Emerging Structural Insights into Glycoprotein Quality Control Coupled with N-Glycan Processing in the Endoplasmic Reticulum
Abstract
:1. Introduction
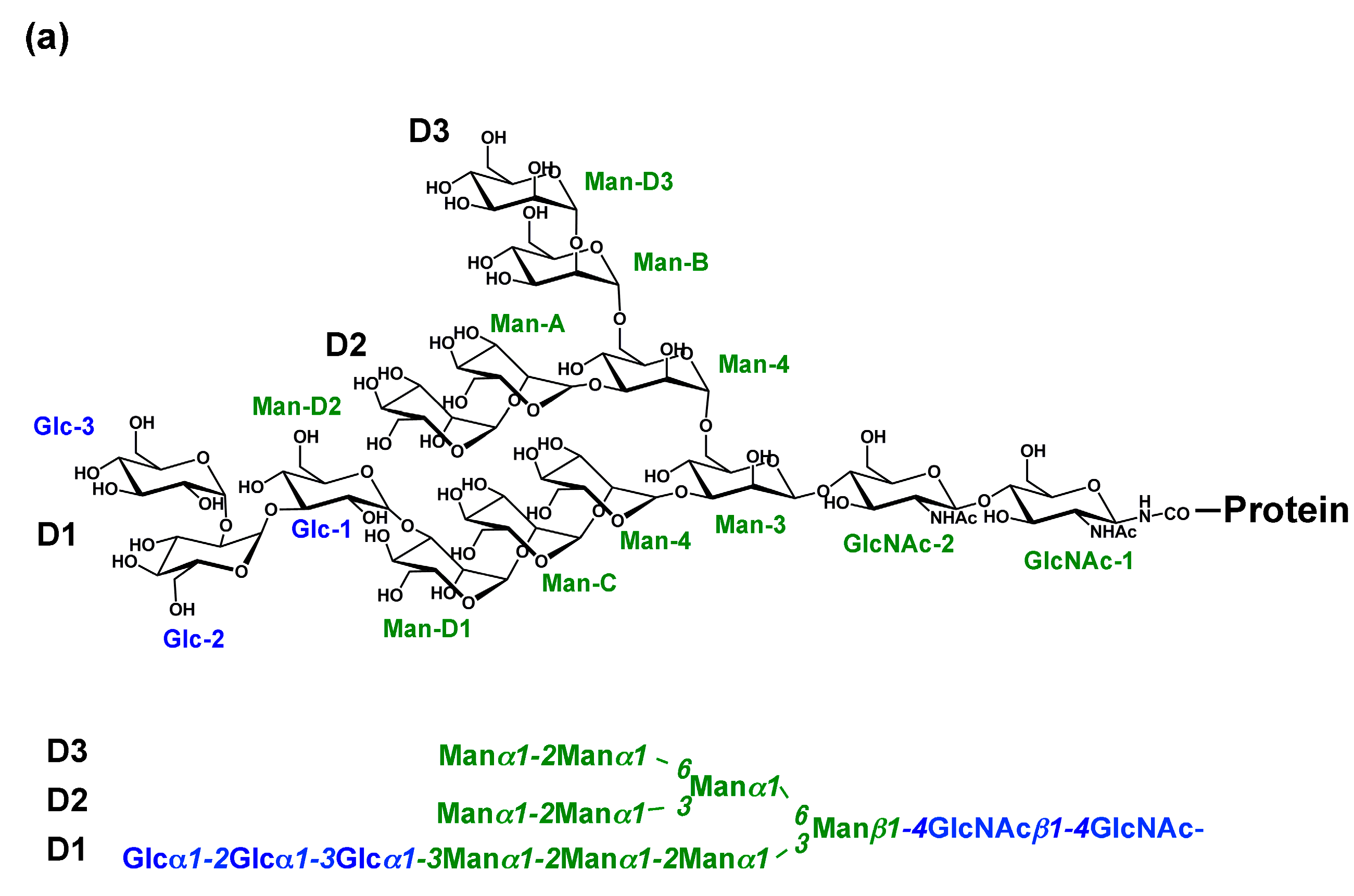
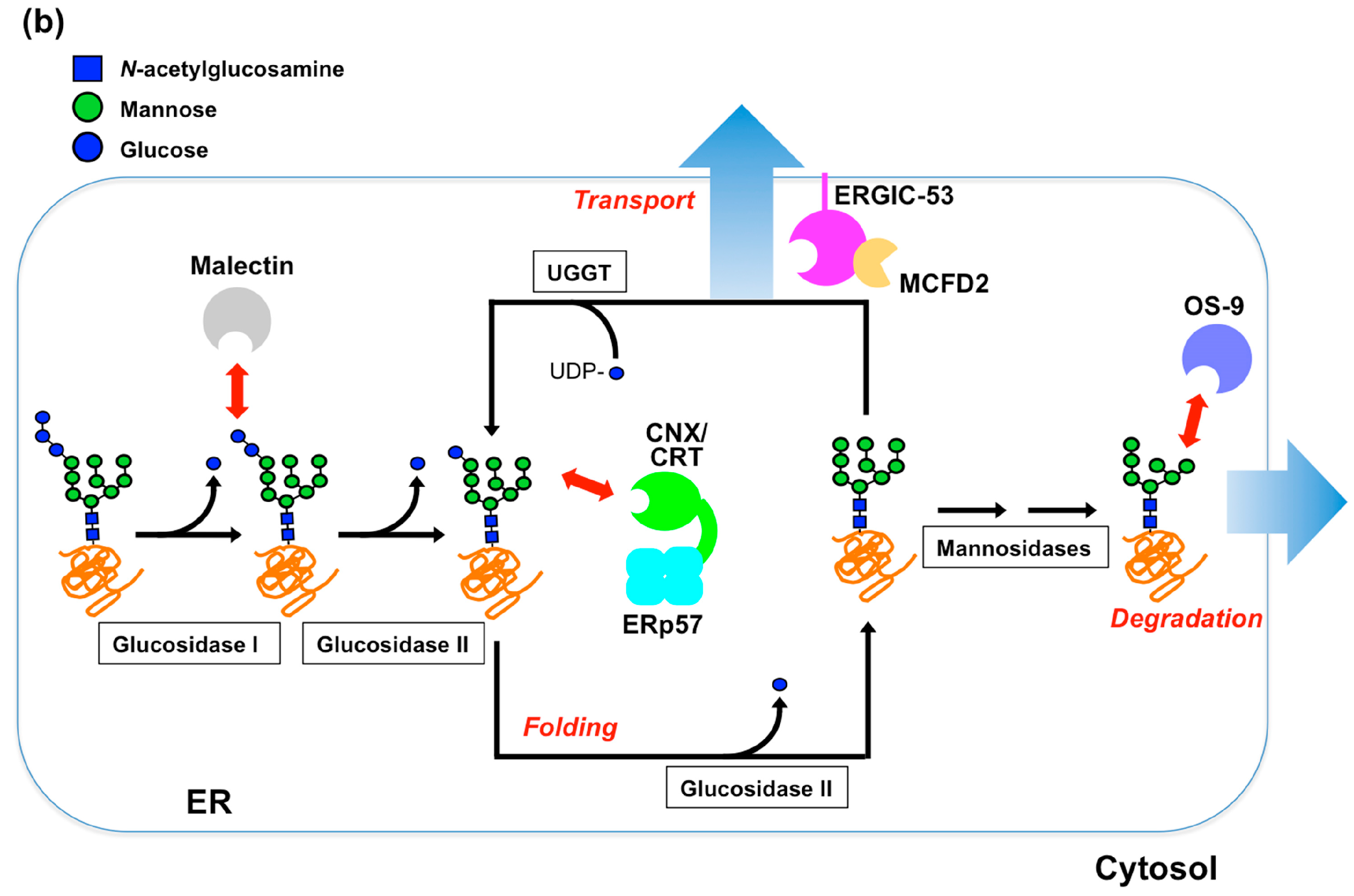
2. Glycoprotein Folding Assisted by Lectin/Chaperone Complexes

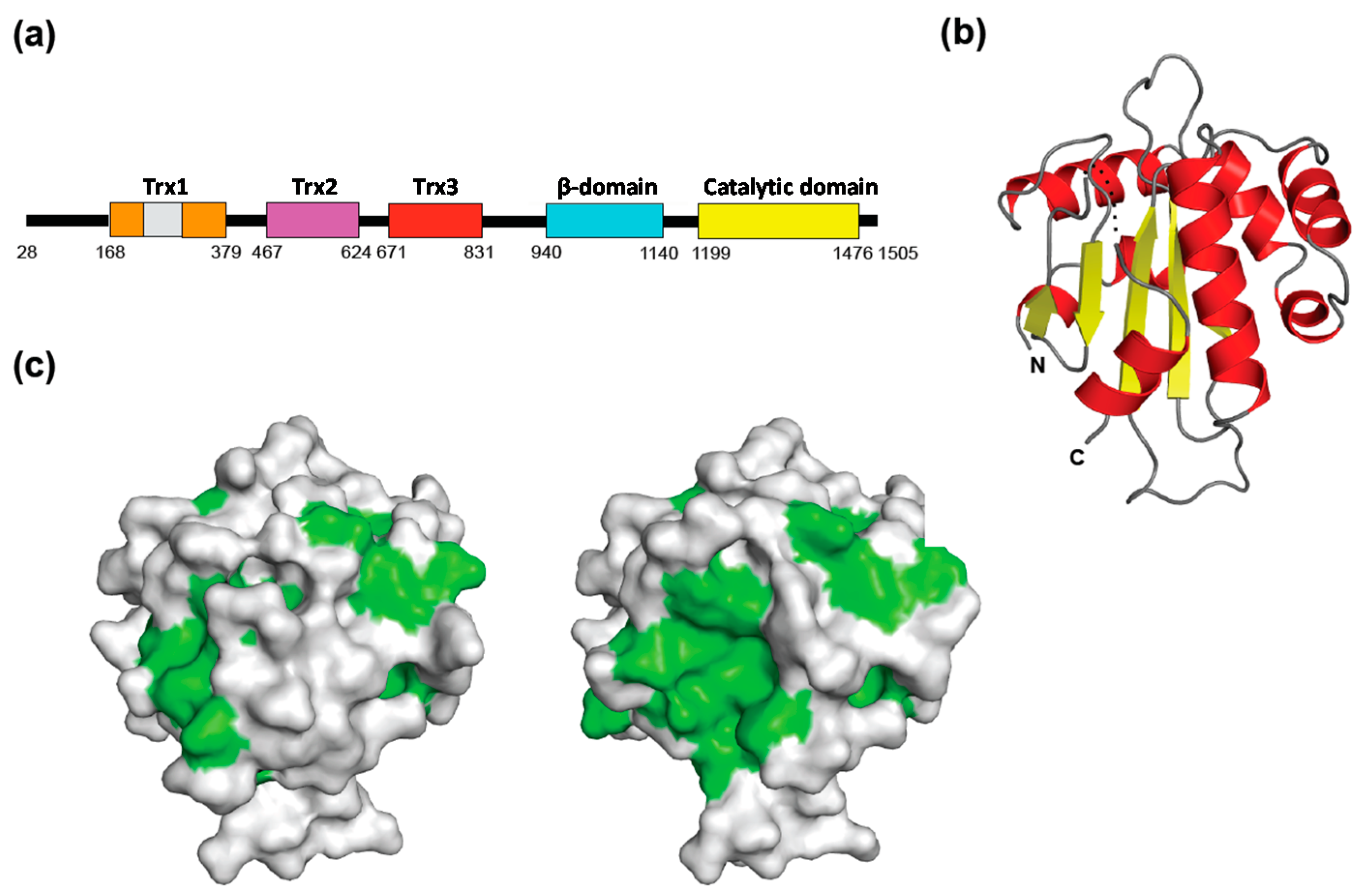
3. Glycoprotein Transport Mediated by Lectins as Cargo Receptors
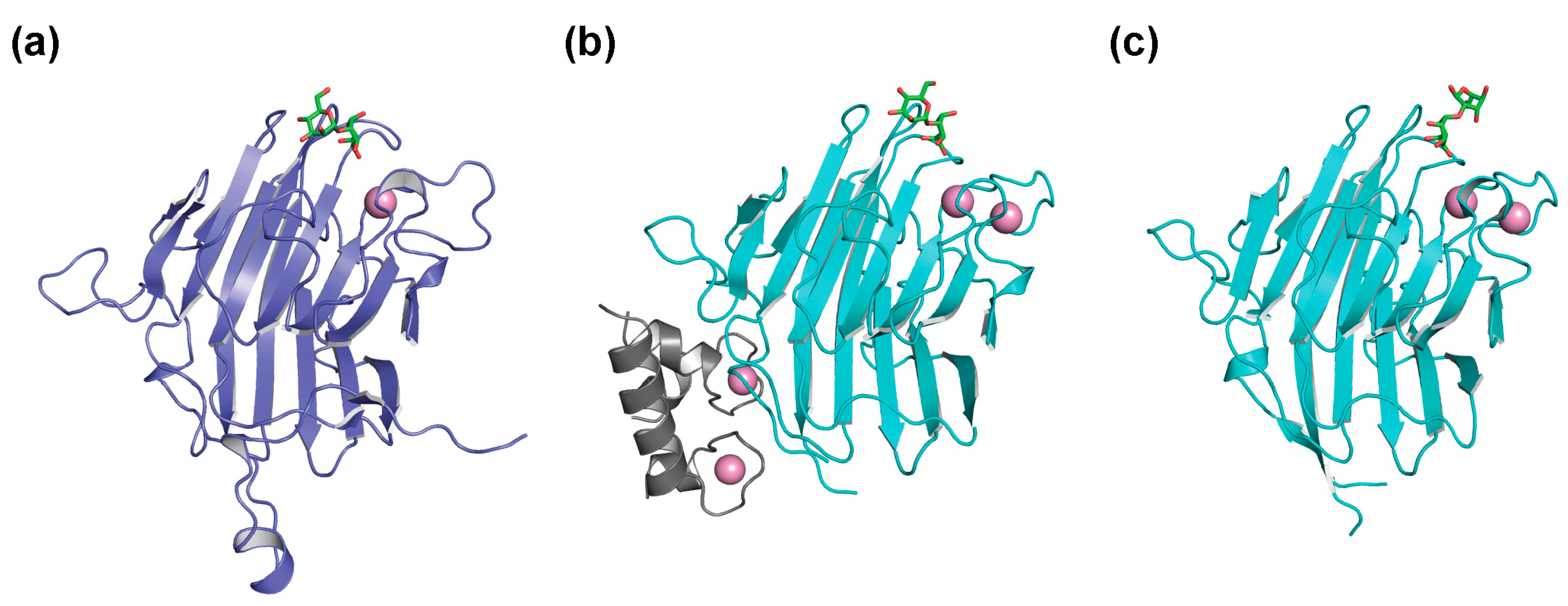

4. Glycoprotein Degradation Mediated by ERAD Lectins
5. Conformational Dynamics of High-Mannose-Type Oligosaccharides
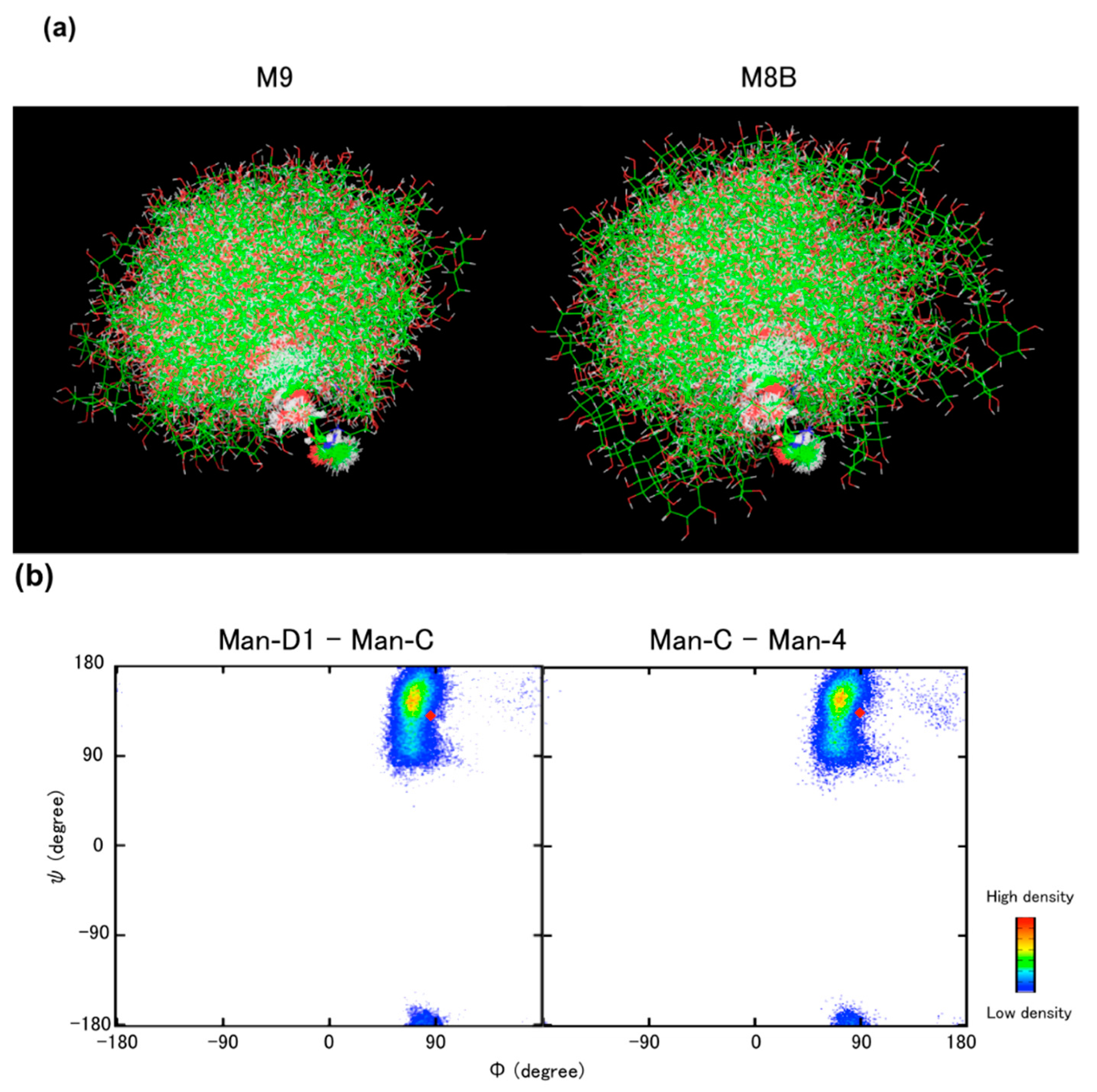
6. Concluding Remarks and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vliegenthart, J.F.G.; Dornald, L.; Halveek, H.V. High-resolution, 1H-nuclear magnetic resonance spectroscopy as a tool on the structural analysis of carbohydrates related to glycoproteins. Adv. Carbohydr. Chem. Biochem. 1983, 41, 209–374. [Google Scholar]
- Aebi, M.; Bernasconi, R.; Clerc, S.; Molinari, M. N-glycan structures: Recognition and processing in the ER. Trends Biochem. Sci. 2010, 35, 74–82. [Google Scholar]
- Kamiya, Y.; Satoh, T.; Kato, K. Molecular and structural basis for N-glycan-dependent determination of glycoprotein fates in cells. Biochim. Biophys. Acta 2012, 1820, 1327–1337. [Google Scholar]
- Kato, K.; Kamiya, Y. Structural views of glycoprotein-fate determination in cells. Glycobiology 2007, 17, 1031–1044. [Google Scholar]
- Lederkremer, G.Z. Glycoprotein folding, quality control and ER-associated degradation. Curr. Opin. Struct. Biol. 2009, 19, 515–523. [Google Scholar]
- Deprez, P.; Gautschi, M.; Helenius, A. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol. Cell 2005, 19, 183–195. [Google Scholar]
- Grinna, L.S.; Robbins, P.W. Substrate specificities of rat liver microsomal glucosidases which process glycoproteins. J. Biol. Chem. 1980, 255, 2255–2258. [Google Scholar]
- Barker, M.K.; Rose, D.R. Specificity of Processing α-glucosidase I is guided by the substrate conformation: Crystallographic and in silico studies. J. Biol. Chem. 2013, 288, 13563–13574. [Google Scholar]
- Asano, N.; Kizu, H.; Oseki, K.; Tomioka, E.; Matsui, K.; Okamoto, M.; Baba, M. N-alkylated nitrogen-in-the-ring sugars: Conformational basis of inhibition of glycosidases and HIV-1 replication. J. Med. Chem. 1995, 38, 2349–2356. [Google Scholar]
- Mehta, A.; Zitzmann, N.; Rudd, P.M.; Block, T.M.; Dwek, R.A. α-glucosidase inhibitors as potential broad based anti-viral agents. FEBS Lett. 1998, 430, 17–22. [Google Scholar]
- Durantel, D.; Alotte, C.; Zoulim, F. Glucosidase inhibitors as antiviral agents for hepatitis B and C. Curr. Opin. Investig. Drugs 2007, 8, 125–129. [Google Scholar]
- D’Alessio, C.; Caramelo, J.J.; Parodi, A.J. UDP-GlC:glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin. Cell Dev. Biol. 2010, 21, 491–499. [Google Scholar]
- Totani, K.; Ihara, Y.; Matsuo, I.; Ito, Y. Substrate specificity analysis of endoplasmic reticulum glucosidase II using synthetic high mannose-type glycans. J. Biol. Chem. 2006, 281, 31502–31508. [Google Scholar]
- Totani, K.; Ihara, Y.; Matsuo, I.; Ito, Y. Effects of macromolecular crowding on glycoprotein processing enzymes. J. Am. Chem. Soc. 2008, 130, 2101–2107. [Google Scholar]
- Trombetta, E.S.; Fleming, K.G.; Helenius, A. Quaternary and domain structure of glycoprotein processing glucosidase II. Biochemistry 2001, 40, 10717–10722. [Google Scholar]
- Hu, D.; Kamiya, Y.; Totani, K.; Kamiya, D.; Kawasaki, N.; Yamaguchi, D.; Matsuo, I.; Matsumoto, N.; Ito, Y.; Kato, K.; et al. Sugar-binding activity of the MRH domain in the ER α-glucosidase II beta subunit is important for efficient glucose trimming. Glycobiology 2009, 19, 1127–1135. [Google Scholar]
- Olson, L.J.; Orsi, R.; Alculumbre, S.G.; Peterson, F.C.; Stigliano, I.D.; Parodi, A.J.; D’Alessio, C.; Dahms, N.M. Structure of the lectin mannose 6-phosphate receptor homology (MRH) domain of glucosidase II, an enzyme that regulates glycoprotein folding quality control in the endoplasmic reticulum. J. Biol. Chem. 2013, 288, 16460–16475. [Google Scholar]
- Satoh, T.; Chen, Y.; Hu, D.; Hanashima, S.; Yamamoto, K.; Yamaguchi, Y. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol. Cell 2010, 40, 905–916. [Google Scholar]
- Caramelo, J.J.; Parodi, A.J. Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 2008, 283, 10221–10225. [Google Scholar]
- Kamiya, Y.; Kamiya, D.; Urade, R.; Suzuki, T.; Kato, K. Sophisticated modes of sugar recognition by intracellular lectins involved in quality control of glycoproteins. In Glycobiology Research Trends; Powell, G., McCabe, O., Eds.; Nova Science Publisher: New York, NY, USA, 2009; pp. 27–40. [Google Scholar]
- Schrag, J.D.; Bergeron, J.J.; Li, Y.; Borisova, S.; Hahn, M.; Thomas, D.Y.; Cygler, M. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell 2001, 8, 633–644. [Google Scholar]
- Kozlov, G.; Pocanschi, C.L.; Rosenauer, A.; Bastos-Aristizabal, S.; Gorelik, A.; Williams, D.B.; Gehring, K. Structural basis of carbohydrate recognition by calreticulin. J. Biol. Chem. 2010, 285, 38612–38620. [Google Scholar]
- Chouquet, A.; Païdassi, H.; Ling, W.L.; Frachet, P.; Houen, G.; Arland, G.J.; Gaboriaud, F. X-ray structure of the human calreticulin globular domain reveals a peptide-binding area and suggests a multi-molecular mechanism. PLoS One 2011, 6, e17886. [Google Scholar]
- Kozlov, G.; Bastos-Aristizabal, S.; Maattanen, P.; Rosenauer, A.; Zheng, F.; Killikelly, A.; Trempe, J.F.; Thomas, D.Y.; Gehring, K. Structural basis of cyclophilin B binding by the calnexin/calreticulin P-domain. J. Biol. Chem. 2010, 285, 35551–35557. [Google Scholar]
- Pollock, S.; Kozlov, G.; Pelletier, M.F.; Trempe, J.F.; Jansen, G.; Sitnikov, D.; Bergeron, J.J.; Gehring, K.; Ekiel, I.; Thomas, D.Y. Specific interaction of ERp57 and calnexin determined by NMR spectroscopy and an ER two-hybrid system. EMBO J. 2004, 23, 1020–1029. [Google Scholar]
- Sakono, M.; Seko, A.; Takeda, Y.; Ito, Y. PDI family protein ERp29 forms 1:1 complex with lectin chaperone calreticulin. Biochem. Biophys. Res. Commun. 2014, 452, 27–31. [Google Scholar]
- Satoh, T.; Cowieson, N.P.; Hakamata, W.; Ideo, H.; Fukushima, K.; Kurihara, M.; Kato, R.; Yamashita, K.; Wakatsuki, S. Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36. J. Biol. Chem. 2007, 282, 28246–28255. [Google Scholar]
- Zheng, C.; Page, R.C.; Das, V.; Nix, J.C.; Wigren, E.; Misra, S.; Zhang, B. Structural characterization of carbohydrate binding by LMAN1 protein provides new insight into the endoplasmic reticulum export of factors V (FV) and VIII (FVIII). J. Biol. Chem. 2013, 288, 20499–20509. [Google Scholar]
- Satoh, T.; Suzuki, K.; Yamaguchi, T.; Kato, K. Structural basis for disparate sugar-binding specificities in the homologous cargo receptors ERGIC-53 and VIP36. PLoS One 2014, 9, e87963. [Google Scholar]
- Taylor, S.C.; Ferguson, A.D.; Bergeron, J.J.; Thomas, D.Y. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat. Struct. Mol. Biol. 2004, 11, 128–134. [Google Scholar]
- Totani, K.; Ihara, Y.; Tsujimoto, T.; Matsuo, I.; Ito, Y. The recognition motif of the glycoprotein-folding sensor enzyme UDP-Glc:glycoprotein glucosyltransferase. Biochemistry 2009, 48, 2933–2940. [Google Scholar]
- Caramelo, J.J.; Castro, O.A.; Alonso, L.G.; de Prat-Gay, G.; Parodi, A.J. UDP-Glc:glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates. Proc. Natl. Acad. Sci. USA 2003, 100, 86–91. [Google Scholar]
- Zhu, T.; Satoh, T.; Kato, K. Structural insight into substrate recognition by the endoplasmic reticulum folding-sensor enzyme: Crystal structure of third thioredoxin-like domain of UDP-glucose:glycoprotein glucosyltransferase. Sci. Rep. 2014, 4, 7322. [Google Scholar] [CrossRef]
- Labunskyy, V.M.; Ferguson, A.D.; Fomenko, D.E.; Chelliah, Y.; Hatfield, D.L.; Gladyshev, V.N. A novel cysteine-rich domain of Sep15 mediates the interaction with UDP-glucose:glycoprotein glucosyltransferase. J. Biol. Chem. 2005, 280, 37839–37845. [Google Scholar]
- Takeda, Y.; Seko, A.; Hachisu, M.; Daikoku, S.; Izumi, M.; Koizumi, A.; Fujikawa, K.; Kajihara, Y.; Ito, Y. Both isoforms of human UDP-glucose:glycoprotein glucosyltransferase are enzymatically active. Glycobiology 2014, 24, 344–350. [Google Scholar]
- Kamiya, Y.; Yamaguchi, Y.; Takahashi, N.; Arata, Y.; Kasai, K.I.; Ihara, Y.; Matsuo, I.; Ito, Y.; Yamamoto, K.; Kato, K. Sugar-binding properties of VIP36, an intracellular animal lectin operating as a cargo receptor. J. Biol. Chem. 2005, 280, 37178–37182. [Google Scholar]
- Kamiya, Y.; Kamiya, D.; Yamamoto, K.; Nyfeler, B.; Hauri, H.P.; Kato, K. Molecular basis of sugar recognition by the human l-type lectins ERGIC-53, VIPL, and VIP36. J. Biol. Chem. 2008, 283, 1857–1861. [Google Scholar]
- Schweizer, A.; Fransen, J.A.; Bachi, T.; Ginsel, L.; Hauri, H.P. Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J. Cell Biol. 1988, 107, 1643–1653. [Google Scholar]
- Molinari, M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 2007, 3, 313–320. [Google Scholar]
- Qin, S.Y.; Kawasaki, N.; Hu, D.; Tozawa, H.; Matsumoto, N.; Yamamoto, K. Subcellular localization of ERGIC-53 under endoplasmic reticulum stress condition. Glycobiology 2012, 22, 1709–1720. [Google Scholar]
- Roth, J.; Ziak, M.; Zuber, C. The role of glucosidase II and endomannosidase in glucose trimming of asparagine-liked oligosaccharides. Biochimie 2003, 85, 287–294. [Google Scholar]
- Kornfeld, R.; Kornfeld, S. Assembly of asparagine-liked oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664. [Google Scholar]
- Moothoo, D.N.; Canan, B.; Field, R.A.; Naismith, J.H. Man α1–2 Man α-OMe-concanavalin A complex reveals a balance of forces involved in carbohydrate recognition. Glycobiology 1999, 9, 539–545. [Google Scholar]
- Zhang, B.; Cunningham, M.A.; Nichols, W.C.; Bernat, J.A.; Seligsohn, U.; Pipe, S.W.; McVey, J.H.; Schulte-Overberg, U.; de Bosch, N.B.; Ruiz-Saez, A.; et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat. Genet 2003, 34, 220–225. [Google Scholar]
- Zhang, B. Recent developments in the understanding of the combined deficiency of FV and FVIII. Br. J. Haematol. 2009, 145, 15–23. [Google Scholar]
- Zhang, B.; Kaufman, R.J.; Ginsburg, D. LMAN1 and MCFD2 form a cargo receptor complex and interact with coagulation factor VIII in the early secretory pathway. J. Biol. Chem. 2005, 280, 25881–25886. [Google Scholar]
- Nyfeler, B.; Zhang, B.; Ginsburg, D.; Kaufman, R.J.; Hauri, H.P. Cargo selectivity of the ERGIC-53/MCFD2 transport receptor complex. Traffic 2006, 7, 1473–1481. [Google Scholar]
- Nishio, M.; Kamiya, Y.; Mizushima, T.; Wakatsuki, S.; Sasakawa, H.; Yamamoto, K.; Uchiyama, S.; Noda, M.; McKay, A.R.; Fukui, K.; et al. Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency. Proc. Natl. Acad. Sci. USA 2010, 107, 4034–4039. [Google Scholar]
- Wigren, E.; Bourhis, J.M.; Kursula, I.; Guy, J.E.; Lindqvist, Y. Crystal structure of the LMAN1-CRD/MCFD2 transport receptor complex provides insight into combined deficiency of factor V and factor VIII. FEBS Lett. 2010, 584, 878–882. [Google Scholar]
- Chen, Y.; Hojo, S.; Matsumoto, N.; Yamamoto, K. Regulation of Mac-2BP secretion is mediated by its N-glycan binding to ERGIC-53. Glycobiology 2013, 23, 904–916. [Google Scholar]
- Klaus, J.P.; Eisenhauer, P.; Russo, J.; Mason, A.B.; Do, D.; King, B.; Taatjes, D.; Cornillez-Ty, C.; Boyson, J.E.; Thali, M.; et al. The intracellular cargo receptor ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles. Cell Host Microbe 2013, 14, 522–534. [Google Scholar]
- Jakob, C.A.; Bodmer, D.; Spirig, U.; Battig, P.; Marcil, A.; Dignard, D.; Bergeron, J.J.; Thomas, D.Y.; Aebi, M. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001, 2, 423–430. [Google Scholar]
- Hosokawa, N.; Wada, I.; Hasegawa, K.; Yorihuzi, T.; Tremblay, L.O.; Herscovics, A.; Nagata, K. A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001, 2, 415–422. [Google Scholar]
- Vallée, F.; Lipari, F.; Yip, P.; Sleno, B.; Herscovics, A.; Howell, P.L. Crystal structure of a class I α1,2-mannosidase involved in N-glycan processing and endoplasmic reticulum quality control. EMBO J. 2000, 19, 581–588. [Google Scholar]
- Vallée, F.; Karaveg, K.; Herscovics, A.; Moremen, K.W.; Howell, P.L. Structural basis for catalysis and inhibition of N-glycan processing class I α1,2-mannosidases. J. Biol. Chem. 2000, 275, 41287–41298. [Google Scholar]
- Karaveg, K.; Moremen, K.W. Energetics of substrate binding and catalysis by class 1 (glycosylhydrolase family 47) α-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control. J. Biol. Chem. 2005, 280, 29837–29848. [Google Scholar]
- Karaveg, K.; Siriwardena, A.; Tempel, W.; Liu, Z.J.; Glushka, J.; Wang, B.C.; Moremen, K.W. Mechanism of class 1 (glycosylhydrolase family 47) α-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control. J. Biol. Chem. 2005, 280, 16197–16207. [Google Scholar]
- Hirao, K.; Natsuka, Y.; Tamura, T.; Wada, I.; Morito, D.; Natsuka, S.; Romero, P.; Sleno, B.; Tremblay, L.O.; Herscovics, A.; et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 2006, 281, 9650–9658. [Google Scholar]
- Olivari, S.; Galli, C.; Alanen, H.; Ruddock, L.; Molinari, M. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J. Biol. Chem. 2005, 280, 2424–2428. [Google Scholar]
- Quan, E.M.; Kamiya, Y.; Kamiya, D.; Denic, V.; Weibezahn, J.; Kato, K.; Weissman, J.S. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell 2008, 32, 870–877. [Google Scholar]
- Clerc, S.; Hirsch, C.; Oggier, D.M.; Deprez, P.; Jakob, C.; Sommer, T.; Aebi, M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 2009, 184, 159–172. [Google Scholar]
- Hosokawa, N.; Kamiya, Y.; Kamiya, D.; Kato, K.; Nagata, K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J. Biol. Chem. 2009, 284, 17061–17068. [Google Scholar]
- Hosokawa, N.; Tremblay, L.O.; Sleno, B.; Kamiya, Y.; Wada, I.; Nagata, K.; Kato, K.; Herscovics, A. EDEM1 accelerates the trimming of α1,2-linked mannose on the C branch of N-glycans. Glycobiology 2010, 20, 567–575. [Google Scholar]
- Ninagawa, S.; Okada, T.; Sumitomo, Y.; Kamiya, Y.; Kato, K.; Horimoto, S.; Ishikawa, T.; Takeda, S.; Sakuma, T.; Yamamoto, T.; et al. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J. Cell Biol. 2014, 206, 347–356. [Google Scholar]
- Gauss, R.; Kanehara, K.; Carvalho, P.; Ng, D.T.; Aebi, M. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol. Cell 2011, 42, 782–793. [Google Scholar]
- Helenius, A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol. Biol. Cell 1994, 5, 253–265. [Google Scholar]
- Mikami, K.; Yamaguchi, D.; Tateno, H.; Hu, D.; Qin, S.Y.; Kawasaki, N.; Yamada, M.; Matsumoto, N.; Hirabayashi, J.; Ito, Y.; et al. The sugar-binding ability of human OS-9 and its involvement in ER-associated degradation. Glycobiology 2010, 20, 310–321. [Google Scholar]
- Bernasconi, R.; Pertel, T.; Luban, J.; Molinari, M. A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum: Inhibiting secretion of misfolded protein conformers and enhancing their disposal. J. Biol. Chem. 2008, 283, 16446–16454. [Google Scholar]
- Christianson, J.C.; Shaler, T.A.; Tyler, R.E.; Kopito, R.R. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008, 10, 272–282. [Google Scholar]
- Yamaguchi, D.; Hu, D.; Matsumoto, N.; Yamamoto, K. Human XTP3-B binds to α1-antitrypsin variant nullHong Kong via the C-terminal MRH domain in a glycan-dependent manner. Glycobiology 2010, 20, 348–355. [Google Scholar]
- Fujimori, T.; Kamiya, Y.; Nagata, K.; Kato, K.; Hosokawa, N. Endoplasmic reticulum lectin XTP3-B inhibits endoplasmic reticulum-associated degradation of a misfolded α1-antitrypsin variant. FEBS J. 2013, 280, 1563–1575. [Google Scholar]
- Seidler, P.M.; Shinsky, S.A.; Hong, F.; Li, Z.; Cosgrove, M.S.; Gewirth, D.T. Characterization of the Grp94/OS-9 chaperone-lectin complex. J. Mol. Biol. 2014, 426, 3590–3605. [Google Scholar]
- Dersh, D.; Jones, S.M.; Eletto, D.; Christianson, J.C.; Argon, Y. OS-9 facilitates turnover of nonnative GRP94 marked by hyperglycosylation. Mol. Biol. Cell 2014, 25, 2220–2234. [Google Scholar]
- Hanna, J.; Schutz, A.; Zimmermann, F.; Behlke, J.; Sommer, T.; Heinemann, U. Structural and biochemical basis of Yos9 protein dimerization and possible contribution to self-association of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation ubiquitin-ligase complex. J. Biol. Chem. 2012, 287, 8633–8640. [Google Scholar]
- Burkhalter, N.F.; Dimick, S.M.; Toone, E.J. Protein-carbohydrate interaction: Fundamental considerations. In Carbohydrates in Chemistry and Biology, Part II; Ernst, B., Hart, G.W., Sinaÿ, P., Eds.; Wiley-VCH: Weinhein, Germany, 2000; Volume 2, pp. 863–914. [Google Scholar]
- Kamiya, Y.; Yanagi, K.; Kitajima, T.; Yamaguchi, T.; Chiba, Y.; Kato, K. Application of metabolic 13C labeling in conjunction with high-field nuclear magnetic resonance spectroscopy for comparative conformational analysis of high mannose-type oligosaccharides. Biomolecules 2013, 3, 108–123. [Google Scholar]
- Yamaguchi, T.; Kamiya, Y.; Choo, Y.-M.; Yamamoto, S.; Kato, K. Terminal spin labeling of a high-mannose-type oligosaccharide for quantitative NMR analysis of its dynamic conformation. Chem. Lett. 2013, 42, 544–546. [Google Scholar]
- Wooten, E.W.; Bazzo, R.; Edge, C.J.; Zamze, S.; Dwek, R.A.; Rademacher, T.W. Primary sequence dependence of conformation in oligomannose oligosaccharides. Eur. Biophys. J. 1990, 18, 139–148. [Google Scholar]
- Wormald, M.R.; Petrescu, A.J.; Pao, Y.L.; Glithero, A.; Elliott, T.; Dwek, R.A. Conformational studies of oligosaccharides and glycopeptides: Complementarity of NMR, X-ray crystallography, and molecular modelling. Chem. Rev. 2002, 102, 371–386. [Google Scholar]
- Balaji, P.V.; Qasba, P.K.; Rao, V.S. Molecular dynamics simulations of high-mannose oligosaccharides. Glycobiology 1994, 4, 497–515. [Google Scholar]
- Petrescu, A.J.; Butters, T.D.; Reinkensmeier, G.; Petrescu, S.; Platt, F.M.; Dwek, R.A.; Wormald, M.R. The solution NMR structure of glucosylated N-glycans involved in the early stages of glycoprotein biosynthesis and folding. EMBO J. 1997, 16, 4302–4310. [Google Scholar]
- Zhang, Y.; Yamaguchi, T.; Satoh, T.; Yagi-Utsumi, M.; Kamiya, Y.; Sakae, Y.; Okamoto, Y.; Kato, K. Conformational dynamics of oligosaccharides characterized by paramagnetism-assisted NMR spectroscopy in conjunction with molecular dynamics simulation. Adv. Exp. Med. Biol. 2015, 842, 217–230. [Google Scholar]
- Yamaguchi, T.; Kato, K. Paramagnetism-assisted nuclear magnetic resonance analysis of dynamic conformations and interactions of oligosaccharides. In Glycoscience: Biology and Medicine; Taniguchi, N., Endo, T., Hart, G.W., Seeberger, P., Wong, C.-H., Eds.; Springer: Tokyo, Japan, 2014; in press. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Sakae, Y.; Zhang, Y.; Yamamoto, S.; Okamoto, Y.; Kato, K. Exploration of conformational spaces of high-mannose-type oligosaccharides by an NMR-validated simulation. Angew. Chem. Int. Ed. 2014, 53, 10941–10944. [Google Scholar]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satoh, T.; Yamaguchi, T.; Kato, K. Emerging Structural Insights into Glycoprotein Quality Control Coupled with N-Glycan Processing in the Endoplasmic Reticulum. Molecules 2015, 20, 2475-2491. https://doi.org/10.3390/molecules20022475
Satoh T, Yamaguchi T, Kato K. Emerging Structural Insights into Glycoprotein Quality Control Coupled with N-Glycan Processing in the Endoplasmic Reticulum. Molecules. 2015; 20(2):2475-2491. https://doi.org/10.3390/molecules20022475
Chicago/Turabian StyleSatoh, Tadashi, Takumi Yamaguchi, and Koichi Kato. 2015. "Emerging Structural Insights into Glycoprotein Quality Control Coupled with N-Glycan Processing in the Endoplasmic Reticulum" Molecules 20, no. 2: 2475-2491. https://doi.org/10.3390/molecules20022475
APA StyleSatoh, T., Yamaguchi, T., & Kato, K. (2015). Emerging Structural Insights into Glycoprotein Quality Control Coupled with N-Glycan Processing in the Endoplasmic Reticulum. Molecules, 20(2), 2475-2491. https://doi.org/10.3390/molecules20022475





