Discrimination and Nitric Oxide Inhibitory Activity Correlation of Ajwa Dates from Different Grades and Origin
Abstract
:1. Introduction
2. Results and Discussion
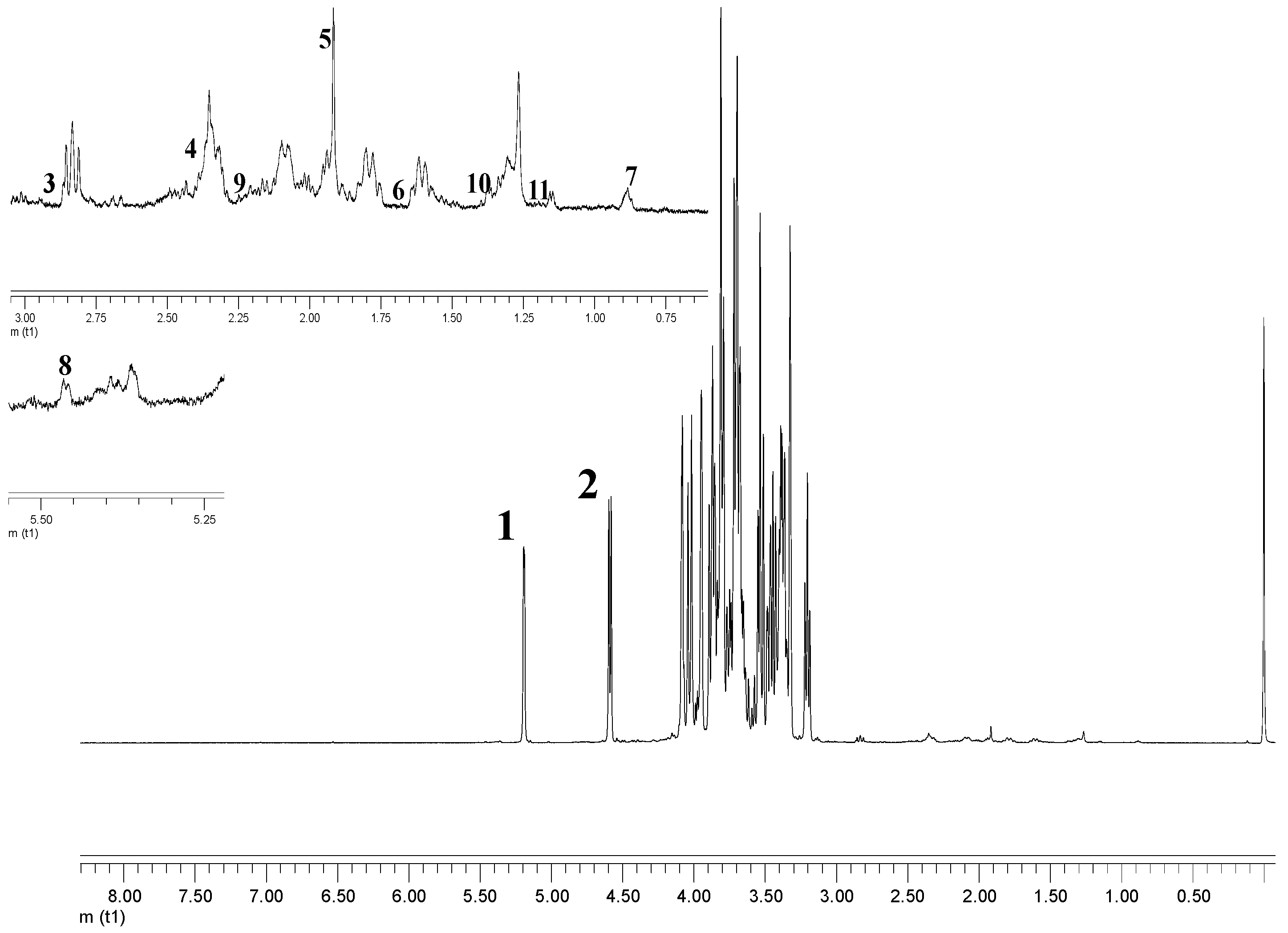
2.1. Spectral Features and Assignment of Metabolites
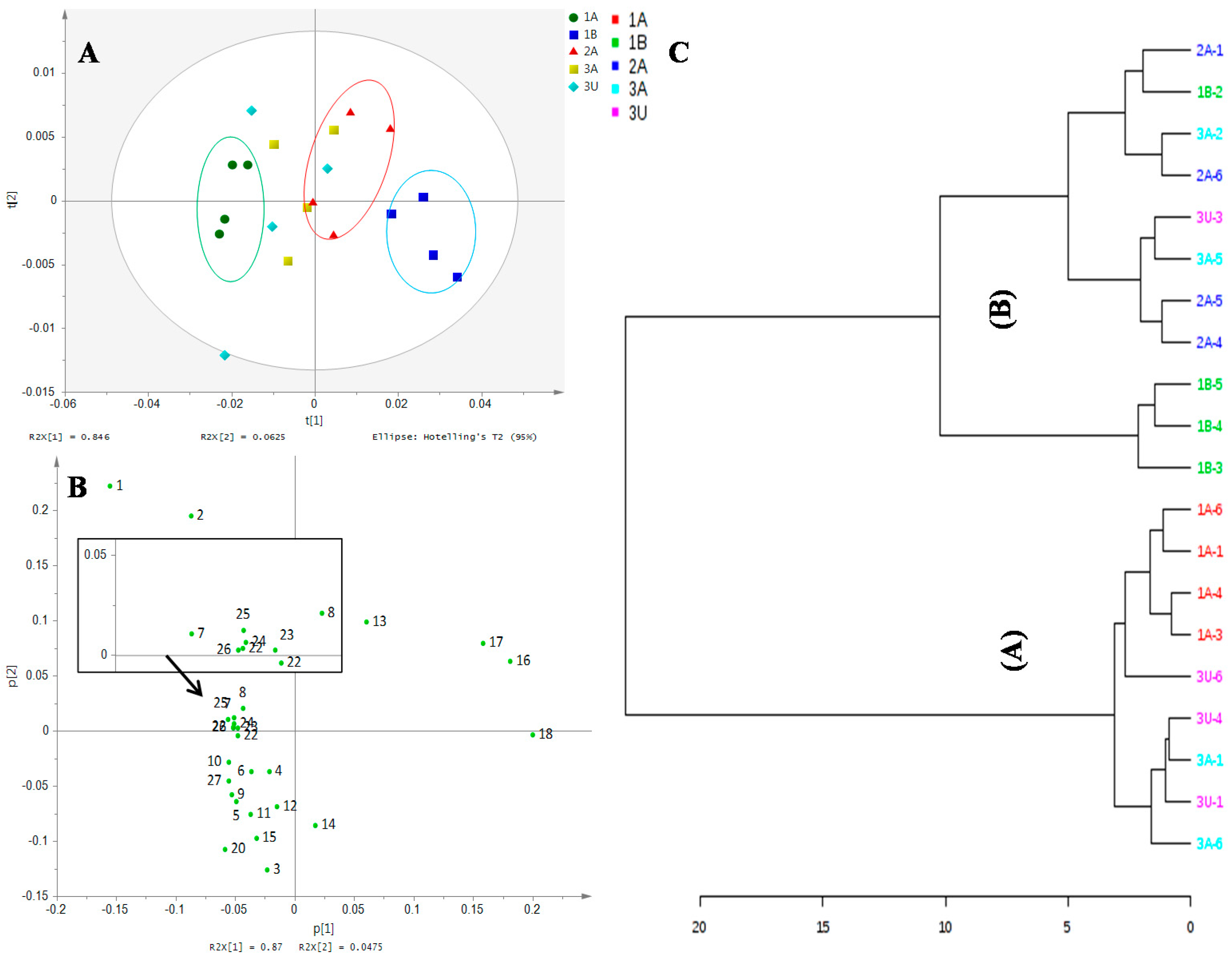
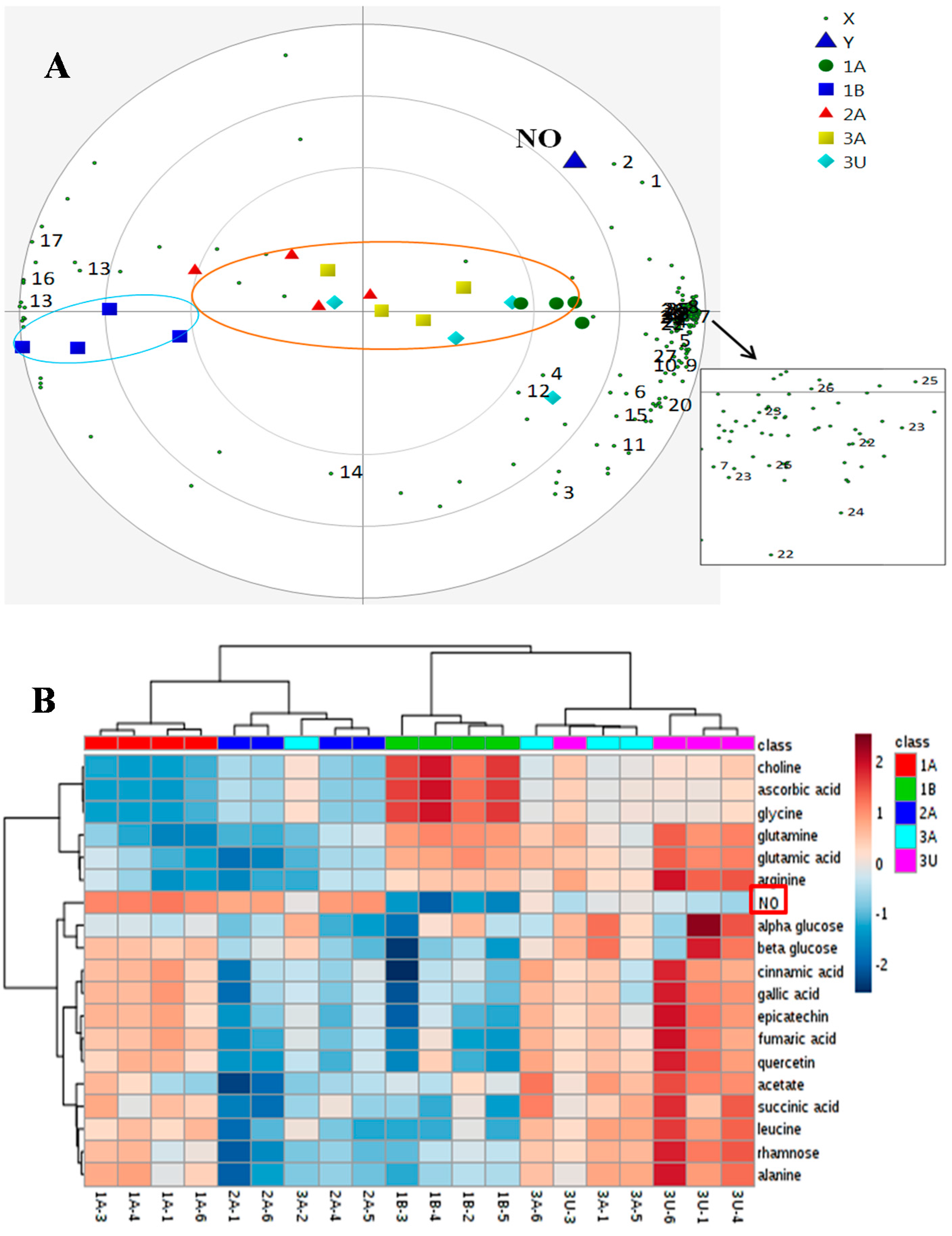
2.2. Discrimination of Ajwa Samples on Unsupervised PCA of the NMR Finger Print
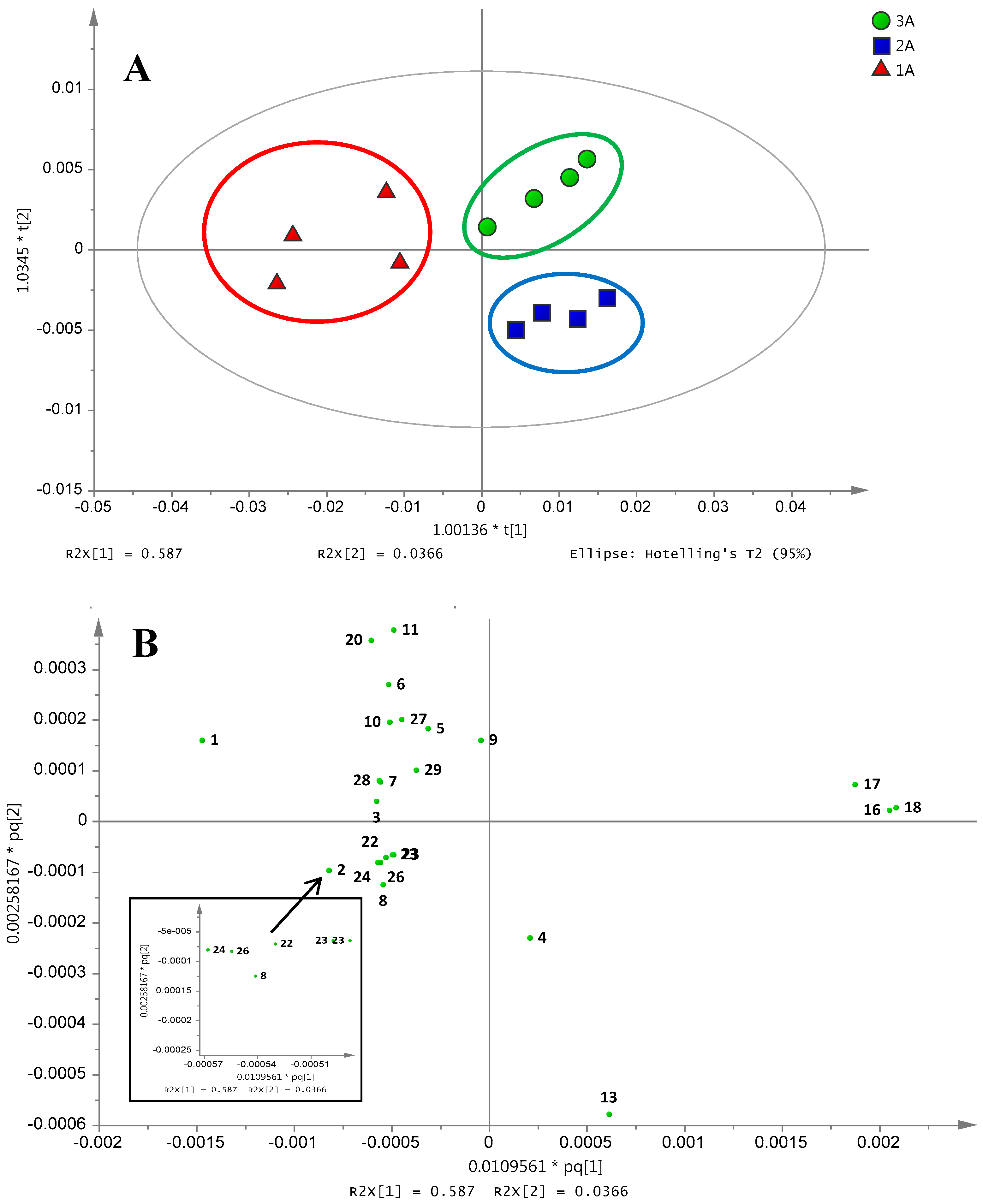
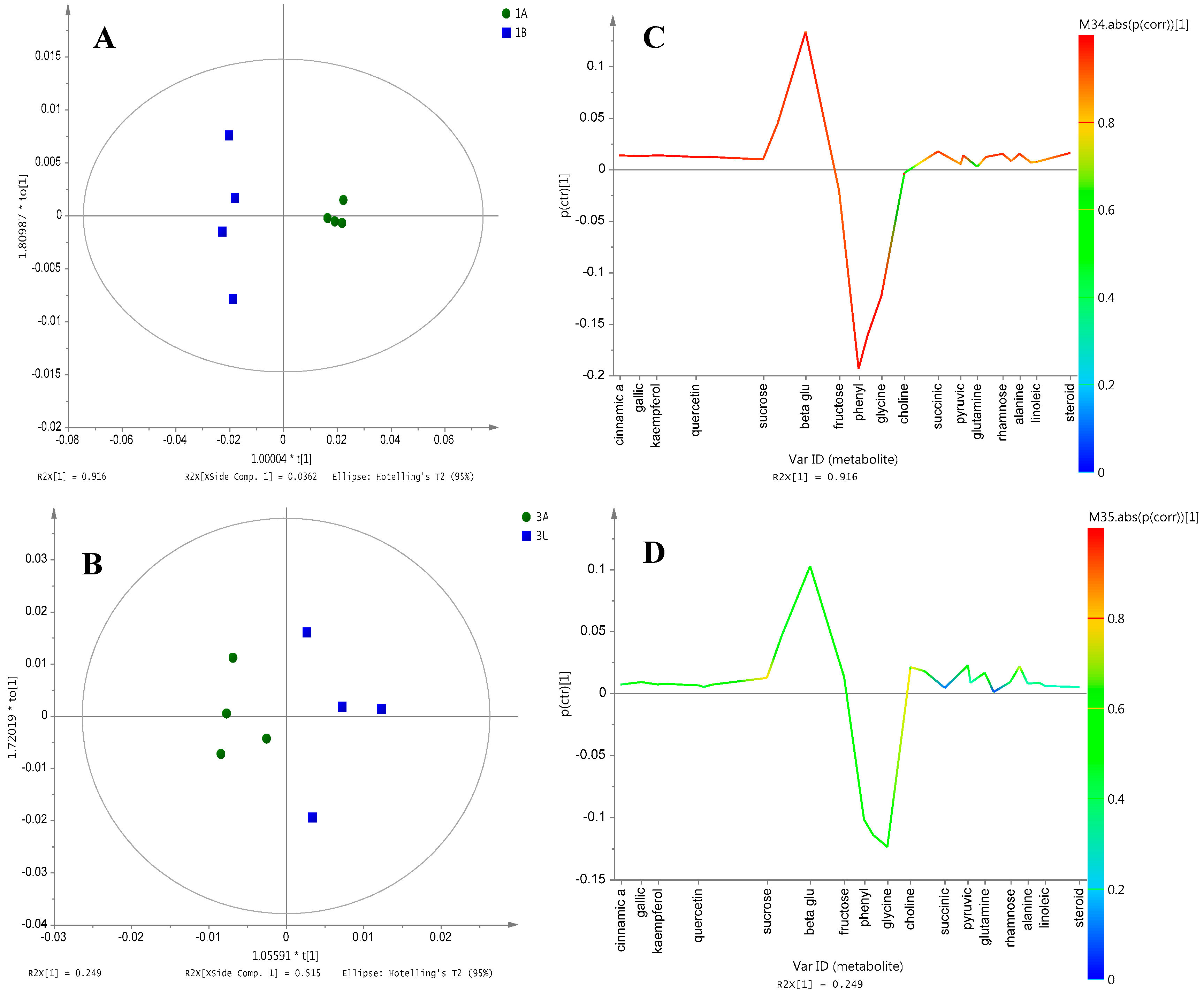
2.3. Differentiation of Ajwa Grades from al-Aliah Farm
2.4. Differentiation of Ajwa Based on Farm Origins
2.5. NO Inhibitory Activity of the Different Grades of Ajwa Dates
2.6. Correlation between Metabolite Profiles and NO Inhibitory Activity
3. Materials and Methods
3.1. Materials
3.2. Fruit Materials
3.3. Sample Preparation
3.4. NMR Measurement
3.5. Nitric Oxide Inhibitory Activity
3.5.1. Measurement of Nitrite
3.5.2. Cell Viability
3.5.3. Data Analysis
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Amira, E.L.; Arem, G.; Flamini, B.; Saafi Emna, M.; Issaoui, N.; Zayene, A.; Mohamed, H.; Noureddine, H.A.; Lotfi, A. Chemical and aroma volatile compositions of date palm (Phoenix dactylifera L.) fruits at three maturation stages. Food Chem. 2011, 127, 1744–1754. [Google Scholar] [CrossRef]
- Puri, A.; Sahai, R.; Singh, K.L.; Saxena, R.P.; Tandon, J.S.; Saxena, K.C. Immunostimulant activity of dry fruits and plant materials used in Indian traditional medical system for mothers after child birth and invalids. J. Ethnopharmacol. 2000, 71, 89–92. [Google Scholar] [CrossRef]
- Vayalil, P.K. Date fruits (Phenix dactylifera Linn): An emerging medicinal food. Food Sci. Nutr. 2008, 52, 249–271. [Google Scholar]
- Ku, K.M.; Choi, J.N.; Kim, J.; Kim, J.K.; Yoo, L.G.; Lee, S.J.; Hong, Y.S.; Lee, C.H. Metabolomics analysis reveals the compositional differences of shade grown tea (Camellia sinensis L.). J. Agric. Food Chem. 2010, 58, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, A.; Nyberg, N.T.; Molgaard, P.; Asii, J.; Jaroszewski, J.W. 1H-NMR metabolic fingerprinting of saffron extracts. Metabolomics 2010, 6, 511–517. [Google Scholar] [CrossRef]
- Zhang, C.H.; Aldosari, S.A.; Vidyasagar, P.S.P.V.; Nair, K.M.; Nair, M.G. Antioxidant and anti-inflammatory assays confirm bioactive compounds in Ajwa date fruit. J. Agric. Food. Chem. 2013, 61, 5834–5840. [Google Scholar] [CrossRef] [PubMed]
- Hseu, Y.C.; Wu, F.Y.; Wu, J.J.; Chen, J.Y.; Chang, W.H.; Lu, F.J.; Lai, F.C.; Yang, H.L. Anti-inflammatory potential of Antrodia Camphorata through inhibition of iNOS, COX-2 and cytokines via the NF-kappaB pathway. Int. Immunopharmacol. 2005, 5, 1914–1925. [Google Scholar] [CrossRef] [PubMed]
- Terra, X.; Vall, J.; Vitrac, X.; Merrillon, J.M.; Arola, L.; Ardevol, A.; Blade, C.; Fernandez-Larrea, J.; Pujadas, G.; Salvado, J.; et al. Grape seed procyanidins acts as anti-inflammatory agents in endoxin-stimulated RAW 264.7 macrophages by inhibiting NF-κB signalling pathway. J. Agric. Food Chem. 2007, 55, 4357–4365. [Google Scholar] [CrossRef] [PubMed]
- Abdu, A.B. The protective role of Ajwa date against the hepatotoxicity induced by ochratoxin A. Egypt. J. Nat. Toxins 2011, 8, 1–15. [Google Scholar]
- Ragab, A.R.; Elkablawy, M.A.; Sheik, B.Y.; Baraka, H.N. Antioxidant and tissue protective studies on Ajwa extract: Dates from Al Madinah Al-Munawarah, Saudi Arabia. J. Environ. Anal. Toxicol. 2013, 3, 1–8. [Google Scholar] [CrossRef]
- Bassem, Y.S.; Elsaed, W.M.; Samman, A.H. Ajwa dates as a protective agent against liver toxicity in rat. Eur. Sci. J. 2014, 3, 358–368. [Google Scholar]
- Krasensky, J.; Jonak, C. Drought, salt and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jung, Y.; Song, B.; Bong, Y.S.; Ryu, D.H.; Lee, K.S.; Hwang, G.S. Discrimination of cabbage (Brassica rapa ssp. pekinensis) cultivars grown in different geographical areas using 1H-NMR-based metabolomics. Food Chem. 2013, 137, 68–75. [Google Scholar]
- Abdrabo, S.S.; Grindlay, G.; Gras, L.; Mora, J. Multi-element analysis of Spanish date palm (Phoenix dactylifera L.) by inductively coupled plasma-based techniques. Discrimination using multivariate statistical analysis. Food Anal. Methods 2015, 8, 1268–1278. [Google Scholar] [CrossRef]
- Maulidiani, M.; Bassem, Y.S.; Mediani, A.; Leong, S.W.; Abas, F.; Lajis, N.H. Differentitation of Nigella sativa seeds from four different origins and their bioactivity correlations based on NMR-metabolomics approach. Phytochem. Lett. 2015, 13, 308–318. [Google Scholar] [CrossRef]
- Kader, A.A.; Hussein, A. Harvesting and Postharvest Handling of Dates; ICARDA: Aleppo, Syria, 2009; pp. 1–18. [Google Scholar]
- Ashraf, Z.; Hamidi-Eshafani, Z. Date and date processing: A review. Food Rev. Int. 2011, 27, 101–131. [Google Scholar] [CrossRef]
- Caboni, P.; Liori, B.; Kumar, A.; Santoru, M.L.; Asthana, S.; Pieroni, E.; Faiz, A.; Era, B.; Cacace, E.; Ruggiero, V.; et al. Metabolomics analysis and modeling suggest a lysophosphocholines-PAF receptor interactions in fibromyalgia. PLoS ONE 2014, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Westerhuis, J.A.; Van Velzen, E.J.J.; Hoefsloot, H.C.J.; Smilde, A.K. Multivariate Paired Data Analysis: Multilevel PLS-DA Versus OPLSDA. Metabolomics 2010, 6, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Mediani, A.; Abas, F.; Khatib, A.; Tan, C.P.; Ismail, I.S.; Shaari, K.; Choi, Y.H.; Lajis, N.H. Phytochemical and biological fatures of Phyllanthus niruri and Phyllanthus urinaria harvested at different growth stages revealed by 1H-NMR based metabolomics. Ind. Crops Prod. 2015, 77, 602–613. [Google Scholar] [CrossRef]
- Charlop-Powers, Z.; Owen, J.G.; Reddy, B.V.B.; Ternei, M.A.; Brady, S.F. Chemical-biogeographic survey of secondary metabolism in soil. Proc. Natl. Acad. Sci. 2013, 111, 3757–3762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Niu, X.; Shi, M.; Pei, G.; Zhang, X.; Chen, L.; Zhang, W. Identification of a transporter Slr0982 involved in ethanol tolerance in cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bryk, J.; Ochoa, J.B.; Correcia, I.T.D.; Munera Seeley, V.; Ppovic, P.J. Effect of citrulline and glutamine on nitric oxide production in RAW 264.7 cells in an arginine-depleted environment. J. Parenter. Enteral. Nutr. 2008, 32, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Yang, S.O.; Kim, S.H.; Kim, H.; Ko, J.S.; Riu, K.Z.; Lee, H.Y.; Choi, H. Classification and prediction of free-radical scavenging activities of dangyuja (Citrus grandis Osbeck) fruit extracts using 1H-NMR spectroscopy and multivariate statistical analysis. J. Pharm. Biomed. Anal. 2009, 49, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Shin, M.R.; Kim, Y.S.; Bae, W.J.; Roh, D.H.; Hwang, Y.S.; Kim, E.C. Anti-inflammatory effects of glutamine on LPS-stimulated human dental pulp cells correlate with activation of MKP-1 and attenuation of the MAPK and NF-κB pathways. Int. Endod. J. 2014, 48, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lee, H.S.; Kim, S.H.; Moon, B.; Lee, C. Antioxidant and anti-inflammatory activities of methanol extracts of Tremella fuciformis and its major phenolic acids. J. Food Sci. 2014, 9, C460–C468. [Google Scholar] [CrossRef] [PubMed]
- Joo, T.; Sowndhararajan, K.; Hong, S.; Lee, J.; Park, S.Y.; Kim, S.; Jhoo, J.W. Inhibition of nitric oxide production in LPS-stimulated RAW 264.7 cells by stem bark of Ulmus pumila L. Saudi J. Biol. Sci. 2014, 21, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.K.; Kim, Y.S.; Natarajan, S.K.; Kim, W.S.; Hwang, J.W.; Jeon, N.J.; Jeong, J.H.; Moon, S.H.; Jeon, B.T.; Park, P.J. Antioxidant and anti-inflammatory effects of Chaenomeles sinensis leaf extracts on LPS-stimulated RAW 264.7 cells. Molecules 2016, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Q.; Ye, X.Q.; Fang, Z.X.; Chen, J.C.; Xu, G.H.; Liu, D.H. Phenolic compounds and antioxidant activity of extracts from ultrasonic treatment of satsuma mandarin (Citrus unshiu Marc.) peels. J. Agric. Food Chem. 2008, 56, 5682–5690. [Google Scholar] [CrossRef] [PubMed]
- Kriaa, W.; Fetoui, H.; Makni, M.; Zeghal, N.; Drira, N.E. Phenolic contents and antioxidant activities of date palm (Phoenix dactylifera L.) leaves. Intl. J. Food. Prop. 2012, 15, 1220–1232. [Google Scholar] [CrossRef]
- Mediani, A.; Abas, F.; Khatib, A.; Maulidiani, H.; Shaari, K.; Choi, Y.H.; Lajis, N.H. 1H-NMR based metabolomics approach to understanding the drying effects on the phytochemicals in Cosmos caudatus. Food Res. Int. 2012, 49, 763–770. [Google Scholar] [CrossRef]
- Abas, F.; Lajis, N.H.; Israf, D.A.; Khozirah, S.; Umi Kalsom, Y. Antioxidant and nitric oxide inhibition activities of selected Malay traditional vegetables. Food Chem. 2006, 95, 566–573. [Google Scholar] [CrossRef]
- Choi, Y.H.; Kim, H.K.; Linthorst, H.J.M.; Hollander, J.G.; Lefeber, A.W.M.; Erkelens, C.; Verpoorte, R. NMR metabolomics to revisit the tobacco mosaic virus infection in Nicoatiana tabacum leaves. J. Nat. Prod. 2006, 69, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Ismail, B.; Haffar, I.; Baalbaki, R.; Mechref, Y.; Henry, J. Physico-chemical characteristics and total quality of five date varieties grown in the United Arab Emirates. Int. J. Food Sci. Technol. 2006, 41, 919–926. [Google Scholar] [CrossRef]
- Georgiev, M.I.; Ali, K.; Alipieva, K.; Verpoorte, R.; Choi, Y.G. Metabolic differentiation and classification of Verbascum species by NMR-based metabolomics. Phytochemistry 2011, 72, 2045–2051. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Park, S.J.; Hyun, S.H.; Yang, S.O.; Lee, J.; Auh, J.H.; Kim, J.H.; Cho, S.M.; Choi, H.K. Biochemical monitoring of black raspberry (Rubus coreanus Miquel) fruits according to maturation stage by 1H-NMR using multiple solvent systems. Food Res. Int. 2011, 44, 1977–1987. [Google Scholar] [CrossRef]
- Maulidiani; Abas, F.; Khatib, A.; Shitan, M.; Shaari, K.; Lajis, N.H. Comparison of partial least squares and artificial neural network for the prediction of antioxidant activity in extract of Pegaga (Centella) varieties from 1H Nuclear Magnetic Resonance spectroscopy. Food Res. Int. 2013, 54, 852–860. [Google Scholar]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. Metaboanalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the extracts are available from the authors.

| Origin | Predicted Y Value for Dummy Variables | ||
|---|---|---|---|
| Aliah 1 | Aliah 2 | Aliah 3 | |
| Aliah 1 | 1.08 | −0.17 | 0.09 |
| Aliah 1 | 0.74 | −0.25 | 0.52 |
| Aliah 1 | 1.12 | 0.12 | −0.23 |
| Aliah 1 | 0.65 | 0.24 | 0.11 |
| Aliah 2 | −0.19 | 0.93 | 0.26 |
| Aliah 2 | 0.15 | 0.94 | −0.09 |
| Aliah 2 | −0.08 | 1.00 | 0.08 |
| Aliah 2 | 0.06 | 0.89 | 0.05 |
| Aliah 3 | 0.32 | 0.19 | 0.48 |
| Aliah 3 | 0.15 | 0.11 | 0.74 |
| Aliah 3 | 0.03 | 0.04 | 0.93 |
| Aliah 3 | −0.03 | −0.04 | 1.07 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdul-Hamid, N.A.; Mediani, A.; Maulidiani, M.; Abas, F.; Ismail, I.S.; Shaari, K.; Lajis, N.H. Discrimination and Nitric Oxide Inhibitory Activity Correlation of Ajwa Dates from Different Grades and Origin. Molecules 2016, 21, 1423. https://doi.org/10.3390/molecules21111423
Abdul-Hamid NA, Mediani A, Maulidiani M, Abas F, Ismail IS, Shaari K, Lajis NH. Discrimination and Nitric Oxide Inhibitory Activity Correlation of Ajwa Dates from Different Grades and Origin. Molecules. 2016; 21(11):1423. https://doi.org/10.3390/molecules21111423
Chicago/Turabian StyleAbdul-Hamid, Nur Ashikin, Ahmed Mediani, M. Maulidiani, Faridah Abas, Intan Safinar Ismail, Khozirah Shaari, and Nordin H. Lajis. 2016. "Discrimination and Nitric Oxide Inhibitory Activity Correlation of Ajwa Dates from Different Grades and Origin" Molecules 21, no. 11: 1423. https://doi.org/10.3390/molecules21111423
APA StyleAbdul-Hamid, N. A., Mediani, A., Maulidiani, M., Abas, F., Ismail, I. S., Shaari, K., & Lajis, N. H. (2016). Discrimination and Nitric Oxide Inhibitory Activity Correlation of Ajwa Dates from Different Grades and Origin. Molecules, 21(11), 1423. https://doi.org/10.3390/molecules21111423






