Quantitative Analysis of Differential Proteome Expression in Epithelial-to-Mesenchymal Transition of Bladder Epithelial Cells Using SILAC Method
Abstract
1. Introduction
2. Results and Discussions
2.1. SILAC Cell Model for Quantification of Proteome in TGF-β-Induced EMT of Bladder Cells
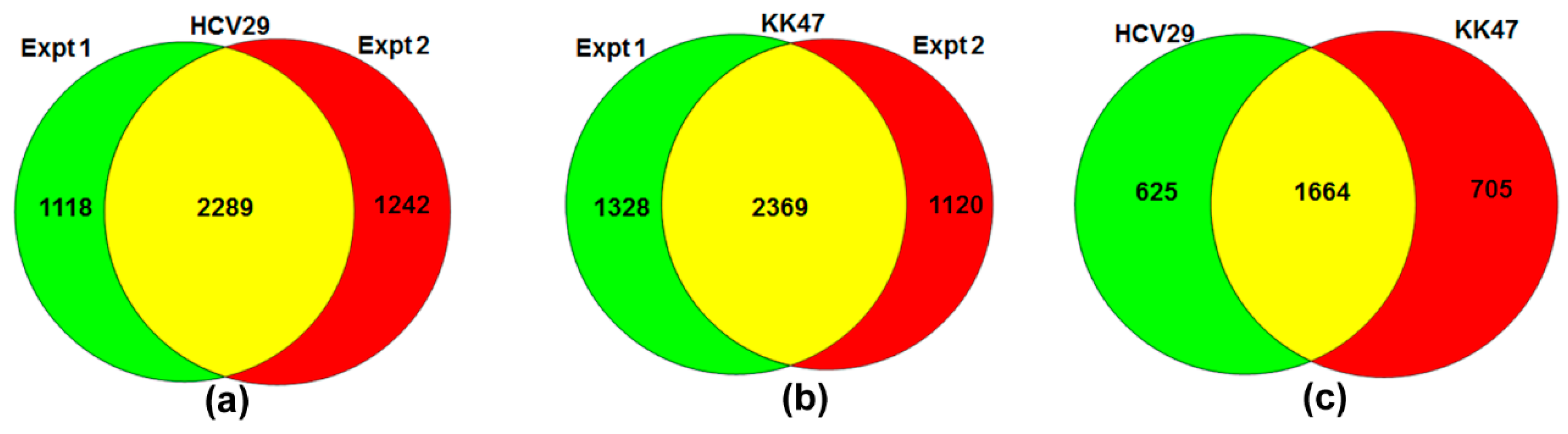
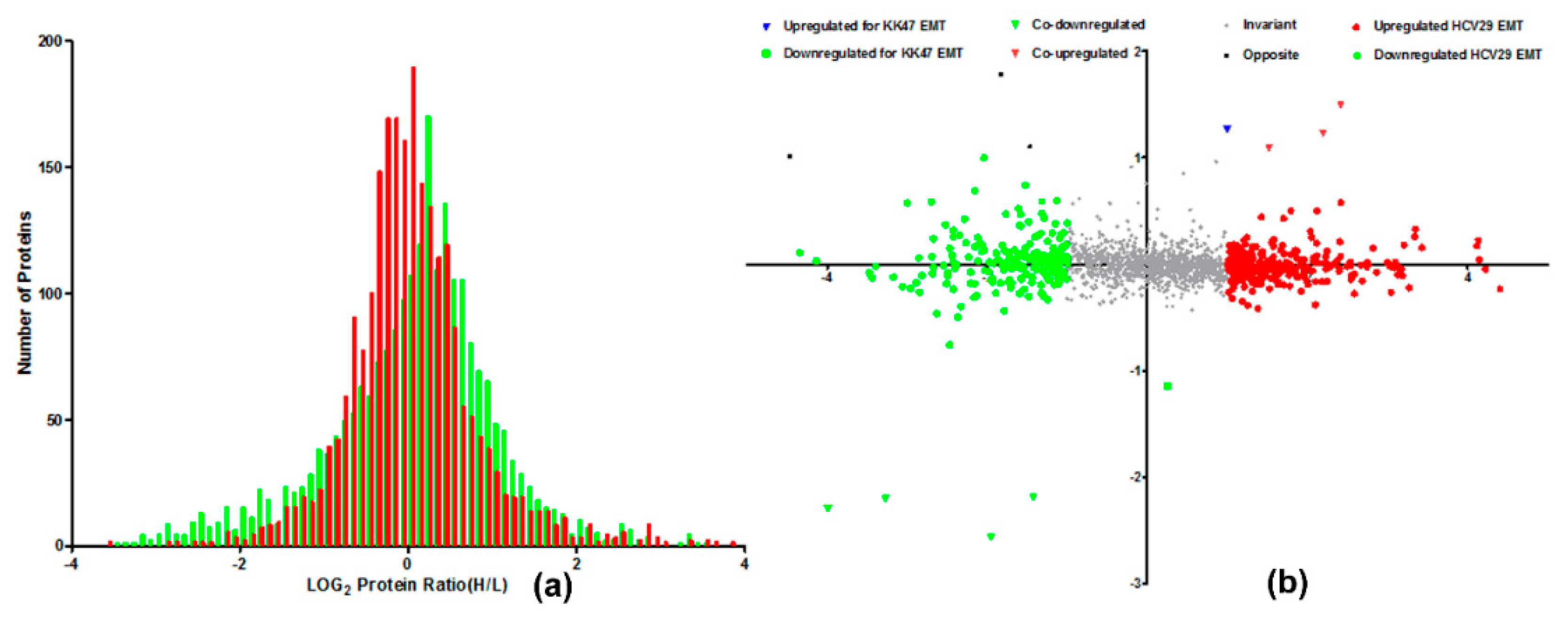
| Cell Line | log2 Mean | log2 SD | Z-Scores a | ||
|---|---|---|---|---|---|
| ±1.960σ | ±2.576σ | ±3.291σ | |||
| HCV29 | −0.146 | 1.298 | 48, 106 | 16, 40 | 5, 5 |
| KK47 | −0.005 | 0.209 | 56, 24 | 32, 12 | 18, 9 |
| Both cell lines | 3, 4 | 3, 4 | 3, 4 | ||
2.2. Functional Classification and Pathway Analysis of Identified Proteins
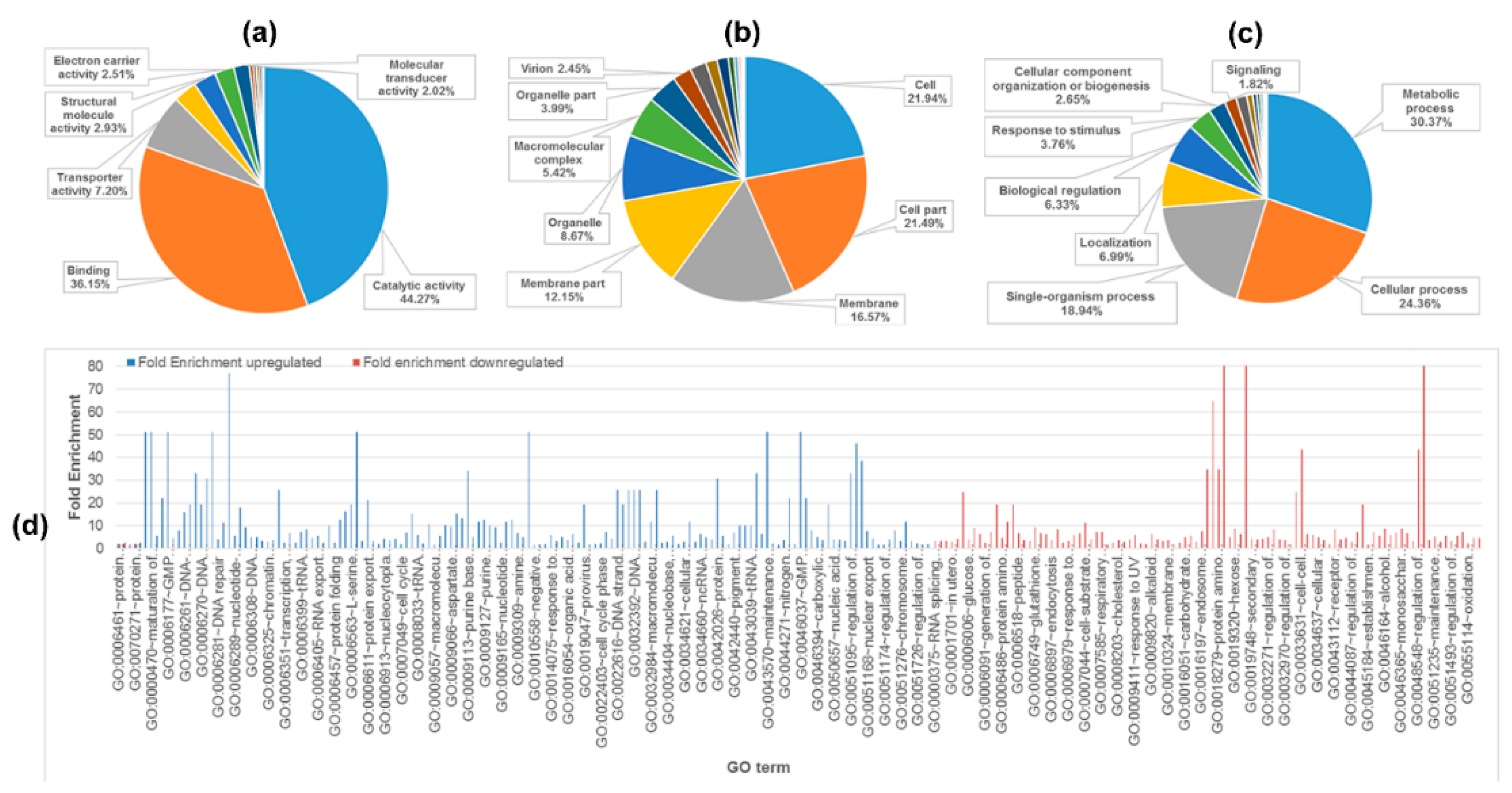
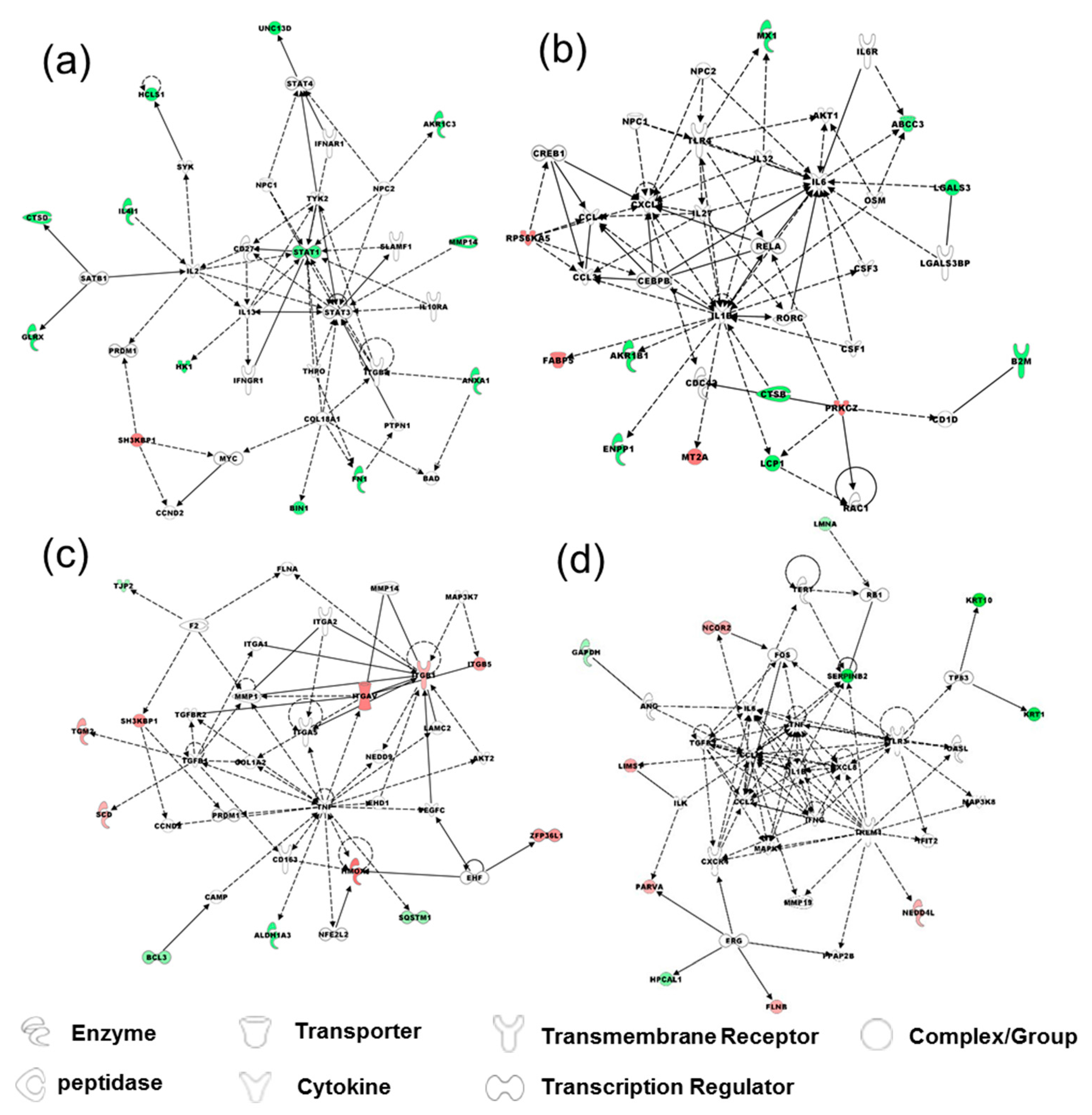
2.3. Comparison of Proteomes of TGF-β-Induced EMT in Bladder Cells vs. Bladder Cancer Cells
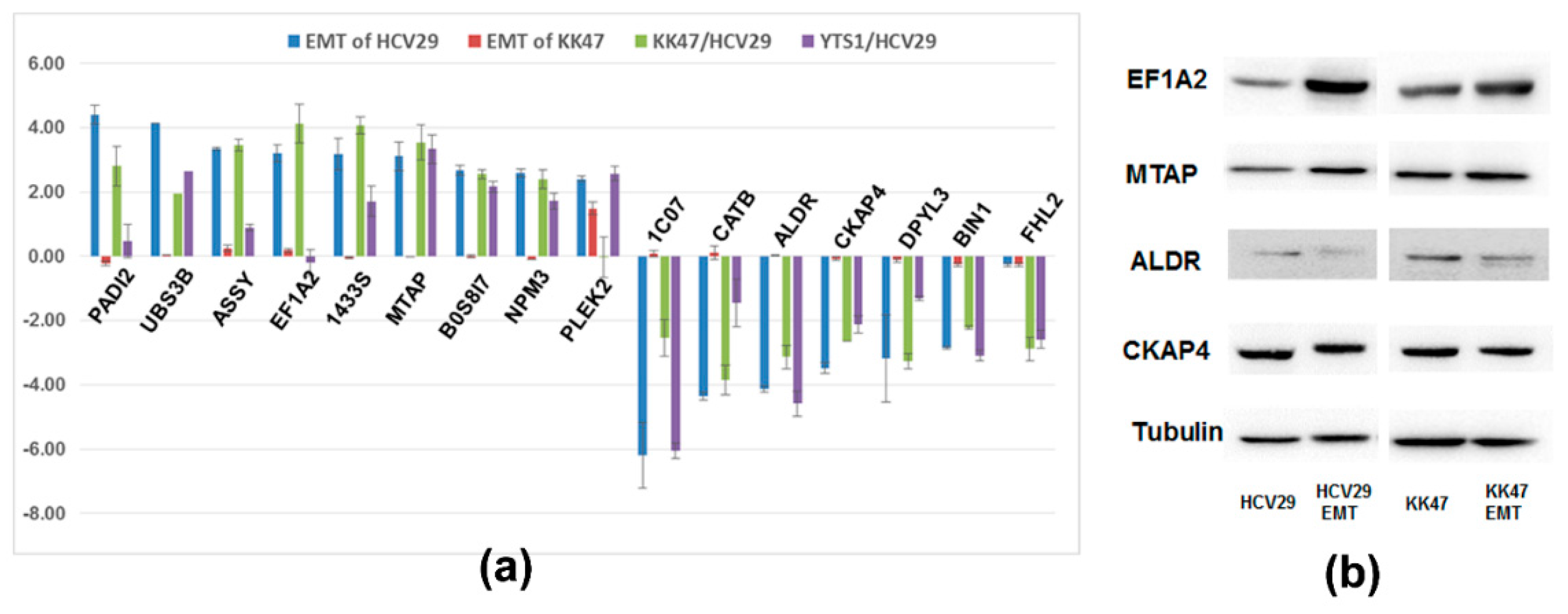
3. Experimental Section
3.1. Cell Culture
3.2. Cell Lysis and Protein Extraction
3.3. In-Solution Digestion
3.4. LC-MS/MS Analysis
3.5. Data Analysis
3.6. Functional Annotation and Ingenuity Pathways Analysis
3.7. Differential Analysis of Proteomes of TGF-β-Induced EMT in Bladder Cells vs. Bladder Cancer Cells and Validation by Western Blot
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Jing, R.; Hu, H.; Sun, C.; Huang, T.; Deng, W.; Li, J.; Yu, J.; Liu, Y.; Zhang, C. Serum glycoprotein profiling by lectin affinity microarray to distinguish the various stages of primary liver carcinogenesis. Zhonghua Gan Zang Bing Za Zhi 2014, 22, 358–363. [Google Scholar] [PubMed]
- Kang, X.; Wang, N.; Pei, C.; Sun, L.; Sun, R.; Chen, J.; Liu, Y. Glycan-related gene expression signatures in human metastatic hepatocellular carcinoma cells. Exp. Ther. Med. 2012, 3, 415–422. [Google Scholar] [PubMed]
- McConkey, D.J.; Choi, W.; Marquis, L.; Martin, F.; Williams, M.B.; Shah, J.; Svatek, R.; Das, A.; Adam, L.; Kamat, A.; et al. Role of epithelial-to-mesenchymal transition (EMT) in drug sensitivity and metastasis in bladder cancer. Cancer Metastasis Rev. 2009, 28, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cai, Z.; Yang, F.; Luo, J.; Satoh, M.; Arai, Y.; Li, D. Enhanced antitumor efficacy of integrin-targeted oncolytic adenovirus AxdAdB3-F/RGD on bladder cancer. Urology 2014, 83, 508.e13–508.e19. [Google Scholar] [CrossRef] [PubMed]
- Goodison, S.; Rosser, C.J.; Urquidi, A.V. Urinary proteomic profiling for diagnostic bladder cancer biomarkers. Expert Rev. Proteom. 2009, 6, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Rosanas-Urgell, A.; Martin-Jaular, L.; Ricarte-Filho, J.; Ferrer, M.; Kalko, S.; Kimura, E.; del Portillo, H.A. Expression of non-TLR pattern recognition receptors in the spleen of BALB/c mice infected with Plasmodium yoelii and Plasmodium chabaudi chabaudi AS. Mem. Inst. Oswaldo Cruz 2012, 107, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Bridle, A.R.; Koop, B.F.; Nowak, B.F. Identification of surrogates of protection against yersiniosis in immersion vaccinated atlantic salmon. PLoS ONE 2012, 7, e40841. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Li, B.; Zeng, X.; Hu, H.; Hong, Y.; Ouyang, H.; Zhang, X.; Wang, Z.; Zhu, H.; Lei, P.; et al. Mutation of TGF-β receptor ii facilitates human bladder cancer progression through altered TGF-β1 signaling pathway. Int. J. Oncol. 2013, 43, 1549–1559. [Google Scholar] [PubMed]
- Yamasaki, S.; Matsumoto, M.; Takeuchi, O.; Matsuzawa, T.; Ishikawa, E.; Sakuma, M.; Tateno, H.; Uno, J.; Hirabayashi, J.; Mikami, Y.; et al. C-type lectin mincle is an activating receptor for pathogenic fungus, malassezia. Proc. Natl. Acad. Sci. USA 2009, 106, 1897–1902. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.; Jin, D.; Hou, C.; Hu, J.; Wu, W.; Ni, X.; Wang, D.; Lou, W. Proteomics analysis of serum protein profiling in pancreatic cancer patients by DIGE: Up-regulation of mannose-binding lectin 2 and myosin light chain kinase 2. BMC Gastroenterol. 2010, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Chen, K.T.; Fan, C.W.; Han, C.L.; Chen, Y.J.; Yu, J.S.; Chang, Y.S.; Chien, C.W.; Wu, C.P.; Hung, R.P.; et al. Comparison of membrane fraction proteomic profiles of normal and cancerous human colorectal tissues with gel-assisted digestion and itraq labeling mass spectrometry. FEBS J. 2010, 277, 3028–3038. [Google Scholar] [CrossRef] [PubMed]
- Caceres, N.E.; Aerts, M.; Marquez, B.; Mingeot-Leclercq, M.P.; Tulkens, P.M.; Devreese, B.; van Bambeke, F. Analysis of the membrane proteome of ciprofloxacin-resistant macrophages by stable isotope labeling with amino acids in cell culture (SILAC). PLoS ONE 2013, 8, e58285. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteom. 2002, 1, 376–386. [Google Scholar] [CrossRef]
- Matic, I.; Jaffray, E.G.; Oxenham, S.K.; Groves, M.J.; Barratt, C.L.; Tauro, S.; Stanley-Wall, N.R.; Hay, R.T. Absolute SILAC-compatible expression strain allows sumo-2 copy number determination in clinical samples. J. Proteome Res. 2011, 10, 4869–4875. [Google Scholar] [CrossRef] [PubMed]
- Kruger, M.; Moser, M.; Ussar, S.; Thievessen, I.; Luber, C.A.; Forner, F.; Schmidt, S.; Zanivan, S.; Fassler, R.; Mann, M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 2008, 134, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Y.; Lamond, A.I. A perspective on proteomics in cell biology. Trends Cell Biol. 2014, 24, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, X.; Tan, Z.; Lu, W.; Yang, G.; Guan, F. Alteration of N-glycans and expression of their related glycogenes in the epithelial-mesenchymal transition of HCV29 bladder epithelial cells. Molecules 2014, 19, 20073–20090. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Xu, Z.; Lu, W.; Li, X.; Sun, C.; Guo, J.; Xue, P.; Guan, F. Quantitative analysis of differential proteome expression in bladder cancer vs. normal bladder cells using SILAC method. PLoS ONE 2015, 10, e0134727. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Huang, J.; Sui, M. Targeting arginine metabolism pathway to treat arginine-dependent cancers. Cancer Lett. 2015, 364, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, M.; Endo, C.; Sakurada, A.; Hoshi, F.; Notsuda, H.; Kondo, T. The prognostic significance of eukaryotic elongation factor 1 alpha-2 in non-small cell lung cancer. Anticancer Res. 2014, 34, 651–658. [Google Scholar] [PubMed]
- Yang, S.; Lu, M.; Chen, Y.; Meng, D.; Sun, R.; Yun, D.; Zhao, Z.; Lu, D.; Li, Y. Overexpression of eukaryotic elongation factor 1 alpha-2 is associated with poorer prognosis in patients with gastric cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Aptsiauri, N.; Cabrera, T.; Mendez, R.; Garcia-Lora, A.; Ruiz-Cabello, F.; Garrido, F. Role of altered expression of HLA class I molecules in cancer progression. Adv. Exp. Med. Biol. 2007, 601, 123–131. [Google Scholar] [PubMed]
- He, X.P.; Song, F.J.; Liu, X.Y.; Wang, Z.; Li, X.X.; Liu, F.Y.; Chen, G.; Jiang, W.P. The relationship between KRAS gene mutations and HLA class I antigen downregulation in the metastasis of non-small cell lung cancer. J. Int. Med. Res. 2013, 41, 1473–1483. [Google Scholar] [CrossRef] [PubMed]
- Bendall, S.C.; Hughes, C.; Stewart, M.H.; Doble, B.; Bhatia, M.; Lajoie, G.A. Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol. Cell Proteom. 2008, 7, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Cui, T.; Wang, Y.; Sun, S.; Ma, T.; Wang, T.; Chen, Q.; Li, Z. Selective isolation and analysis of glycoprotein fractions and their glycomes from hepatocellular carcinoma sera. Proteomics 2013, 13, 1481–1498. [Google Scholar] [CrossRef] [PubMed]
- Michael, P.W.; Wolters, D.; Yates, J.R. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001, 19, 242–247. [Google Scholar] [CrossRef]
- Quan, H.; Xue, P.; Liu, S.; Fu, B.; Yang, L.; Huang, Z.; Li, H.; Chen, X.; Di, W. Differentially expressed protein profile of renal tubule cell stimulated by elevated uric acid using SILAC coupled to LC-MS. Cell. Physiol. Biochem. 2011, 27, 91–98. [Google Scholar] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.P.B.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Matic, I.; Hilger, M.; Nagaraj, N.; Selbach, M.; Olsen, J.V.; Mann, M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009, 4, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Berard, A.R.; Cortens, J.P.; Krokhin, O.; Wilkins, J.A.; Severini, A.; Coombs, K.M. Quantification of the host response proteome after mammalian reovirus t1l infection. PLoS ONE 2012, 7, 17. [Google Scholar]
- Coombs, K.M.; Berard, A.; Xu, W.; Krokhin, O.; Meng, X.; Cortens, J.P.; Kobasa, D.; Wilkins, J.; Brown, E.G. Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J. Virol. 2010, 84, 10888–10906. [Google Scholar] [CrossRef] [PubMed]
- Camon, E.; Magrane, M.; Barrell, D.; Binns, D.; Fleischmann, W.; Kersey, P.; Mulder, N.; Oinn, T.; Maslen, J.; Cox, A.; et al. The gene ontology annotation (GOA) project: Implementation of GO in SWISS-PROT, TReMBL, and InterPro. Genome Res. 2003, 13, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Ingenuity Systems. Available online: www.ingenuity.com (accessed on 11 January 2016).
- Sample Availability: Samples of the compounds for cells are available from the authors.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.; Lu, W.; Yu, D.; Sun, C.; Guo, J.; Li, Z.; Guan, F. Quantitative Analysis of Differential Proteome Expression in Epithelial-to-Mesenchymal Transition of Bladder Epithelial Cells Using SILAC Method. Molecules 2016, 21, 84. https://doi.org/10.3390/molecules21010084
Yang G, Lu W, Yu D, Sun C, Guo J, Li Z, Guan F. Quantitative Analysis of Differential Proteome Expression in Epithelial-to-Mesenchymal Transition of Bladder Epithelial Cells Using SILAC Method. Molecules. 2016; 21(1):84. https://doi.org/10.3390/molecules21010084
Chicago/Turabian StyleYang, Ganglong, Wei Lu, Di Yu, Chengwen Sun, Jia Guo, Zheng Li, and Feng Guan. 2016. "Quantitative Analysis of Differential Proteome Expression in Epithelial-to-Mesenchymal Transition of Bladder Epithelial Cells Using SILAC Method" Molecules 21, no. 1: 84. https://doi.org/10.3390/molecules21010084
APA StyleYang, G., Lu, W., Yu, D., Sun, C., Guo, J., Li, Z., & Guan, F. (2016). Quantitative Analysis of Differential Proteome Expression in Epithelial-to-Mesenchymal Transition of Bladder Epithelial Cells Using SILAC Method. Molecules, 21(1), 84. https://doi.org/10.3390/molecules21010084





