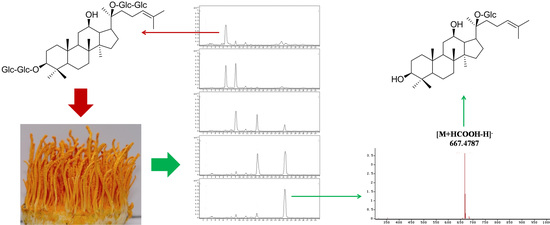
Highly Selective Bioconversion of Ginsenoside Rb1 to Compound K by the Mycelium of Cordyceps sinensis under Optimized Conditions
Abstract
:1. Introduction
| 20(S)-Protopanaxdiol Type | ||||
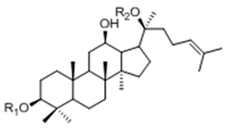 | Ginsenoside | R1 (C-3) | R2 (C-20) | |
| Rb1 | -Glc2-Glc | -Glc6-Glc | ||
| Rd | -Glc2-Glc | -Glc | ||
| Rg3 | -Glc2-Glc | -H | ||
| F2 | -Glc | -Glc | ||
| Rh2 | -Glc | -H | ||
| Compound K | -H | -Glc | ||
| 20(S)-Protopanaxtriol Type | ||||
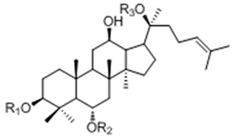 | Ginsenoside | R1 (C-3) | R2 (C-6) | R3 (C-20) |
| Re | -H | -Glc2-Rha | -Glc | |
| Rf | -H | -Glc2-Glc | -H | |
| Rg1 | -H | -Glc | -Glc | |
| Rg2 | -H | -Glc2-Rha | -H | |
| Rh1 | -H | -Glc | -H | |
2. Results and Discussion
2.1. Optimization on Biotransformation Parameters of Ginsenoside Rb1 by C. sinensis
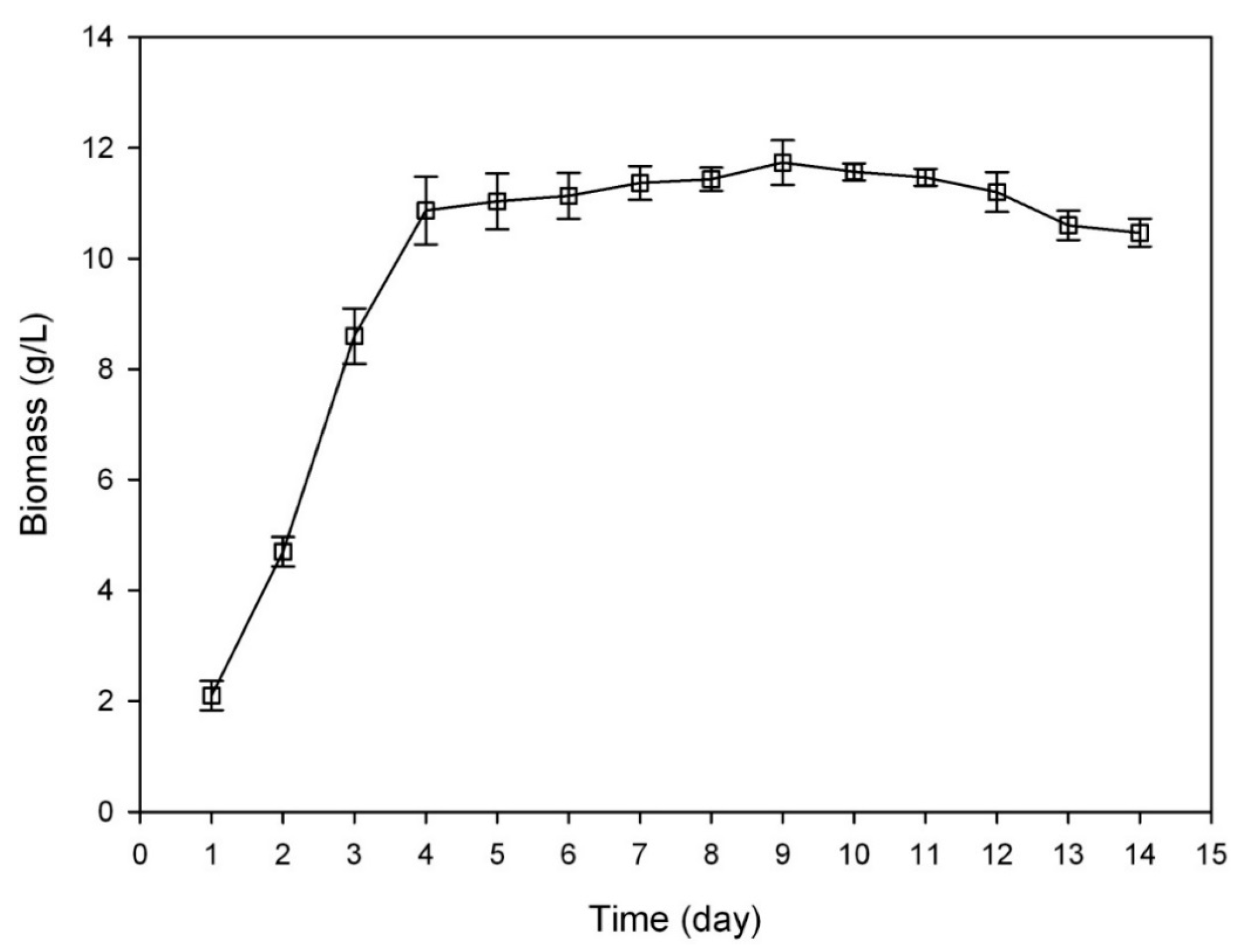
2.1.1. Determination on Substrate-Feeding Time
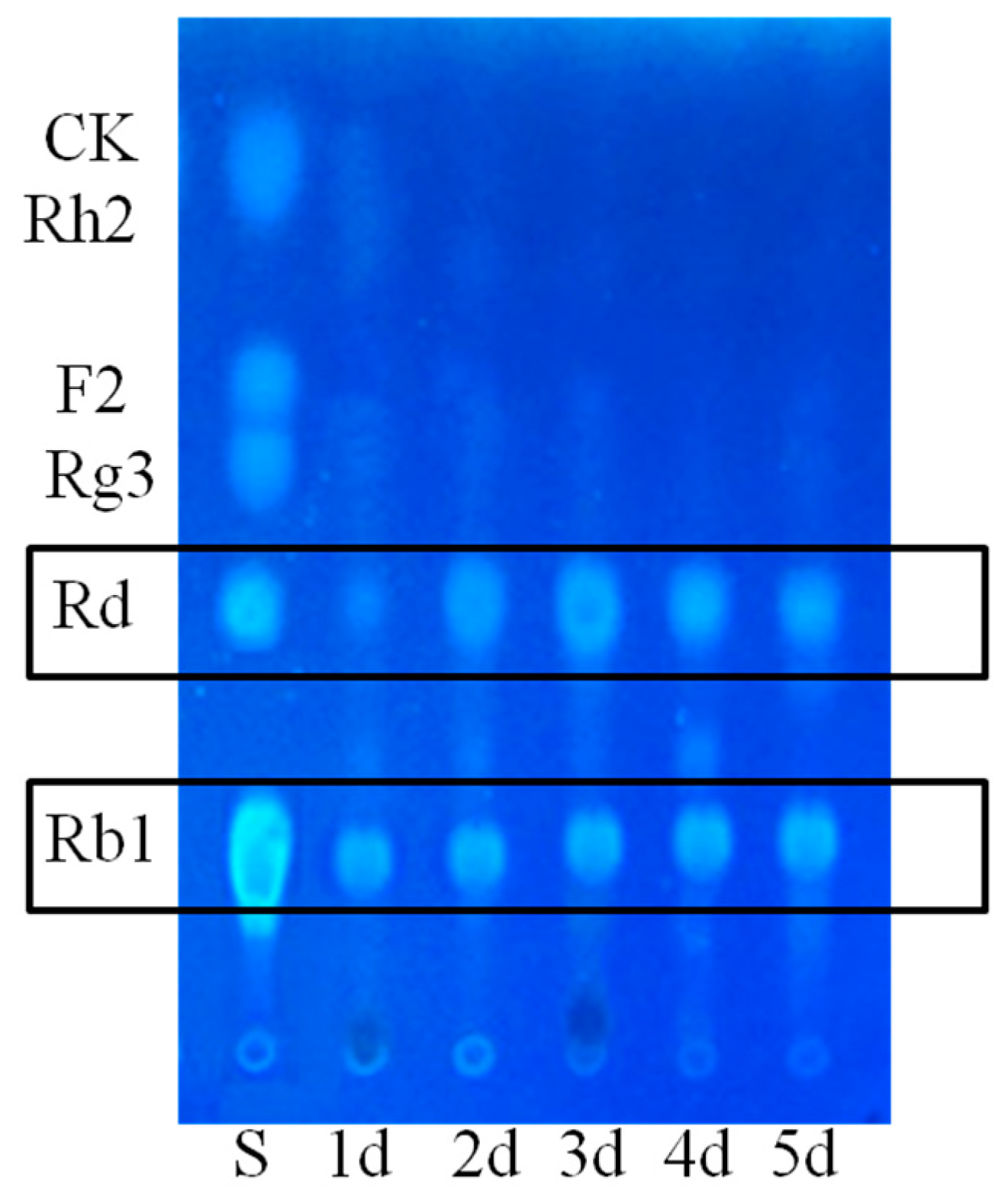
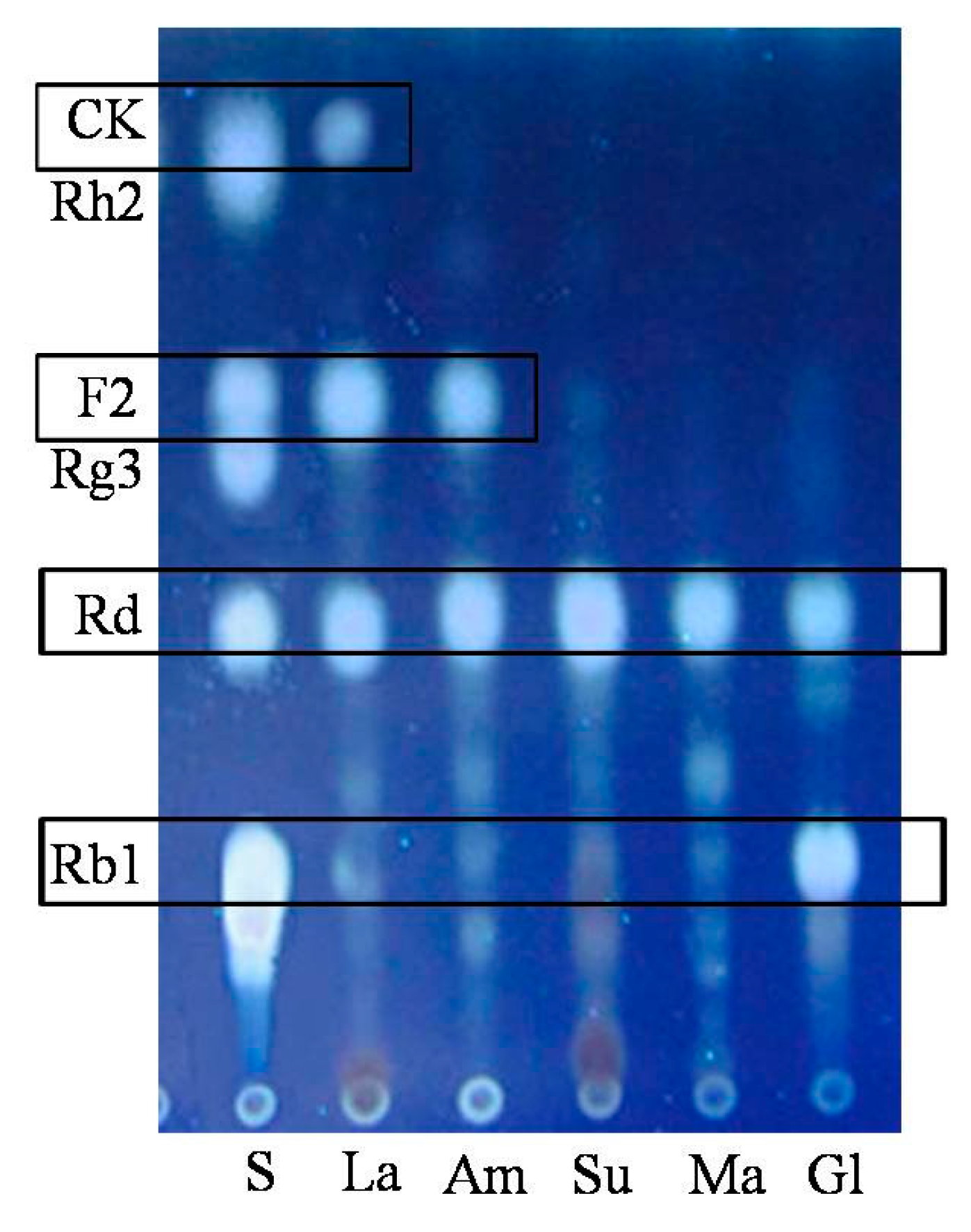
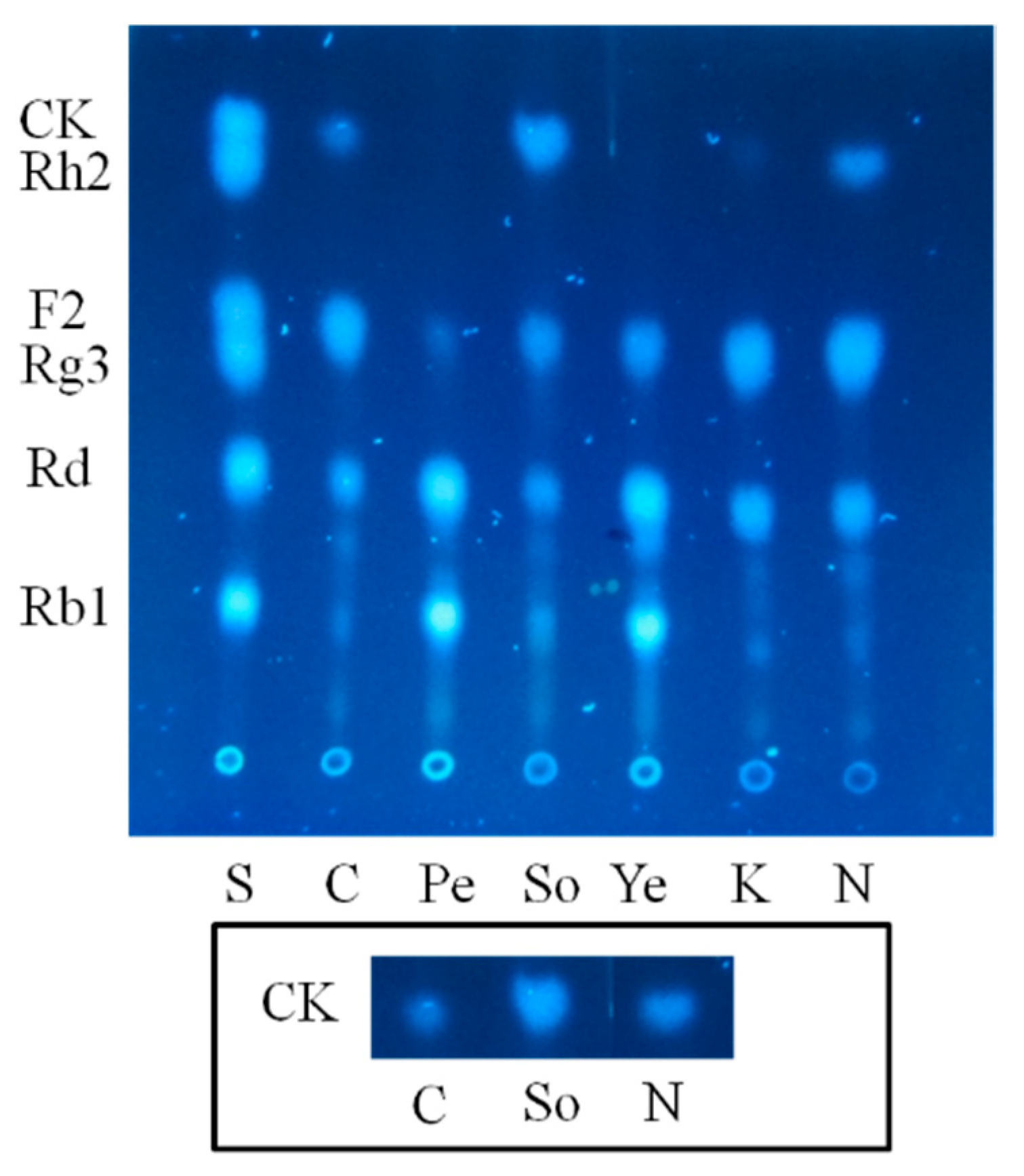
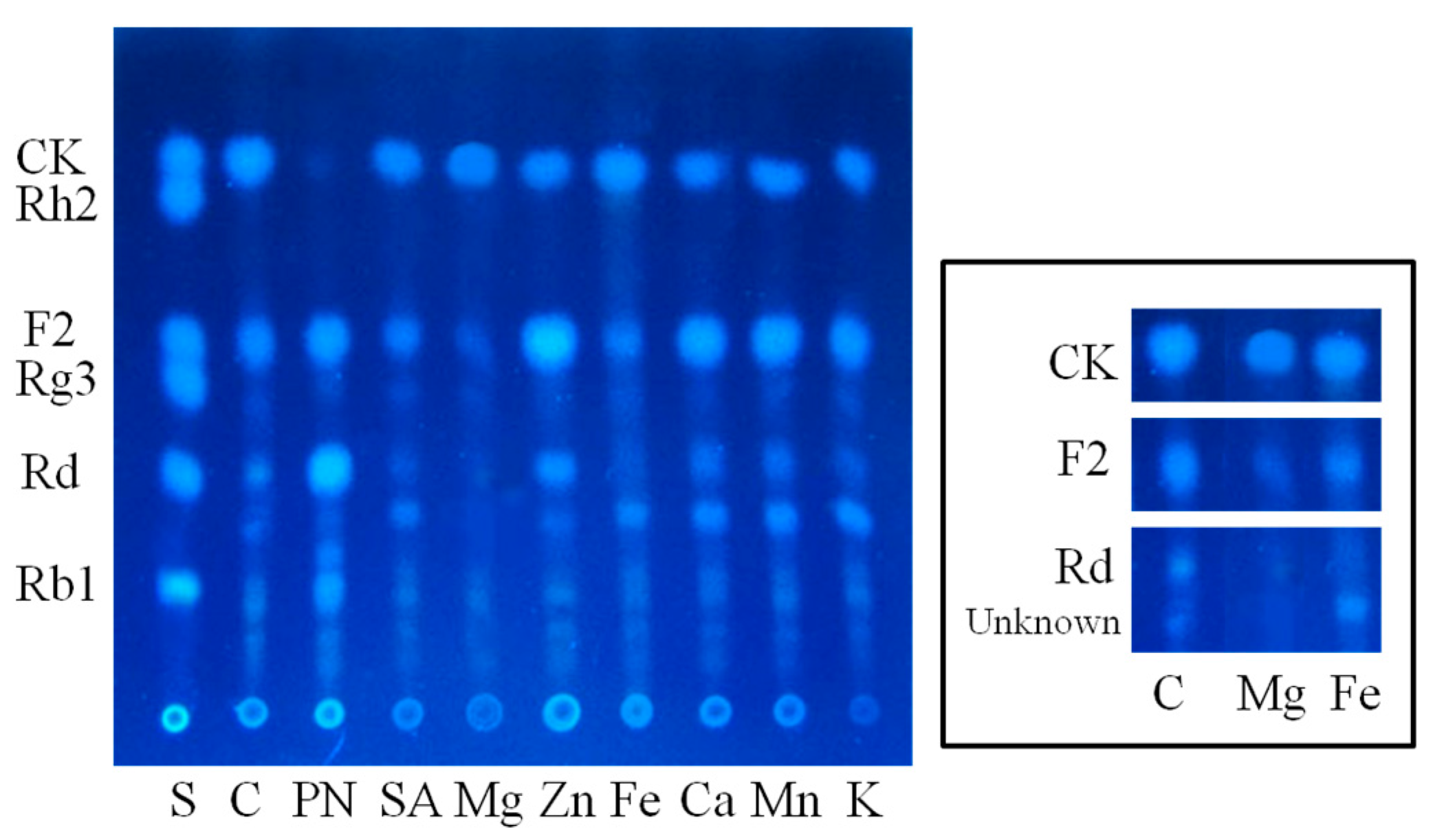
2.1.2. Optimization on Carbon Sources, Nitrogen Sources, Elicitors, Reaction pH and Temperature for the Biotransformation Process
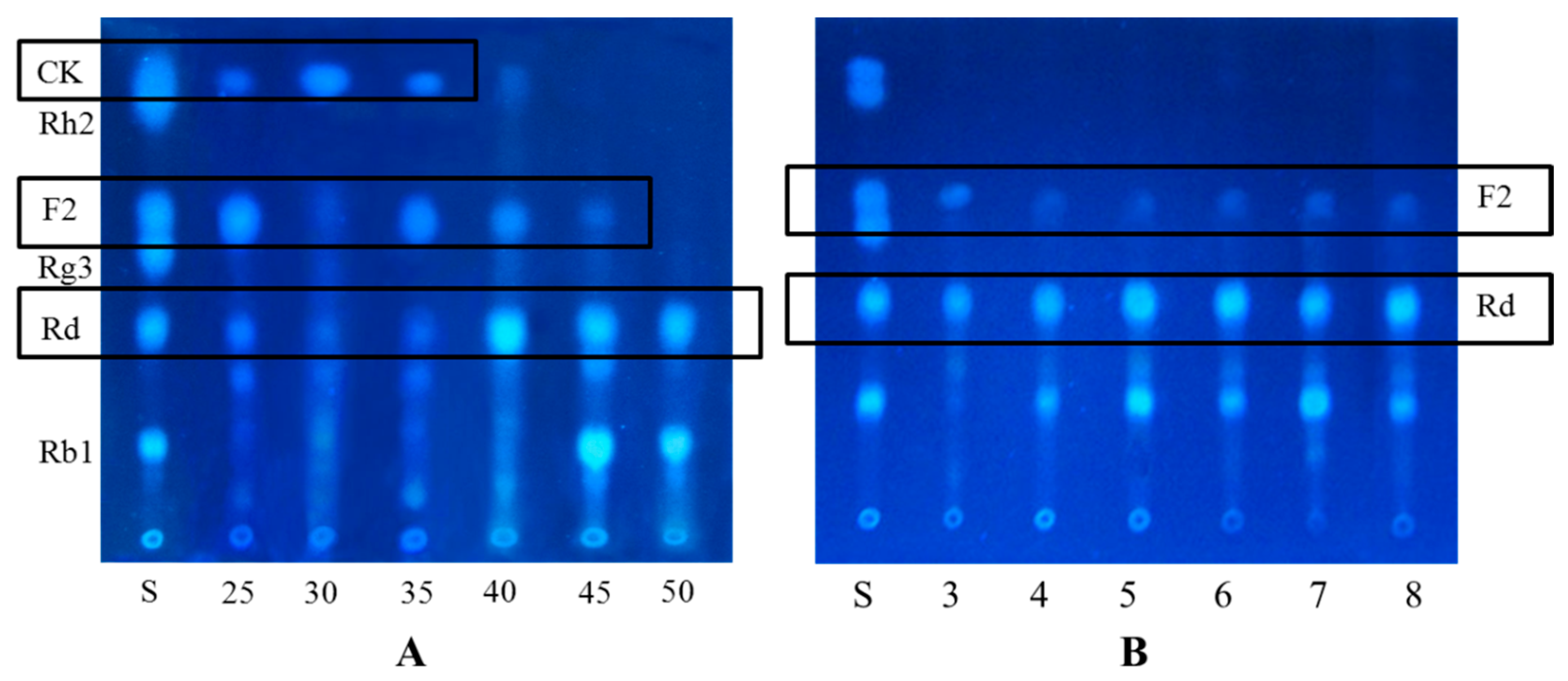
2.2. Biotransformation Pathway of Ginsenoside Rb1 by C. sinensis
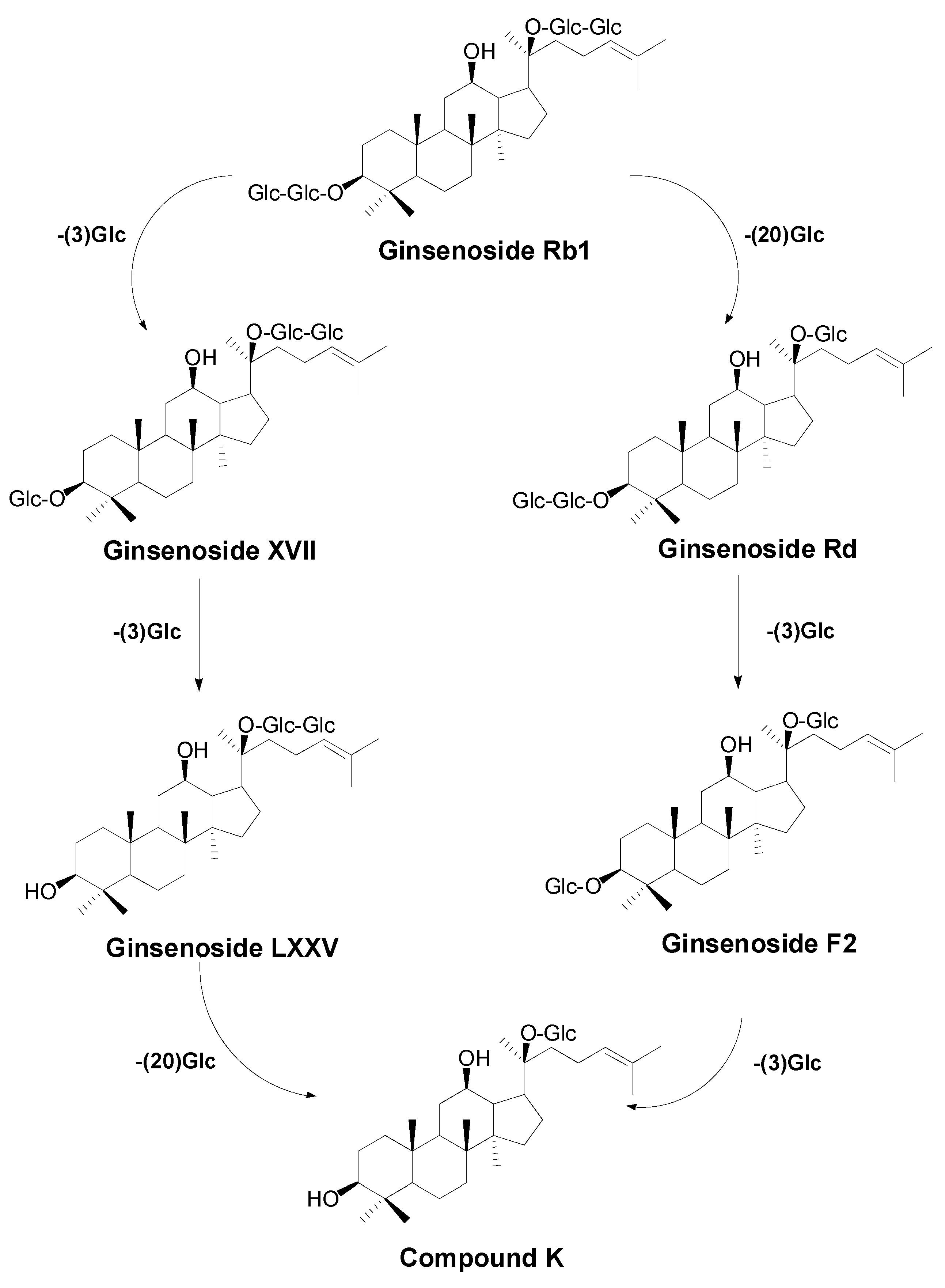
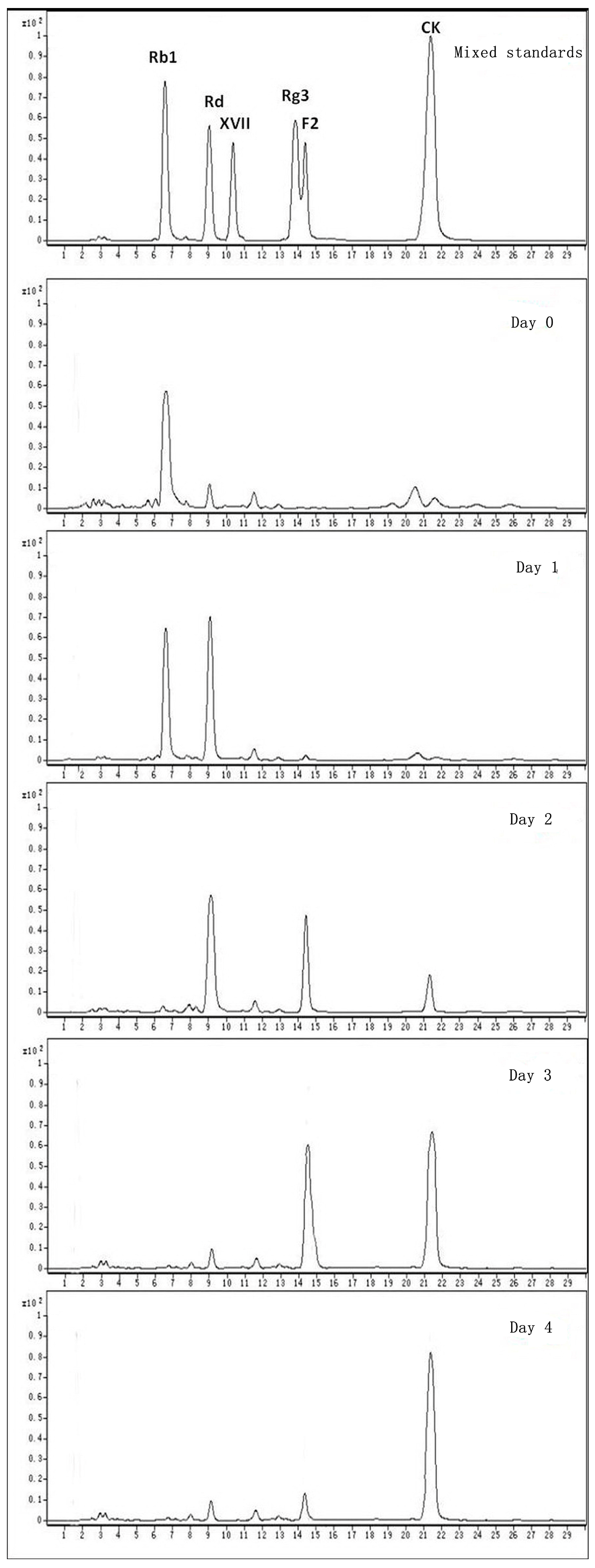
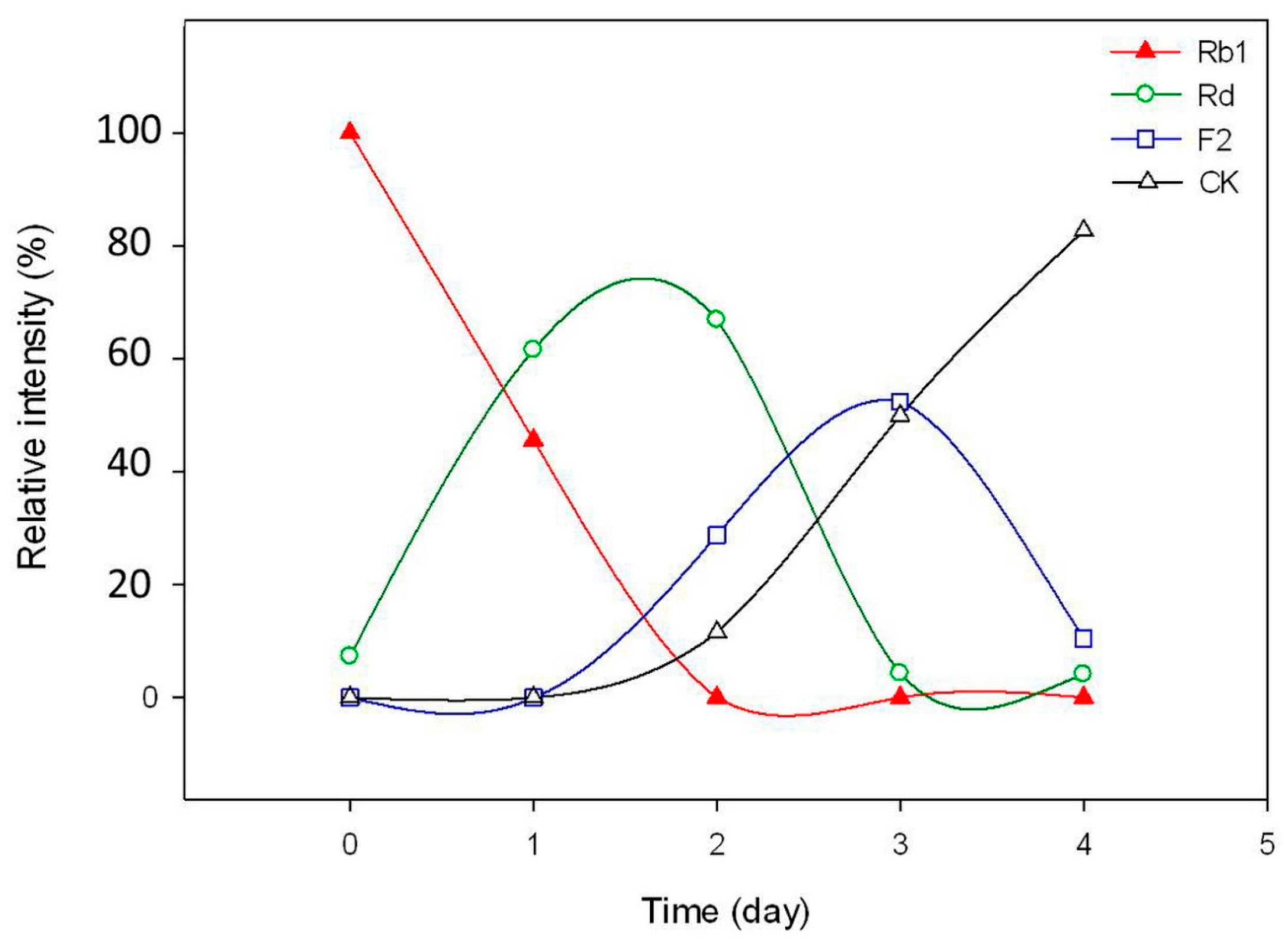
2.3. Calculation on Molar Conversion Rate of Rb1 and Purity of CK
| No. | Day 0 | Day 4 | ||
|---|---|---|---|---|
| Compound | MRM Intensity (×107) | Compound | MRM Intensity (×107) | |
| 1 | Rb1 | 2.3991 | Rb1 | - |
| 2 | Rg1 | 1.1094 | Rg1 | 1.1137 |
| 3 | CK | - | CK | 1.9749 |
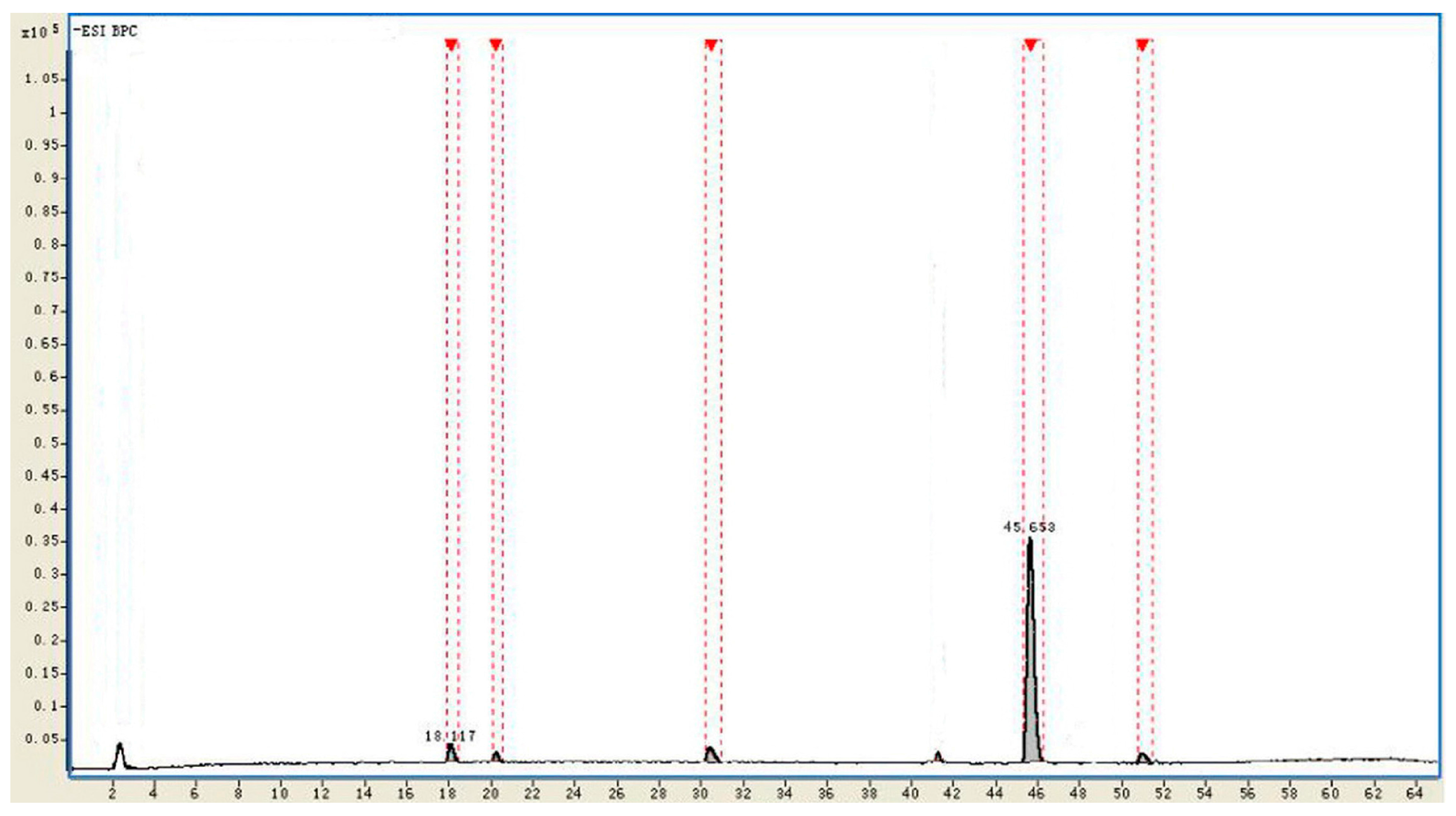
| No. | tR (min) | Identificaiton | Intensity | Relative Intensity (%) | Diff. (ppm) |
|---|---|---|---|---|---|
| 1 | 18.117 | Rd | 215,429 | 3.1 | −1.02 |
| 2 | 20.185 | Unkown | 78,191 | 1.2 | - |
| 3 | 30.809 | F2 | 199,501 | 2.8 | −0.93 |
| 4 | 45.653 | CK | 6,331,023 | 91.4 | −0.61 |
| 5 | 51.291 | Unkown | 105,419 | 1.5 | - |
2.4. Identification of the Biotransformation Metabolites
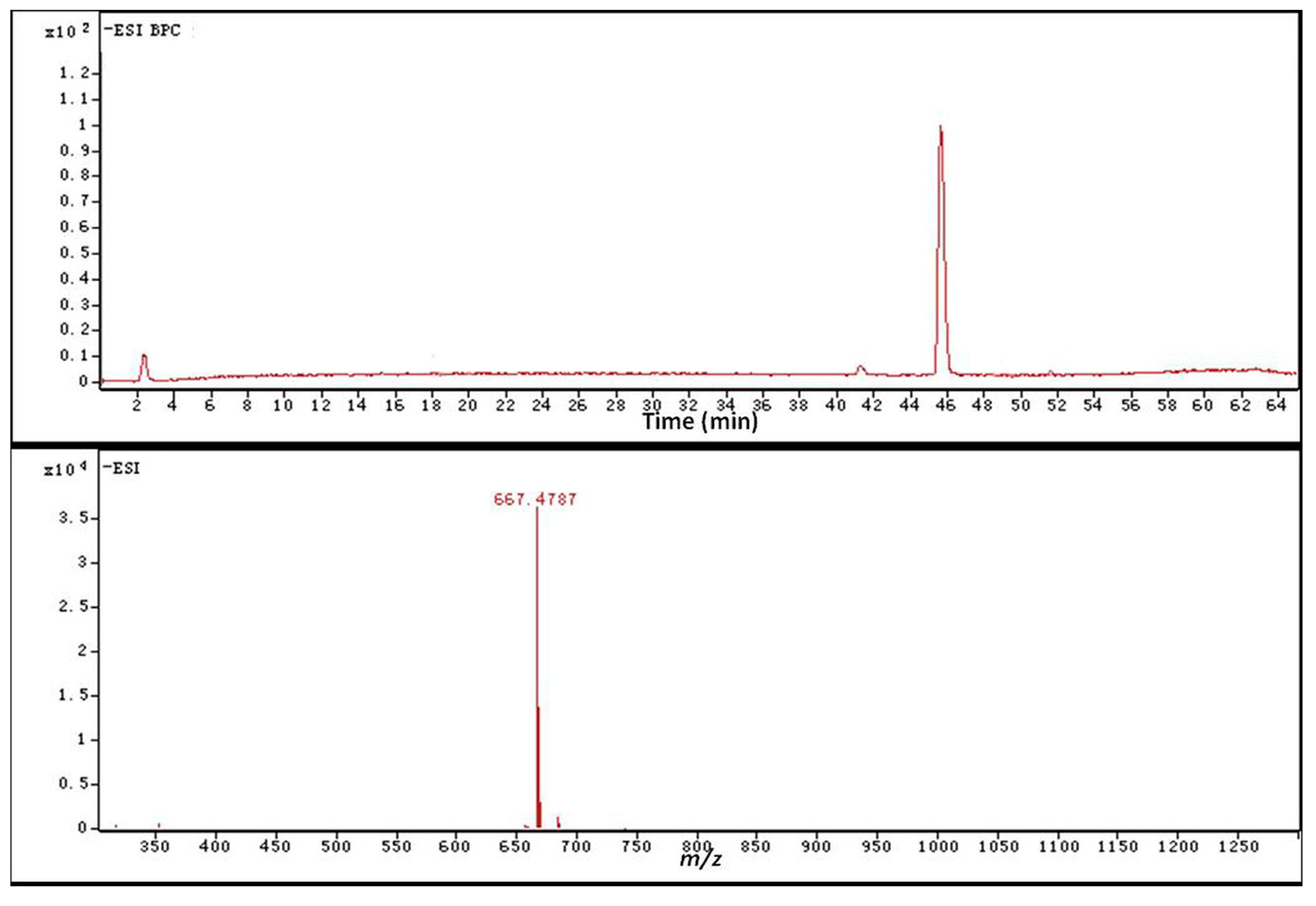
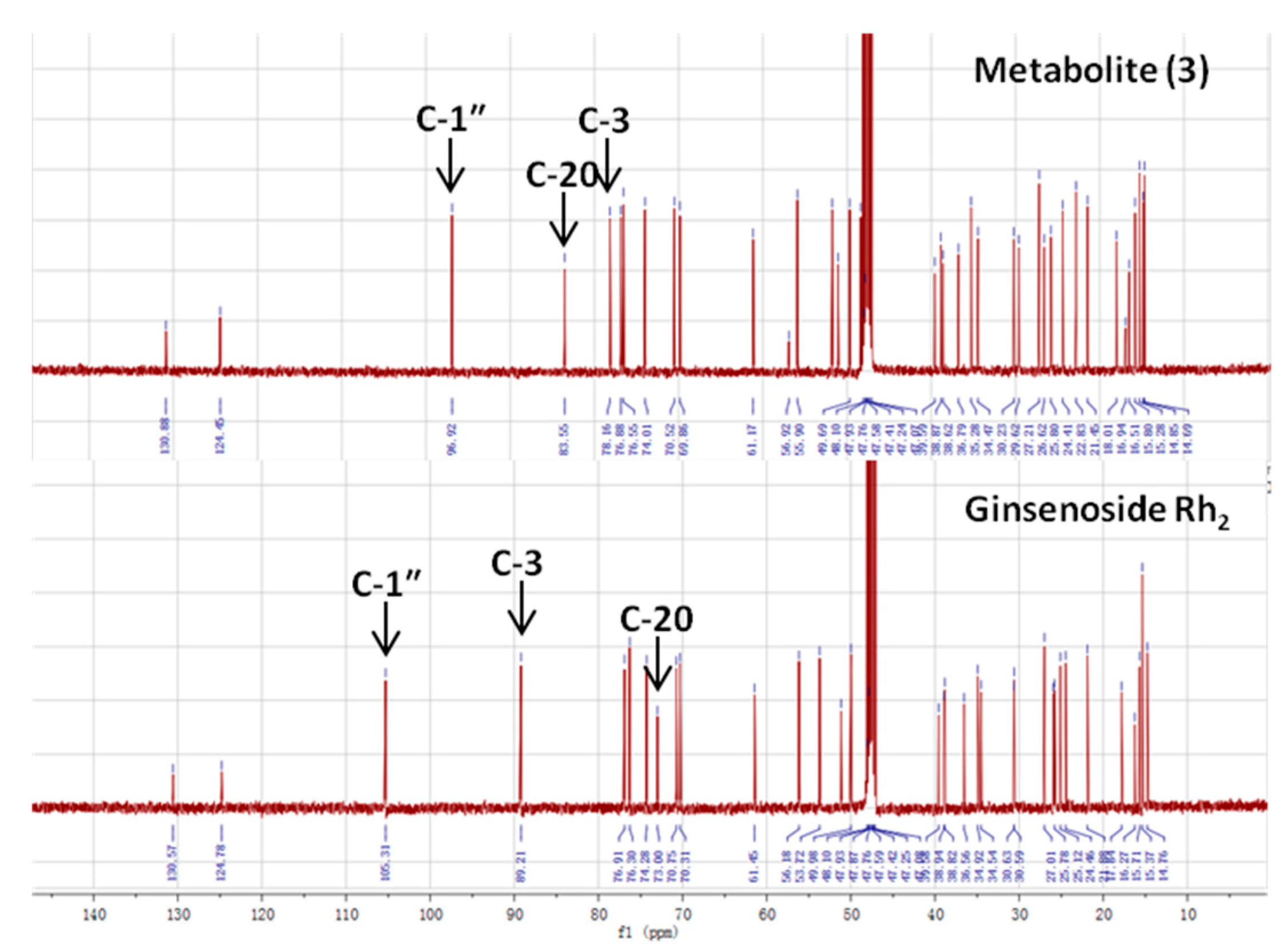
| Carbon Site | 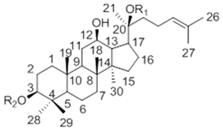 | ||
|---|---|---|---|
| CK R1 = Glc, R2 = H | Metabolite (3) | Rh2 R1 = H, R2 = Glc | |
| 1 | 39.19 | 38.62 | 39.12 |
| 2 | 28.05 | 27.21 | 27.05 |
| 3 | 77.83 | 78.16 | 88.78 |
| 4 | 39.34 | 38.87 | 39.66 |
| 5 | 56.14 | 56.92 | 56.35 |
| 6 | 18.55 | 18.01 | 18.43 |
| 7 | 34.95 | 34.47 | 35.15 |
| 8 | 39.86 | 39.59 | 40.00 |
| 9 | 50.09 | 51.01 | 50.38 |
| 10 | 37.14 | 36.79 | 36.94 |
| 11 | 30.56 | 29.62 | 32.02 |
| 12 | 69.96 | 69.86 | 70.96 |
| 13 | 49.30 | 49.69 | 49.54 |
| 14 | 51.21 | 51.66 | 51.69 |
| 15 | 30.74 | 30.23 | 31.32 |
| 16 | 26.42 | 26.62 | 26.70 |
| 17 | 51.39 | 51.77 | 54.77 |
| 18 | 15.81 | 15.28 | 16.77 |
| 19 | 16.14 | 15.80 | 15.61 |
| 20 | 83.08 | 83.55 | 72.94 |
| 21 | 22.13 | 22.83 | 26.83 |
| 22 | 35.96 | 35.28 | 35.88 |
| 23 | 22.98 | 22.83 | 22.97 |
| 24 | 125.75 | 124.45 | 126.30 |
| 25 | 130.69 | 130.88 | 130.73 |
| 26 | 25.54 | 25.80 | 25.78 |
| 27 | 17.54 | 16.94 | 17.66 |
| 28 | 28.47 | 29.62 | 28.14 |
| 29 | 16.14 | 16.51 | 16.34 |
| 30 | 17.18 | 16.94 | 17.65 |
| Glucosyl C-1″ | 98.10 | 96.92 | 106.92 |
3. Experimental Section
3.1. Materials and Chemicals
3.2. Microorganism and Culture Condition
3.3. Growth Curve of C. sinensis
3.4. Biotransformation of Ginsenoside Rb1 by C. sinensis
3.5. Sample Preparation
3.6. Thin Layer Chromatography Analysis
3.7. HPLC-Q-TOF-MS Analysis
3.8. Determination of the Metabolites
3.9. Parameters Optimization for the Biotransformation of Ginsenoside Rb1
3.9.1. Substrate-Feeding Time
3.9.2. Carbon Sources
3.9.3. Nitrogen Sources
3.9.4. Elicitors
3.9.5. Reaction pH
3.9.6. Biotransformation Temperature
3.10. Time-Course Experiments
- (1)
- Rb1: m/z 1107→945 [M − H]−
- (2)
- Rg1: m/z 799→637 [M − H]−
- (3)
- Rd: m/z 945→783 [M − H]−
- (4)
- F2: m/z 783→621 [M − H]−
- (5)
- CK: m/z 621→459 [M − H]−
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chae, S.; Kang, K.A.; Chang, W.Y.; Kim, M.J.; Lee, S.J.; Lee, Y.S.; Kim, H.S.; Kim, D.H.; Hyun, J.W. Effect of compound K, a metabolite of ginseng saponin, combined with gamma-ray radiation in human lung cancer cells in vitro and in vivo. J. Agric. Food Chem. 2009, 57, 5777–5782. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.S.; Che, C.M.; Leung, K.W. Recent advances in ginseng as cancer therapeutics: A functional and mechanistic overview. Nat. Prod. Rep. 2015, 32, 256–272. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.X.; Yu, N.J.; Yang, X.H. The study of ginsenoside on PPAR gamma expression of mononuclear macrophage in type 2 diabetes. Mol. Biol. Rep. 2010, 37, 2975–2979. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.F.; Chiou, W.F.; Zhang, J.T. Comparison of the pharmacological effects of Panax ginseng and Panax quinquefolium. Acta Pharmacol. Sin. 2008, 29, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.U.; Bae, E.A.; Han, M.J.; Kim, N.J.; Kim, D.H. Hepato protective effect of ginsenoside Rb 1 and compound K on tertbutyl hydroperoxide-induced liver injury. Liver Int. 2005, 25, 1069–1073. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Q.; Luo, X.Y.; Liu, G.Z.; Chen, Y.P.; Wang, Z.C.; Sun, Y.X. In vitro study of the relationship between the structure of ginsenoside and its antioxidative or prooxidative activity in free radical induced hemolysis of human erythrocytes. J. Agric. Food Chem. 2003, 51, 2555–2558. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, M.; Yoo, Y.C.; Matsuzawa, K.; Sato, K.; Saiki, I.; Tono-oka, S.; Samukawa, K.; Azuma, I. Inhibitory effect of tumor metastasis in mice by saponins, ginsenoside Rb2, 20(R)- and 20(S)-ginsenoside Rg3, of red ginseng. Biol. Pharm. Bull. 1995, 18, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zang, L.; Hu, S. Amplified immune response by ginsenoside-based nanoparticles (ginsomes). Vaccine 2009, 27, 2306–2311. [Google Scholar] [CrossRef] [PubMed]
- Stavro, P.M.; Woo, M.; Heim, T.F.; Leiter, L.A.; Vuksan, V. North American ginseng exerts a neutral effect on blood pressure in individuals with hypertension. Hypertension 2005, 46, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Odani, T.; Tanizawa, H.; Takino, Y. Studies on the absorption, distribution, excretion and metabolism of ginseng saponins III. The absorption, distribution and excretion of ginsenoside Rb1 in the rat. Chem. Pharm. Bull. 1983, 31, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.H.; Piao, J.Y.; Min, J.W.; Kim, H.B.; Kim, S.R.; Yang, D.U.; Yang, D.C. Biotransformation of Ginsenoside Rb1 to Prosapogenins, Gypenoside XVII, Ginsenoside Rd, Ginsenoside F2, and Compound K by Leuconostoc mesenteroides DC102. J. Ginseng Res. 2011, 35, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zhou, W.; Shi, X.L. Biotransformation pathways of ginsenoside Rb1 to compound K by β-glucosidases in fungus Paecilomyces bainier sp. 229. Process Biochem. 2010, 45, 1550–1556. [Google Scholar] [CrossRef]
- Hou, J.G.; Xue, J.J.; Sun, M.Q. Highly selective microbial transformation of major ginsenoside Rb1 to gypenoside LXXV by Esteya vermicola CNU120806. J. Appl. Microbiol. 2012, 113, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.H.; Min, J.W.; Jin, Y. Enzymatic Biotransformation of Ginsenoside Rb1 to Compound K by Recombinant β-Glucosidase from Microbacterium esteraromaticum. J. Agric. Food Chem. 2012, 60, 3776–3781. [Google Scholar] [CrossRef] [PubMed]
- Park, C.S.; Yoo, M.H.; Noh, K.W. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Appl. Microbiol. Biotechnol. 2010, 87, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.D.; Yang, Y.Y.; Ouyang, D.S.; Yang, G.P. A review of biotransformation and pharmacology of ginsenoside compound K. Fitoterapia 2015, 100, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Tryfona, T.; Bustard, M.T. Impact of pulsed electric fields on Corynebacterium glutamicum cell membrane permeabilization. J. Biosci. Bioeng. 2008, 105, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Bae, E.A.; Choo, M.K.; Park, E.K.; Park, S.Y.; Shin, H.Y.; Kim, D.H. Metabolism of ginsenoside Rc by human intestinal bacteria and its related antiallergic activity. Biol. Pharm. Bull. 2002, 25, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yan, Q.; Li, J.Y.; Zhang, X.C.; Zhou, P. Biotransformation of Panax notoginseng saponins into ginsenoside compound K production by Paecilomyces bainier sp. 229. J. Appl. Microbiol. 2008, 104, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Ji, G.E. Transformation of ginsenoside Rb1 and Re from Panax ginseng by food microorganisms. Biotechnol. Lett. 2005, 27, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.T.; Yang, M.; Song, Y.; Lu, Z.Q.; Zhang, J.Q.; Huang, H.L.; Wu, L.J.; Guo, D.A. Microbial transformation of ginsenoside Rb1 by Acremonium strictum. Appl. Microbiol. Biotechnol. 2008, 77, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, Y.; Yan, M.; Sun, C.; Zheng, P. Screening of plant pathogenic fungi by ginsenoside compound K production. Zhongguo Zhong Yao Za Zhi 2011, 36, 1596–1598. [Google Scholar] [PubMed]
- Peter, X.C.; Wang, S.N.; Nie, S.P. Properties of C. sinensis: A review. J. Funct. Foods 2013, 5, 550–569. [Google Scholar]
- Li, S.P.; Yang, F.Q.; Tsim, K.W.K. Quality control of C. sinensis, a valued traditional Chinese medicine. J. Pharmaceut. Biomed. 2006, 41, 1571–1584. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Huang, Y.L.; Huang, B.M. C. sinensis mycelium activates PKA and PKC signal pathways to stimulate steroidogenesis in MA-10 mouse Leydig tumor cells. Int. J. Biochem. Cell Biol. 2005, 37, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Shiao, M.S.; Wang, Z.N.; Lin, L.J. Profiles of nucleosides and nitrogen bases in Chinese medicinal fungus C. sinensis and related species. Bot. Bull. Acad. Sin. 1994, 35, 261–267. [Google Scholar]
- Zhu, J.S.; Halpern, G.M.; Jones, K. The Scientific Rediscovery of an Ancient Chinese Herbal Medicine: C. sinensis Part I. J. Altern. Complement. Med. 1998, 4, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.S.; Halpern, G.M.; Jones, K. The Scientific Rediscovery of a Precious Ancient Chinese Herbal Regimen: C. sinensis Part II. J. Altern. Complement. Med. 1998, 4, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Benoit, I.; Zhou, M.; Vivas, D.A.; Downes, D.J.; Todd, R.B.; Kloezen, W.; Post, H.; Heck, A.J.; Maarten Altelaar, A.F.; de Vries, R.P. Spatial differentiation of gene expression in Aspergillus niger colony grown for sugar beet pulp utilization. Sci. Rep. 2015, 5, 13592. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, S.; Khetkorn, W.; Incharoensakdi, A. GABA accumulation in response to different nitrogenous compounds in unicellular cyanobacterium Synechocystis sp. PCC 6803. Curr. Microbiol. 2015, 70, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Samir, K.R.; Syamal, K.R.; Susanta, K.D. Induction and Catabolite Repression of β-Glucosidase Synthesis in Myceliophthora thermophila D-14 (= ATCC 48104). Appl. Environ. Microb. 1998, 8, 2152–2153. [Google Scholar]
- Gunter, E.A.; Kapustina, O.M.; Popeyko, O.V. Induction of beta-1,3-glucanase in callus cultures in vitro. Biochemistry 2008, 73, 826–832. [Google Scholar] [PubMed]
- Christian, P.K. Involvement of a Conidial Endoglucanase and a Plasma-membrane-bound β-Glucosidase in the Induction of Endoglucanase Synthesis by cellulose in Trichoderma reesei. J. Gen. Microb. 1987, 133, 1481–1487. [Google Scholar]
- Park, S.Y.; Bae, E.A.; Sung, J.H.; Lee, S.K.; Kim, D.H. Purification and characterization of ginsenoside Rb1-metabolizing beta-glucosidase from Fusobacterium K-60, a human intestinal anaerobic bacterium. Biosci. Biotechnol. Biochem. 2001, 65, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zhou, X.W.; Zhou, W.; Li, X.W.; Feng, M.Q.; Zhou, P. Purification and properties of a novel beta-glucosidase, hydrolyzing ginsenoside Rb1 to compound K, from Paecilomyces bainier. J. Microbiol. Biotechnol. 2008, 18, 1081–1089. [Google Scholar] [PubMed]
- Zhao, X.; Gao, L.; Wang, J.; Bi, H.; Gao, J.; Du, X.; Zhou, Y.; Tai, G. A novel ginsenoside Rb1-hydrolyzing beta-d-glucosidase from Cladosprium fulvum. Process Biochem. 2009, 44, 612–618. [Google Scholar] [CrossRef]
- Yu, H.; Liu, H.; Zhang, C.; Tan, D.; Lu, M.; Jin, F. Purification and characterization of gypenoside-alpha-l-rhamnosidase hydrolyzing gypenoside-5 into ginsenoside Rd. Process Biochem. 2004, 39, 861–867. [Google Scholar] [CrossRef]
- Su, J.H.; Xu, J.H.; Lu, W.Y.; Lin, G.Q. Enzymatic transformation of ginsenoside Rg3 to Rh2 using newly isolated Fusarium proliferatum ECU 2042. J. Mol. Catal. B Enzym. 2006, 38, 113–118. [Google Scholar] [CrossRef]
- Kim, B.H.; Lee, S.Y.; Cho, H.J.; You, S.N.; Kim, Y.J.; Park, Y.M.; Lee, J.K.; Baik, M.Y.; Park, C.S.; Ahn, S.C. Biotransformation of Korean Panax ginseng by pectinex. Biol. Pharm. Bull. 2006, 29, 2472–2478. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, K.H.; Jee, H.S.; Lee, N.K.; Park, S.H.; Lee, N.W.; Paik, H.D. Optimization of the enzymatic production of 20(S)-ginsenoside Rg3 from white ginseng extract using response surface methodology. New Biotechnol. 2009, 26, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Yoo, M.H.; Yeom, S.J.; Park, C.S.; Lee, K.W.; Oh, D.K. Production of aglycon protopanaxadiol via compound K by a thermostable beta-glycosidase from Pyrococcus furiosus. Appl. Microbiol. Biotechnol. 2011, 89, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.E.; Park, E.K.; Kim, E.J. Cytotoxicity of Compound K (IH-901) and Ginsenoside Rh2, Main Biotransformants of Ginseng Saponins by Bifidobacteria, against some Tumor Cells. J. Ginseng Res. 2003, 27, 129–134. [Google Scholar]
- Atopkina, L.N.; Denisenko, V.A. Synthesis of 20S-protopanaxadiol 20-O-β-d-glucopyranoside, a metabolite of Panax ginseng glycosides, and compounds related to it. Chem. Nat. Compd. 2006, 42, 452–458. [Google Scholar] [CrossRef]
- Steffen, G.; Stefan, B.; Manfred, S.Z.; Sergio, R.; Danieta, M.; Laura, F. Biocatalytic Generation of Molecular Diversity: Modification of Ginsenoside Rb1 by 1,4-Galactosyltransferase and Candida antarctica Lipase. Helv. Chim. Acta 2002, 85, 1943–1959. [Google Scholar]
- Shibata, S.; Tanaka, O.; Soma, K.; Ando, T.; Lida, Y.; Nakamura, H. Studies on saponins and sapogenins of ginseng. The structure of panaxatriol. Tetrahedron Lett. 1965, 42, 207–213. [Google Scholar] [CrossRef]
- Wan, J.Y.; Liu, P.; Wang, H.Y.; Qi, L.W.; Wang, C.Z.; Li, P.; Yuan, C.S. Biotransformation and metabolic profile of American ginseng saponins with human intestinal microflora by liquid chromatography quadrupole time-of-flight mass spectrometry. J. Chromatogr. A 2013, 1286, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.-N.; Yan, B.-X.; Xu, W.-D.; Qiu, Y.; Guo, Y.-L.; Qiu, Z.-D. Highly Selective Bioconversion of Ginsenoside Rb1 to Compound K by the Mycelium of Cordyceps sinensis under Optimized Conditions. Molecules 2015, 20, 19291-19309. https://doi.org/10.3390/molecules201019291
Wang W-N, Yan B-X, Xu W-D, Qiu Y, Guo Y-L, Qiu Z-D. Highly Selective Bioconversion of Ginsenoside Rb1 to Compound K by the Mycelium of Cordyceps sinensis under Optimized Conditions. Molecules. 2015; 20(10):19291-19309. https://doi.org/10.3390/molecules201019291
Chicago/Turabian StyleWang, Wei-Nan, Bing-Xiong Yan, Wen-Di Xu, Ye Qiu, Yun-Long Guo, and Zhi-Dong Qiu. 2015. "Highly Selective Bioconversion of Ginsenoside Rb1 to Compound K by the Mycelium of Cordyceps sinensis under Optimized Conditions" Molecules 20, no. 10: 19291-19309. https://doi.org/10.3390/molecules201019291
APA StyleWang, W.-N., Yan, B.-X., Xu, W.-D., Qiu, Y., Guo, Y.-L., & Qiu, Z.-D. (2015). Highly Selective Bioconversion of Ginsenoside Rb1 to Compound K by the Mycelium of Cordyceps sinensis under Optimized Conditions. Molecules, 20(10), 19291-19309. https://doi.org/10.3390/molecules201019291