Multivalent Aptamers: Versatile Tools for Diagnostic and Therapeutic Applications
Abstract
:1. Introduction
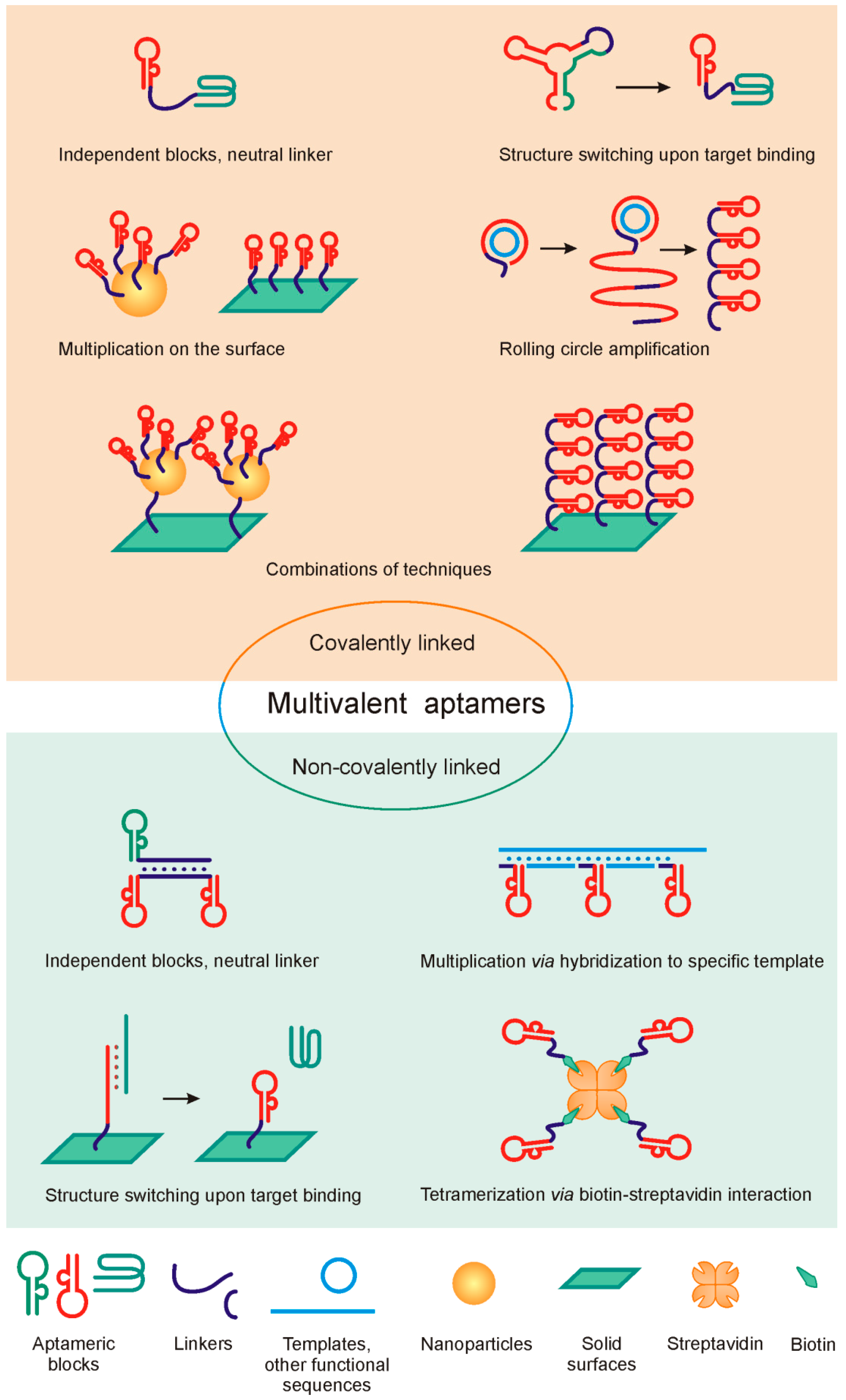
2. Design of Multivalent Aptamers: General Principles
3. Analytical Applications of Multivalent Aptamers
3.1. Aptasensors Based on Multivalent Aptamers
3.2. Multivalent Aptamers as Analytical Probes
3.3. Multivalent Aptamers for Cell Studies
4. Multivalent Aptamers as a Basis for Therapeutic Agents
4.1. Anti-Thrombotic Aptamers
4.2. Anti-Inflammatory Aptamers
4.3. Anticancer Aptamers
4.3.1. Aptamers Binding to Cell-Surface Targets
4.3.2. Aptamers Binding to Soluble Proteins
4.4. Antiviral Aptamers
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ozer, A.; Pagano, J.M.; Lis, J.T. New technologies provide quantum changes in the scale, speed, and success of SELEX methods and aptamer characterization. Mol. Ther. Nucleic Acids 2014, 3, e183. [Google Scholar] [CrossRef] [PubMed]
- Shigdar, S.; Macdonald, J.; O’Connor, M.; Wang, T.; Xiang, D.; Al Shamaileh, H.; Qiao, L.; Wei, M.; Zhou, S.F.; Zhu, Y.; et al. Aptamers as theranostic agents: Modifications, serum stability and functionalisation. Sensors 2013, 13, 13624–13637. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zu, Y. A Highlight of recent advances in aptamer technology and its application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Donovan, M.J.; Jiang, J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013, 113, 2842–2862. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Huang, P.-J.J.; Ding, J.; Liu, J. Aptamer-based biosensors for biomedical diagnostics. Analyst 2014, 139, 2627–2640. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.-H.; Zhang, T.; Luo, H.; Yen, T.; Chen, P.-W.; Han, Y.; Lo, Y.-H. Nucleic acid aptamers: An emerging tool for biotechnology and biomedical sensing. Sensors 2015, 15, 16281–16313. [Google Scholar] [CrossRef] [PubMed]
- Darmostuk, M.; Rimpelová, S.; Gbelcová, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liang, C.; Lv, Q.; Li, D.; Xu, X.; Lui, B.; Lu, A.; Zhang, G. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int. J. Mol. Sci. 2016, 17, 358. [Google Scholar] [CrossRef] [PubMed]
- Maier, K.E.; Levy, M. From selection hits to clinical leads: Progress in aptamer discovery. Mol. Ther. Methods Clin. Dev. 2016, 5, 16014. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; Povsic, T.J.; Sullenger, B.A.; Becker, R.C. Translation and clinical development of antithrombotic aptamers. Nucleic Acid Ther. 2016, 26, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Shigdar, S.; Qiao, G.; Wang, T.; Kouzani, A.Z.; Zhou, S.-F.; Kong, L.; Li, Y.; Pu, C.; Duan, W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: The next generation of cancer medicine. Theranostics 2015, 5, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G. Predicting the uncertain future of aptamer-based diagnostics and therapeutics. Molecules 2015, 20, 6866–6887. [Google Scholar] [CrossRef] [PubMed]
- Camorani, S.; Cerchia, L. Oligonucleotide aptamers for glioma targeting: An update. Cent. Nerv. Syst. Agents Med. Chem. 2015, 15, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Hoffman, B.E.; Lis, J.T. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl. Acad. Sci. USA 1999, 96, 10033–10038. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Lis, J.T. Multivalent RNA Aptamers and Their Expression in Multicellular Organisms. U.S. Patent N 6458559 B1, 1 October 2002. [Google Scholar]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for improving aptamer binding affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Taira, K.; Sode, K.; Ikebukuro, K. Improvement of aptamer affinity by dimerization. Sensors 2008, 8, 1090–1098. [Google Scholar] [CrossRef]
- Le, T.T.; Scott, S.; Cass, A.E.G. Streptavidin binding bifunctional aptamers and their interaction with low molecular weight ligands. Anal. Chim. Acta 2013, 761, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Dupont, D.M.; Bjerregaard, N.; Verpaalen, B.; Andreasen, P.A.; Jensen, J.K. Building a molecular trap for a serine protease from aptamer and peptide modules. Bioconjug. Chem. 2016, 27, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Umehara, T.; Fukuda, K.; Nishikawa, F.; Kohara, M.; Hasegawa, T.; Nishikawa, S. Rational design of dual-functional aptamers that inhibit the protease and helicase activities of HCV NS3. J. Biochem. 2005, 137, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, J.; Shlyahovsky, B.; Li, D.; Willner, I. Parallel analysis of two analytes in solutions or on surfaces by using a bifunctional aptamer: Applications for biosensing and logic gate operations. ChemBioChem 2008, 9, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Goda, T.; Higashi, D.; Matsumoto, A.; Hoshi, T.; Sawaguchi, T.; Miyahara, Y. Dual aptamer-immobilized surfaces for improved affinity through multiple target binding in potentiometric thrombin biosensing. Biosens. Bioelectron. 2015, 73, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.K.; Eckert, M.A.; Ali, M.M.; Riazifar, H.; Pone, E.J.; Liu, L.; Zhao, W. Facile supermolecular aptamer inhibitors of L-selectin. PLoS ONE 2015, 10, e0123034. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Oh, S.S.; Nie, J.; Stewart, R.; Radeke, M.J.; Coffey, P.J.; Thomson, J.A.; Soh, H.T. Array-based discovery of aptamer pairs. Anal. Chem. 2015, 87, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Boltz, A.; Piater, B.; Toleikis, L.; Guenther, R.; Kolmar, H.; Hock, B. Bi-specific aptamers mediating tumor cell lysis. J. Biol. Chem. 2011, 286, 21896–21905. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Wulffen, B.; Pötzsch, B.; Mayer, G. Multidomain targeting generates a high-affinity thrombin-inhibiting bivalent aptamer. ChemBioChem 2007, 8, 2223–2226. [Google Scholar] [CrossRef] [PubMed]
- Soule, E.E.; Bompiani, K.M.; Woodruff, R.S.; Sullenger, B.A. Targeting two coagulation cascade proteases with a bivalent aptamer yields a potent and antidote-controllable anticoagulant. Nucleic Acid Ther. 2016, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ali, M.M.; Eckert, M.A.; Kang, D.K.; Chen, Y.Y.; Sender, L.S.; Fruman, D.A.; Zhao, W. A polyvalent aptamer system for targeted drug delivery. Biomaterials 2013, 34, 9728–9735. [Google Scholar] [CrossRef] [PubMed]
- Riese, S.B.; Buscher, K.; Enders, S.; Kuehne, C.; Tauber, R.; Dernedde, J. Structural requirements of mono- and multivalent L-selectin blocking aptamers for enhanced receptor inhibition in vitro and in vivo. Nanomedicine 2016, 12, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Burke, D.H.; Willis, J.H. Recombination, RNA evolution, and bifunctional RNA molecules isolated through Chimeric SELEX. RNA 1998, 4, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, K.M.; Xiao, Y.; Tom Soh, H. Selection is more intelligent than design: Improving the affinity of a bivalent ligand through directed evolution. Nucleic Acids Res. 2012, 40, 11777–11783. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, V.M.; Estroff, L.A.; Whitesides, G.M. Multivalency in Ligand Design. In Fragment-Based Approaches in Drug Discovery; Jahnke, W., Erlanson, D.A., Eds.; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2006; pp. 11–53. [Google Scholar]
- Xu, D.; Shi, H. Composite RNA aptamers as functional mimics of proteins. Nucleic Acids Res. 2009, 37, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shepard, J.R.E.; Shi, H. An RNA-based transcription activator derived from an inhibitory aptamer. Nucleic Acids Res. 2010, 38, 2378–2386. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Lis, J.T.; Shi, H. A systematic study of the features critical for designing a high avidity multivalent aptamer. Nucleic Acid Ther. 2013, 23, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Di Giusto, D.A.; King, G.C. Construction, stability, and activity of multivalent circular anticoagulant aptamers. J. Biol. Chem. 2004, 279, 46483–46489. [Google Scholar] [CrossRef] [PubMed]
- Di Giusto, D.A.; Knox, S.M.; Lai, Y.; Tyrelle, G.D.; Aung, M.T.; King, G.C. Multitasking by multivalent circular DNA aptamers. ChemBioChem 2006, 7, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, M.N.; Kolpashchikov, D.M. Modular aptameric sensors. J. Am. Chem. Soc. 2004, 126, 9266–9270. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Shimada, I.; Kimura, R.; Hyuga, M. Light-up fluorophore–DNA aptamer pair for label-free turn-on aptamer sensors. Chem. Commun. 2016, 52, 4041–4044. [Google Scholar] [CrossRef] [PubMed]
- Bing, T.; Liu, X.; Cheng, X.; Cao, Z.; Shangguan, D. Bifunctional combined aptamer for simultaneous separation and detection of thrombin. Biosens. Bioelectron. 2010, 25, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cao, Y.; Chen, G.; Li, G. Regulation of thrombin activity with a bifunctional aptamer and hemin: Development of a new anticoagulant and antidote pair. ChemBioChem 2009, 10, 2171–2176. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cao, Z.; Tan, W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Natl. Acad. Sci. USA 2008, 105, 5664–5669. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Heyduk, T. Bivalent ligands with long nanometer-scale flexible linkers. Biochemistry 2009, 48, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P.R.; Ruggiero, A.; Gardner, J.R.; Kuryavyi, V.; Maguire, W.F.; Heaney, M.L.; McDevitt, M.R.; Patel, D.J.; Scheinberg, D.A. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res. 2011, 39, 2458–2469. [Google Scholar] [CrossRef] [PubMed]
- Vorobyeva, M.; Timoshenko, V.; Vorobjev, P.; Venyaminova, A. Aptamers against immunologic targets: Diagnostic and therapeutic prospects. Nucleic Acid Ther. 2016, 26, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Meng, L.; Zhang, X.; Chen, Y.; Zhu, G.; Liu, H.; Xiong, X.; Sefah, K.; Tan, W. Engineering polymeric aptamers for selective cytotoxicity. J. Am. Chem. Soc. 2011, 133, 13380–13386. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Zhang, X.B.; Lv, Y.; Gong, L.; Wang, R.; Zhu, X.; Yang, R.; Tan, W. Functional DNA-containing nanomaterials: Cellular applications in biosensing, imaging, and targeted therapy. Acc. Chem. Res. 2014, 47, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Han, D.; Chen, T.; Peng, L.; Zhu, G.; You, M.; Qiu, L.; Sefah, K.; Zhang, X.; Tan, W. Building a multifunctional aptamer-based dna nanoassembly for targeted cancer therapy. J. Am. Chem. Soc. 2013, 135, 18644–18650. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.L.; Chang, H.T.; Chen, C.T.; Wei, S.C.; Shiang, Y.C.; Huang, C.C. Highly efficient control of thrombin activity by multivalent nanoparticles. Chem. A Eur. J. 2011, 17, 10994–11000. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.S.; Wei, S.C.; Chang, H.T.; Lin, H.J.; Huang, C.C. Gold nanoparticles modified with self-assembled hybrid monolayer of triblock aptamers as a photoreversible anticoagulant. J. Control. Release 2016, 221, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hu, H.; Zhao, Y.; Chen, X.; Li, W.; Qiang, W.; Xu, D. Multifunctional aptamer-silver conjugates as theragnostic agents for specific cancer cell therapy and fluorescence-enhanced cell imaging. Anal. Chem. 2015, 87, 3736–3745. [Google Scholar] [CrossRef] [PubMed]
- Stephanopoulos, N.; Tong, G.J.; Hsiao, S.C.; Francis, M.B. Dual-surface modified virus capsids for targeted delivery of photodynamic agents to cancer cells. ACS Nano 2010, 4, 6014–6020. [Google Scholar] [CrossRef] [PubMed]
- Tong, G.J.; Hsiao, S.C.; Carrico, Z.M.; Francis, M.B. Viral capsid DNA aptamer conjugates as multivalent cell-targeting vehicles. J. Am. Chem. Soc. 2009, 131, 11174–11178. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Jung, H.; Kim, S.A.; Mok, H. Multivalent comb-type aptamer-siRNA conjugates for efficient and selective intracellular delivery. Chem. Commun. 2014, 50, 6765–6767. [Google Scholar] [CrossRef] [PubMed]
- Santulli-Marotto, S.; Nair, S.K.; Rusconi, C.; Santulli-Marotto, S.; Nair, S.K.; Rusconi, C.; Sullenger, B.; Gilboa, E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003, 63, 7483–7489. [Google Scholar] [PubMed]
- McNamara, J.O.; Kolonias, D.; Pastor, F.; Mittler, R.S.; Chen, L.; Giangrande, P.H.; Sullenger, B.; Gilboa, E. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J. Clin. Investig. 2008, 118, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Dollins, C.M.; Nair, S.; Boczkowski, D.; Lee, J.; Layzer, J.M.; Gilboa, E.; Sullenger, B.A. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 2008, 15, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Pastor, F.; Kolonias, D.; McNamara Ii, J.O.; Gilboa, E. Targeting 4-1BB Costimulation to disseminated tumor lesions with bi-specific oligonucleotide aptamers. Mol. Ther. 2011, 19, 1878–1886. [Google Scholar] [CrossRef] [PubMed]
- Tahiri-Alaoui, A.; Frigotto, L.; Manville, N.; Ibrahim, J.; Romby, P.; James, W. High affinity nucleic acid aptamers for streptavidin incorporated into bi-specific capture ligands. Nucleic Acids Res. 2002, 30, e45. [Google Scholar] [CrossRef] [PubMed]
- Parekh, P.; Kamble, S.; Zhao, N.; Zeng, Z.; Portier, B.P.; Zu, Y. Immunotherapy of CD30-expressing lymphoma using a highly stable ssDNA aptamer. Biomaterials 2013, 34, 8909–8917. [Google Scholar] [CrossRef] [PubMed]
- Pratico, E.D.; Sullenger, B.A.; Nair, S.K. Identification and characterization of an agonistic aptamer against the T cell costimulatory receptor, OX40. Nucleic Acid Ther. 2013, 23, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, S.; Lee, C.H.; Chuang, T.L.; Hsueh, P.R.; Lai, H.C.; Lin, C.W. Amplified surface plasmon resonance immunosensor for interferon-Gamma based on a streptavidin-incorporated aptamer. Biosens. Bioelectron. 2012, 37, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.-L.; Chang, C.-C.; Chu-Su, Y.; Wei, S.-C.; Zhao, X.-H.; Hsueh, P.-R.; Lin, C.-W. Disposable surface plasmon resonance aptasensor with membrane-based sample handling design for quantitative interferon-gamma detection. Lab Chip 2014, 14, 2968–2977. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, B.; Wei, H.; Wang, Y.; Wang, E. Biosensor based on an integrated aptamer. Anal. Chem. 2008, 80, 5110–5117. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Tian, D.; Cui, H. Electrochemiluminescence biosensor for the assay of small molecule and protein based on bifunctional aptamer and chemiluminescent functionalized gold nanoparticles. Anal. Chim. Acta 2012, 715, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Chen, J.; Nie, L.; Nie, Z.; Yao, S. Sensitive bifunctional aptamer-based electrochemical biosensor for small molecules and protein. Anal. Chem. 2009, 81, 9972–9978. [Google Scholar] [CrossRef] [PubMed]
- Shui, B.; Ozer, A.; Zipfel, W.; Sahu, N.; Singh, A.; Lis, J.T.; Shi, H.; Kotlikoff, M.I. RNA aptamers that functionally interact with green fluorescent protein and its derivatives. Nucleic Acids Res. 2012, 40, e39. [Google Scholar] [CrossRef] [PubMed]
- Steiner, J.M.; Sartor, M.; Sanchez, A.B.; Messmer, D.; Freed, A.; Esener, S.; Messmer, B.T. DeNAno: Selectable deoxyribonucleic acid nanoparticle libraries. J. Biotechnol. 2010, 145, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, E.; Kang, Y.Y.; Mok, H. Multivalent aptamer-RNA based fluorescent probes for carrier-free detection of cellular microRNA-34a in mucin1-expressing cancer cells. Chem. Commun. 2015, 51, 9038–9041. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Chang, H.T.; Tan, W. Cancer cell targeting using multiple aptamers conjugated on nanorods. Anal. Chem. 2008, 80, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Ocsoy, I.; Gulbakan, B.; Shukoor, M.I.; Xiong, X.; Chen, T.; Powell, D.H.; Tan, W. Aptamer-conjugated multifunctional nanoflowers as a platform for targeting, capture, and detection in laser desorption ionization mass spectrometry. ACS Nano 2013, 7, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.; Chen, T.; Tan, W.; Fan, Z.H. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano 2013, 7, 7067–7076. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sheng, W.; Fan, Z.H. An ensemble of aptamers and antibodies for multivalent capture of cancer cells. Chem. Commun. 2014, 50, 6722–6725. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Cui, C.H.; Bose, S.; Guo, D.; Shen, C.; Wong, W.P.; Halvorsen, K.; Farokhzad, O.C.; Teo, G.S.L.; Phillips, J.A.; et al. Bioinspired multivalent DNA network for capture and release of cells. Proc. Natl. Acad. Sci. USA 2012, 109, 19626–19631. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Jin, G.; Fang, X. Investigation of the interaction between a bivalent aptamer and thrombin by AFM. Langmuir 2012, 28, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Mallik, P.K.; Nishikawa, K.; Millis, A.J.T.; Shi, H. Commandeering a biological pathway using aptamer-derived molecular adaptors. Nucleic Acids Res. 2010, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Maier, K.E.; Jangra, R.K.; Shieh, K.R.; Cureton, D.K.; Xiao, H.; Snapp, E.L.; Whelan, S.P.; Chandran, K.; Levy, M. A new transferrin receptor aptamer inhibits new world hemorrhagic fever mammarenavirus entry. Mol. Ther. Nucleic Acids 2016, 5, e321. [Google Scholar] [CrossRef] [PubMed]
- Raffler, N.A.; Rivera-Nieves, J.; Ley, K. L-selectin in inflammation, infection and immunity. Drug Discov. Today Ther. Strateg. 2005, 2, 213–220. [Google Scholar] [CrossRef]
- Gilboa, E.; McNamara, J.; Pastor, F. Use of oligonucleotide aptamer ligands to modulate the function of immune receptors. Clin. Cancer Res. 2013, 19, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Rudnick, S.I.; Adams, G.P. Affinity and avidity in antibody-based tumor targeting. Cancer Biother. Radiopharm. 2009, 24, 155–161. [Google Scholar] [CrossRef] [PubMed]

| Target | Backbone | Aptamer Domains | Connection | Advantage of Multivalency | Reference |
|---|---|---|---|---|---|
| Biosensors | |||||
| ATP, theophylline, FMN | DNA, RNA | MG + ATP, MG+ theophylline, MG + FMN | Covalent; secondary structure, 2–4 bp stem | Fluorescent detection of the analyte | [41] |
| Thrombin, ATP | DNA | Thrombin+ dapoxyl, ATP + dapoxyl | Covalent; secondary structure, 1–4 bp stem | Fluorescent detection of the analyte | [42] |
| Thrombin | DNA | TBA15, streptavidin | Covalent; 4 nt pyrimidine sequence | Immobilization via streptavidin upon binding of thrombin | [43] |
| INF-γ | DNA | INF-γ, streptavidin | Covalent; no linker or (dT)5 or (dT)10 | Amplified SPR detection of INF-γ | [65] |
| Cocaine, AMP | DNA | Cocaine, AMP | Covalent; (dT)9 | Simultaneous detection of two analytes, functional assembly for logic gate “OR” operation | [24] |
| Thrombin, ATP | DNA | TBA15, ATP | Covalent; no linker | Label-free EIS detection of two analytes | [67] |
| Thrombin, adenosine | DNA | TBA15, adenosine | Covalent; no linker | Detection of two analytes | [68] |
| Lysozyme, adenosine | DNA | Lysozyme, adenosine | Non-covalent; assembled by hybridization of linker sequences | Detection of two analytes | [69] |
| Thrombin | DNA | TBA15, TBA29 | Covalent; no linker or (dT)5 or (dT)10 | Enhancement of the overall binding ability | [25] |
| Analytical Probes | |||||
| Streptavidin, MG, theophylline | RNA | Streptavidin + MG, Streptavidin + theophylline | Covalent, no linker | Streptavidin immobilization of aptamers | [21] |
| Thrombin | DNA | TBA15, TBA27 | Covalent; unspecified 8-unit spacer | AFM study of unbinding dynamics and dissociation energy landscape | [78] |
| Human angiopoetin-2 | DNA | Two aptamers to distinct epitopes | Covalent; (dT)25 | ~200-fold affinity enhancement | [27] |
| Imaging | |||||
| GFP | RNA | GFP | Covalent; 5S rRNA three-way junction | Enhanced binding and fluorescence modulation | [70] |
| Human dendritic cells | DNA | Library of multiplied random blocks | RCA | Selectable library of multivalent nanoparticles | [71] |
| Mucine-1 | DNA | MUC1 | Non-covalent; multiplication via hybridization with multimeric template | Efficient internalization | [72] |
| CCRF-CCM cells | DNA | Sgc8c | Covalent; SH-mediated nanorod surface multiplication | Co-stimulation of T-cells in vitro Tumor rejection in vivo | [73] |
| K-562 cells | DNA | KK1HO8 | Covalent; SH-mediated nanorod surface multiplication | Enhanced cell imaging and targeting | [73] |
| Affinity Cell Capture | |||||
| CD4, streptavidin | RNA | SA19, CD4 aptamer | Non-covalent; via CopA-CopT interactions | Affinity capture of CD4+ cells | [62] |
| CCRF-CCM cells, ATP | DNA | Sgc8c, ATP | Covalent; on Au@MgO nanoflowers | Intracellular capture of ATP for subsequent MALDI analysis | [74] |
| CCRF-CCM cells or Ramos cells | DNA | Sgc8c or TD05 | Covalent; via spherical AuNPs multiplied on microfluidic channel | High efficiency, throughput, and purity of cell capture from blood samples | [75] |
| CCRF-CCM cells | DNA | Sgc8c | Covalent; multiplication on microfluidic channel | Enhanced capture efficiency | [76] |
| CCRF-CCM cells | DNA | Sgc8c | Covalent; RCA multiplied aptamers immobilized on microfluidic channels | Highly efficient specific isolation of target cells from blood samples | [77] |
| Target | Backbone | Aptamer Domains | Connection | Advantage of Multivalency | Reference |
|---|---|---|---|---|---|
| Anticoagulants | |||||
| Thrombin | DNA | TBA15, TBA29 | Covalent; (dA)15 | ~2-fold KD decrease 1; prolonged clotting time | [29] |
| Thrombin | DNA | TBA15, TBA29 | Covalent; (dT)20 | 10-fold KD decrease 2; ~3-fold increase of clotting time | [19,20] |
| Thrombin | DNA | TBA15, TBA29 | Covalent; PEG, (Spacer 18)8 | ~62-fold KA increase 3; ~9-fold increase of clotting time 3 | [45] |
| Thrombin | DNA | TBA15, TBA29 | Covalent; PEG, (Spacer 18)10 | ~100-fold KD decrease 1, ~2.5-fold increase of clotting time 1 | [46] |
| Thrombin | DNA | TBA15, TBA29 | Covalent; in vitro selected 35 nt sequence | ~200-fold KD decrease 1; markedly improved inhibition of fibrinogen cleavage | [34] |
| Thrombin, hemin | DNA | TBA15, hemin deozyribozyme | Covalent; shared 6-nt sequence | 3-fold increase of clotting time, restored upon hemin addition | [44] |
| Prothrombin, factor IXa | 2′-F-RNA | R9D-14t, 11F7t | Covalent; (rA)3 | Clotting time nearly the same as for the mixture of aptamers. Bivalent molecule is preferable for drug development. Effect reversed by oligonucleotide antidote | [30] |
| Thrombin | DNA (circular form) | TBA15, TBA29 | Covalent; DNA hairpin | High serum and plasma stability; 2–3 fold increase of clotting time Effect reversed by oligonucleotide antidote | [39,40] |
| Thrombin | DNA | TBA15, TBA29 | Covalent; attachment to AuNP; 15 TBA15 and 15 TBA29 per NP | 100–10,000-fold KD decrease 3; 10-fold increase of clotting time. Superior to commercial anticoagulants. Effect reversed by oligonucleotide antidote | [52] |
| Thrombin | DNA | TBA15, TBA29 | Non-covalent; Attachment to AuNP by means of anchoring (dA)20 tail; 30 TBA15 and 30TBA29 per NP | 10–1000-fold KD decrease 3; Superior to commercial anticoagulants in clotting test. Superior to heparin in rat bleeding test. Effect reversed by green light irradiation | [53] |
| Anti-inflammatory | |||||
| l-Selectin | DNA | LD201* | Covalent; (dA)9; Trimer | 10-fold increase of IC50 for l-selectin-ligand interaction. Inhibition of target cells’ homing in vivo | [32] |
| l-Selectin | DNA | LD201 | Covalent; (dT)20; ~30 aptamer units per molecule | 103-fold higher affinity to l-selectin. More strong binding with l-selectin on cell surface. Inhibition of target cells’ homing in vivo | [26] |
| Anti-cancer | |||||
| CCRF-CEM cells (PTK7) | DNA | Sgc8c | Covalent; attached to MS2 capsid; up to 60 aptamer units | Target cell internalization. Addressed delivery of porphryin for photodynamic therapy | [55,56] |
| CCRF-CEM cells (PTK7) | DNA | Sgc8c | Covalent; PolyA linker with 3 GC repeats; 30–40 aptamer units. Loaded by doxorubicin | 40-fold KD improvement; More efficient cell internalization. Cytotoxicity against CCRF-CEM cells | [31] |
| CCRF-CEM cells (PTK7) K562 cells Ramos cells (IgM heavy mu chain) | DNA | Sgc8c or T2-KK1B10, or TD05 | Covalent; polyacrylamide backbone; ~90 aptamer units | Improved binding affinity towards target cells; simultaneous cell imaging and cell killing | [49] |
| CCRF-CEM cells (PTK7) K562 cells | DNA | Sgc8c or T2-KK1B10 | Covalent (polyacrilamide backbone) + non covalent (oligonucleotide connectors; Loaded by doxorubicin and anti-MDR1 oligonucleotide | Selective cytotoxicity, including drug-resistant cell line | [50,51] |
| CCRF-CEM cells (PTK7) Ramos cells (IgM heavy mu chain) | DNA | Sgc8c or TD05 | Covalent; Conjugated with AgNP. Loaded with fluorescent dye | Cytotoxicity, cell imaging | [54] |
| MCF-7 cells (MUC1) | DNA | MUC1 aptamer | Non-covalent; comb-like construct. Conjugates of aptamer and sense siRNA strand hybridized to multimerized antisense strand | Specific cell binding and internalization. Inhibition of target gene expression | [57] |
| CTLA-4 T-cell receptor | 2′-F-RNA | Del60 | Non-covalent; tetramer assembled on dsDNA linker | Enhanced bioactivity. Inhibition of tumor growth in vivo | [58] |
| 4-1BB T-cell receptor | 2′-F-RNA | 12–23 | Non-covalent; dimer assembled by hybridization of linker sequences | Co-stimulation of T-cells in vitro. Tumor rejection in vivo | [59] |
| OX40 T-cell receptor | 2′-F-RNA | 9.8 | Non-covalent; dimer assembled on DNA scaffold | Co-stimulation of T-cells in vitro. Tumor rejection in vivo | [60] |
| 4-1BB T-cell receptor PSMA | 2′-F-RNA | 12–23 xPSM-A10 | Covalently linked 12–23 dimer; hybridized with xPSM-A10 through linker sequences | Inhibition of PSMA-positive tumor growth in vivo upon systemic delivery | [61] |
| CD30 T-cell receptor | DNA | C2 | Non-covalent; biotin-streptavidin interactions; tetramer | Induction of receptor oligomerization and apoptosis of target cells | [63] |
| CD16α receptor of NK cells c-Met receptor of PBMC cells | DNA | CD16- α aptamer, C-met aptamer (different combinations) | Covalent; (dA)15, PEG or ”original” oligonucleotide linkers | Simultaneous binding of both target proteins. Target cell lysis | [28] |
| Urokinase-type plasminogen activator | 2′-F-RNA/peptide | upanap-12, upanap126, upain-1 | Covalent; zero linker between nucleic acid aptamers; 3′-conjugate with peptide aptamer | Complete inhibition of UPa processing and catalytic activities | [22] |
| Heat shock protein HSF1 | RNA | AptHSF-RA1 | Covalent; oligonucleotide linker | 100-fold improvement of binding affinity | [38] |
| Opsonin C3b/iC3b, GFP | RNA | AptC3-1, AptGFP-AP3 | Covalent; double-strand oligonucleotide linker | Specific opsonization of GFP (model protein) into phagocytic cells | [79] |
| Antiviral | |||||
| NS3 protein of hepatitis C virus | RNA | NEO-III-14U or G9-II-20U, #5 | Covalent; U40–U50 linkers | Inhibition of both helicase and protease activities of NS3 protein | [23] |
| Human transferrin receptor | 2′-F-RNA | Waz | Non-covalent; biotin-streptavidin interactions | ~10-fold increase of EC50 for inhibition of NWM infection in human cells | [80] |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vorobyeva, M.; Vorobjev, P.; Venyaminova, A. Multivalent Aptamers: Versatile Tools for Diagnostic and Therapeutic Applications. Molecules 2016, 21, 1613. https://doi.org/10.3390/molecules21121613
Vorobyeva M, Vorobjev P, Venyaminova A. Multivalent Aptamers: Versatile Tools for Diagnostic and Therapeutic Applications. Molecules. 2016; 21(12):1613. https://doi.org/10.3390/molecules21121613
Chicago/Turabian StyleVorobyeva, Mariya, Pavel Vorobjev, and Alya Venyaminova. 2016. "Multivalent Aptamers: Versatile Tools for Diagnostic and Therapeutic Applications" Molecules 21, no. 12: 1613. https://doi.org/10.3390/molecules21121613
APA StyleVorobyeva, M., Vorobjev, P., & Venyaminova, A. (2016). Multivalent Aptamers: Versatile Tools for Diagnostic and Therapeutic Applications. Molecules, 21(12), 1613. https://doi.org/10.3390/molecules21121613





