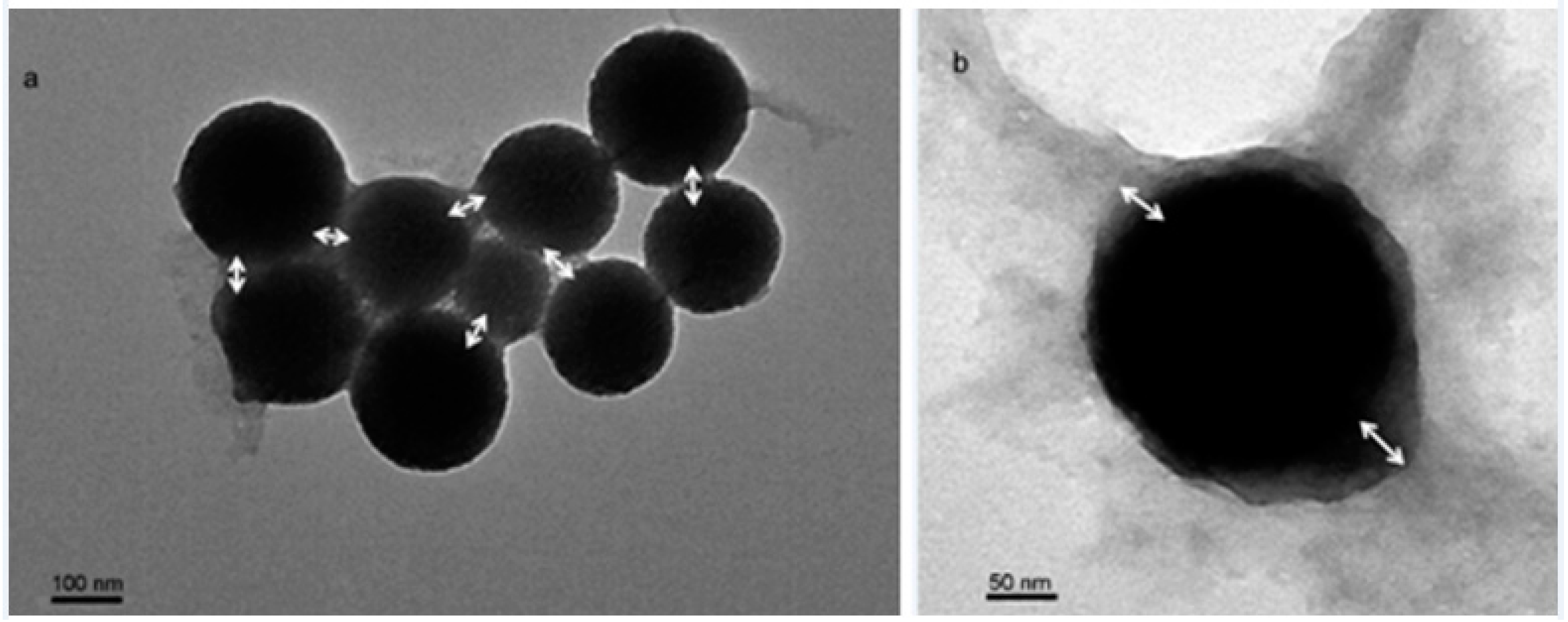

| Stage of Synthesis | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Silica Nanoparticles | APTES and Monomer | Cibacron Blue | FCSNP | |
| Size ± SD (nm) | 138.9 ± 5.6 | 218.4 ± 4.7 | 211.5 ± 18.1 | 243.9 ± 11.6 |
| Zeta potential ± SD (mV) | −47 ± 0.7 | −34.2 ± 1.5 | −54.6 ± 0.5 | −38.1 ± 0.9 |
| Peptide Sequence | Theoretical Mass (Da) | Experimental m/z | Error (Da) | |
|---|---|---|---|---|
| Wash* with 50 mM phosphate buffer | Consecutive elutions ** with 50% isopropanol, 50% Methanol and ACN + NH4OH *** | |||
| 1. YSSKPDIVG | 965.49 | −0.4352 | ||
| 965.06 | (M + H)1+ | |||
| Not eluted | ||||
| 483.2519 | 0.5562 | |||
| (M + 2H)2+ | ||||
| 2. LQQAIAAS | ||||
| NIPLSPLLFQ | ||||
| QSPALSLVQ | 3646.32 | 1216.3632 | 1216.3632 | 0.2304 |
| SLVQ TIR | (M + 3H)3+ | (M + 3H)3+ | ||
| Model Protein | Capacity of FCSNP (µg/mg) |
|---|---|
| Aprotinin | 3.4 ± 0.15 |
| Myoglobin | 3.1 ± 0.26 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez-Leon, S.G.; Sarabia-Sainz, J.A.-i.; Montfort, G.R.-C.; Guzman-Partida, A.M.; Robles-Burgueño, M.D.R.; Vazquez-Moreno, L. Novel Synthesis of Core-Shell Silica Nanoparticles for the Capture of Low Molecular Weight Proteins and Peptides. Molecules 2017, 22, 1712. https://doi.org/10.3390/molecules22101712
Hernandez-Leon SG, Sarabia-Sainz JA-i, Montfort GR-C, Guzman-Partida AM, Robles-Burgueño MDR, Vazquez-Moreno L. Novel Synthesis of Core-Shell Silica Nanoparticles for the Capture of Low Molecular Weight Proteins and Peptides. Molecules. 2017; 22(10):1712. https://doi.org/10.3390/molecules22101712
Chicago/Turabian StyleHernandez-Leon, Sergio G., Jose Andre-i Sarabia-Sainz, Gabriela Ramos-Clamont Montfort, Ana M. Guzman-Partida, Maria Del Refugio Robles-Burgueño, and Luz Vazquez-Moreno. 2017. "Novel Synthesis of Core-Shell Silica Nanoparticles for the Capture of Low Molecular Weight Proteins and Peptides" Molecules 22, no. 10: 1712. https://doi.org/10.3390/molecules22101712
APA StyleHernandez-Leon, S. G., Sarabia-Sainz, J. A.-i., Montfort, G. R.-C., Guzman-Partida, A. M., Robles-Burgueño, M. D. R., & Vazquez-Moreno, L. (2017). Novel Synthesis of Core-Shell Silica Nanoparticles for the Capture of Low Molecular Weight Proteins and Peptides. Molecules, 22(10), 1712. https://doi.org/10.3390/molecules22101712