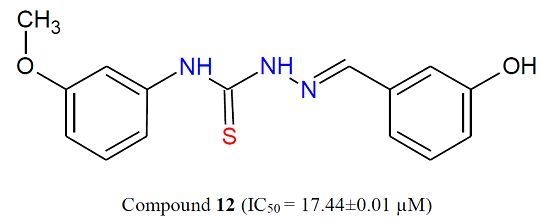
Lead Optimization of 2-Cyclohexyl-N-[(Z)-(3-methoxyphenyl/3-hydroxyphenyl) methylidene]hydrazinecarbothioamides for Targeting the HER-2 Overexpressed Breast Cancer Cell Line SKBr-3
Abstract
:1. Introduction
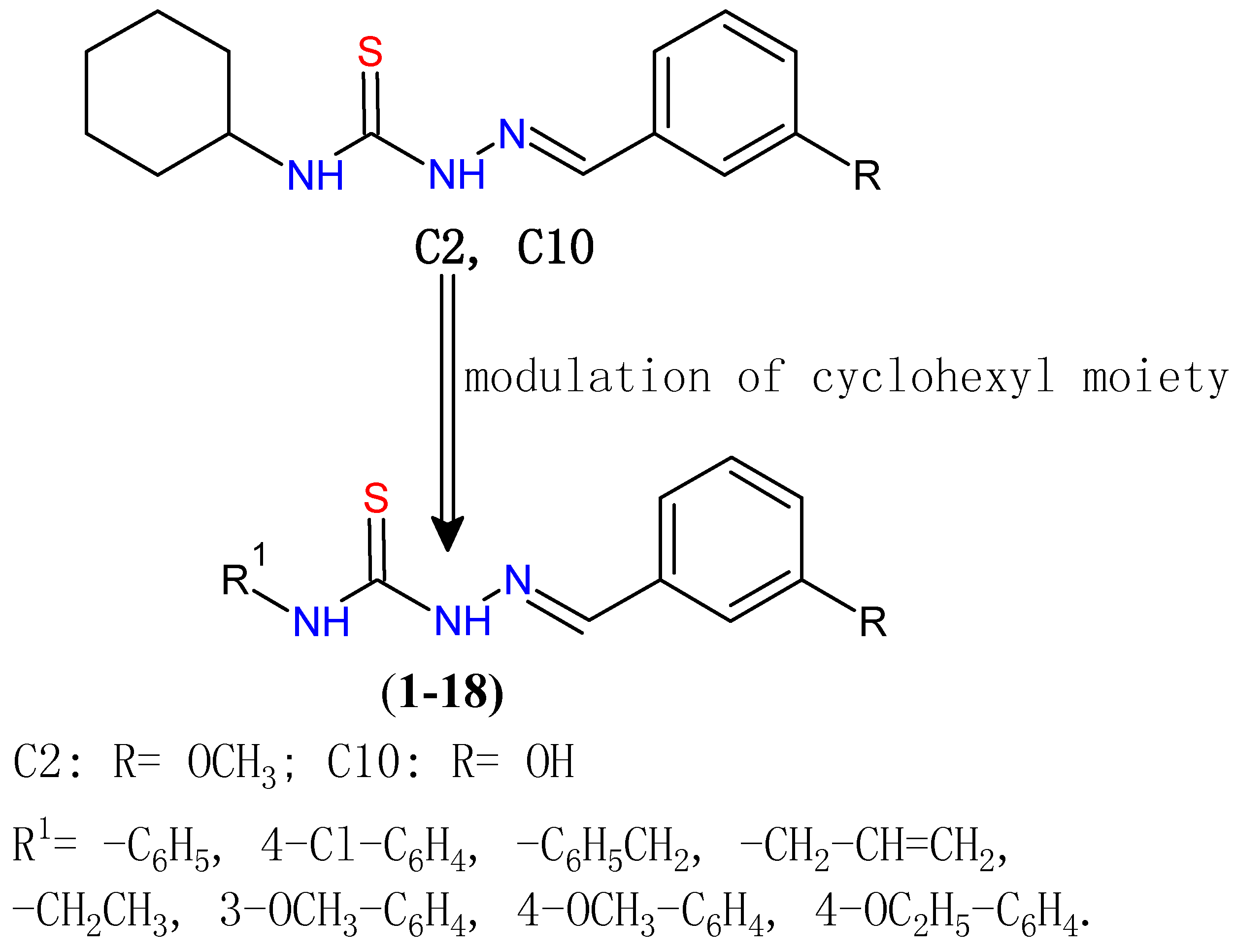
2. Results and Discussion
2.1. Chemistry
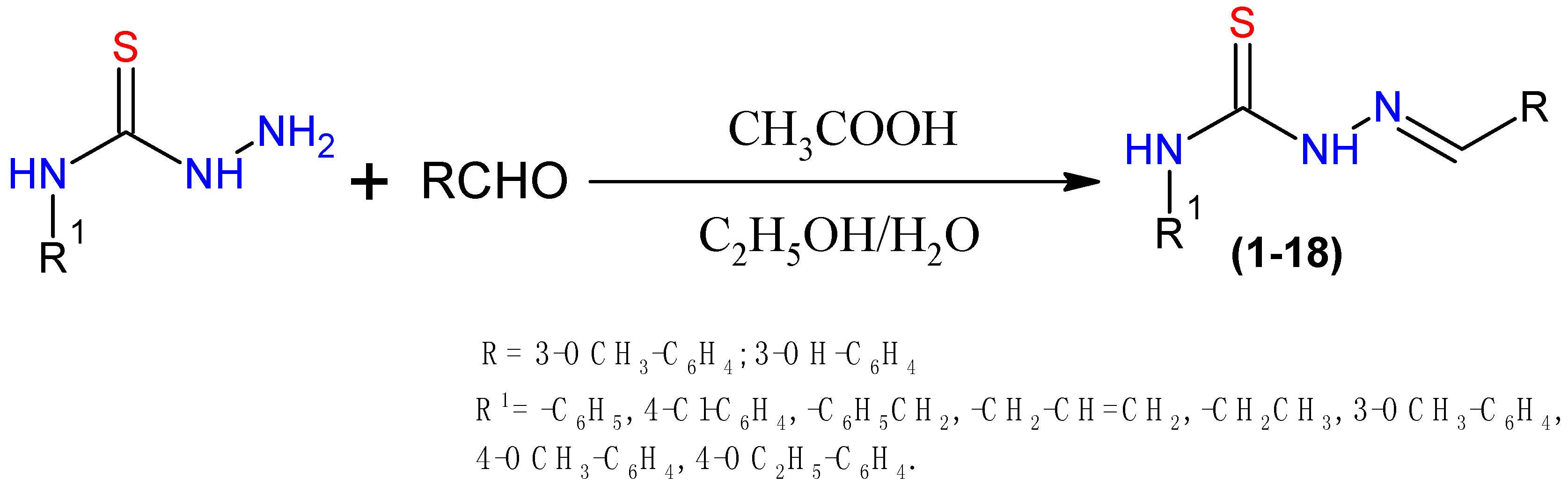
| Compounds | R | R1 | Molecular Formula | Yield % | Mp (°C) | CLog P |
|---|---|---|---|---|---|---|
| 1 | 3-Methoxyphenyl | Phenyl | C15H15N3OS | 70 | 161–162 | 4.42 |
| 2 | 3-Methoxyphenyl | 4-Chlorophenyl | C15H14ClN3OS | 75 | 197–198 | 5.41 |
| 3 | 3-Methoxyphenyl | Prop-2-en | C12H15N3OS | 72 | 115–116 | 2.70 |
| 4 | 3-Methoxyphenyl | Benzyl | C16H17N3OS | 68 | 127–129 | 3.65 |
| 5 | 3-Methoxyphenyl | Ethyl | C11H15N3OS | 72 | 122–123 | 2.40 |
| 6 | 3-Methoxyphenyl | 3-Methoxyphenyl | C16H17N3O2S | 65 | 148–150 | 4.68 |
| 7 | 3-Methoxyphenyl | 4-Methoxyphenyl | C16H17N3O2S | 70 | 156–157 | 4.37 |
| 8 | 3-Methoxyphenyl | 4-Ethoxyphenyl | C17H19N3O2S | 68 | 160–161 | 4.91 |
| 9 | 3-Methoxyphenyl | 3-Chlorophenyl | C15H14ClN3OS | 70 | 138–140 | 5.45 |
| 10 | 3-Hydroxyphenyl | Phenyl | C14H13N3OS | 78 | 212–213 | 3.95 |
| 11 | 3-Hydroxyphenyl | 4-Chlorophenyl | C14H12ClN3OS | 65 | 222–224 | 4.94 |
| 12 | 3-Hydroxyphenyl | 3-Methoxyphenyl | C15H15N3O2S | 70 | 185–186 | 4.21 |
| 13 | 3-Hydroxyphenyl | Prop-2-en | C11H13N3OS | 60 | 94–97 | 2.22 |
| 14 | 3-Hydroxyphenyl | Benzyl | C15H15N3OS | 65 | 130–132 | 3.17 |
| 15 | 3-Hydroxyphenyl | 4-Methoxyphenyl | C15H15N3O2S | 72 | 182–183 | 3.90 |
| 16 | 3-Hydroxyphenyl | 4-Ethoxyphenyl | C16H17N3O2S | 70 | 200–202 | 4.43 |
| 17 | 3-Hydroxyphenyl | 3-Chlorophenyl | C14H12ClN3OS | 75 | 198–200 | 4.98 |
| 18 | 3-Hydroxyphenyl | Ethyl | C10H13N3OS | 60 | 152–154 | 1.93 |
2.2. Anti-Proliferative in Vitro Activity
| Compounds | a IC50 (µM) |
|---|---|
| SKBr-3 | |
| 1 | 45.67 ± 0.18 |
| 2 | 23.69 ± 0.06 |
| 3 | 17.89 ± 0.03 |
| 4 | 34.52 ± 0.18 |
| 5 | 23.00 ± 0.16 |
| 6 | 24.55 ± 0.02 |
| 7 | 25.00 ± 0.03 |
| 8 | 23.76 ± 0.01 |
| 9 | 27.00 ± 0.03 |
| 10 | 22.32 ± 0.02 |
| 11 | 26.49 ± 0.02 |
| 12 | 17.44 ± 0.01 |
| 13 | 29.26 ± 0.05 |
| 14 | 27.74 ± 0.03 |
| 15 | 53.29 ± 0.33 |
| 16 | 24.75 ± 0.08 |
| 17 | 18.28 ± 0.05 |
| 18 | 22.33 ± 0.03 |
| 5-FU | 38.58 ± 0.04 |

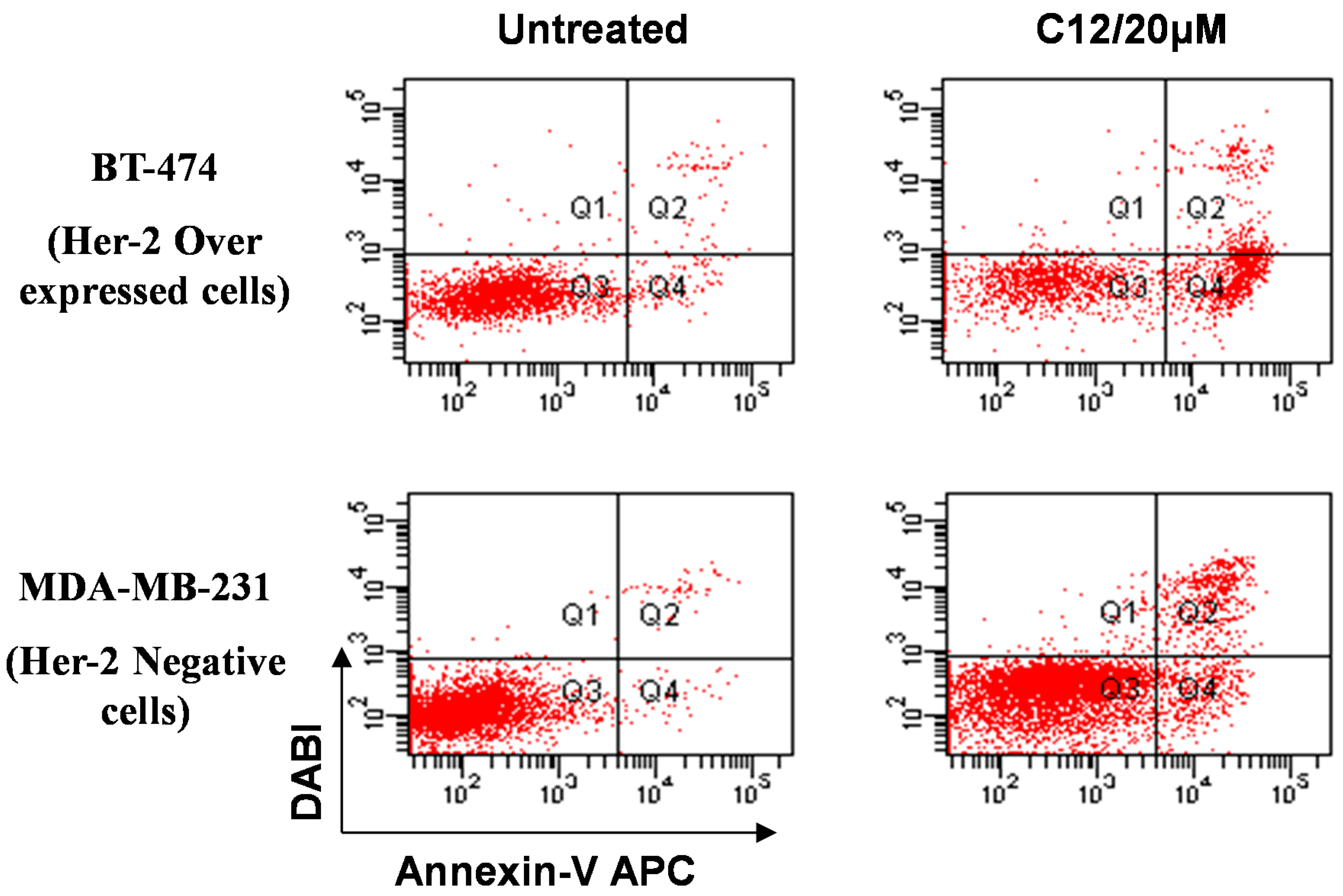
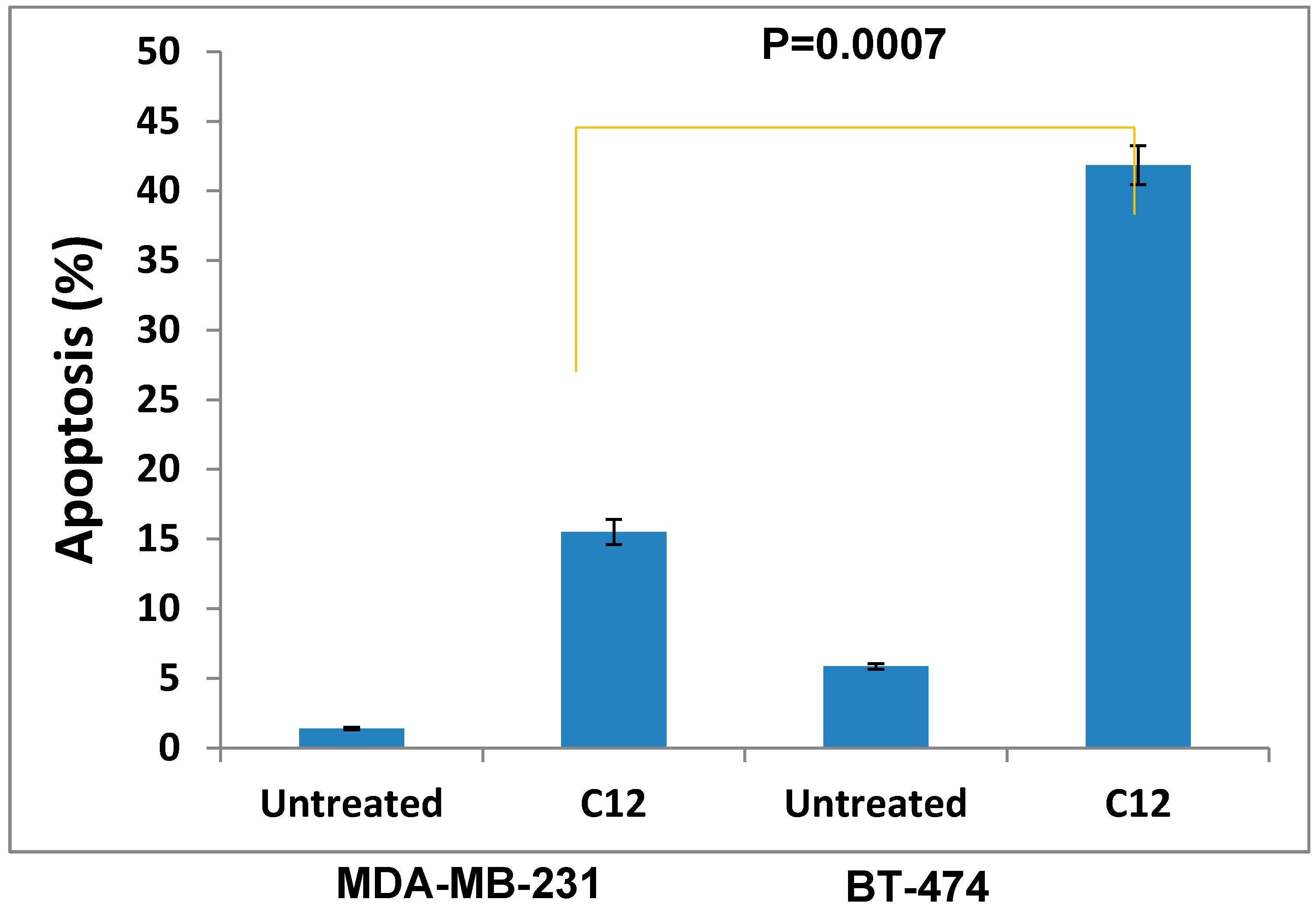
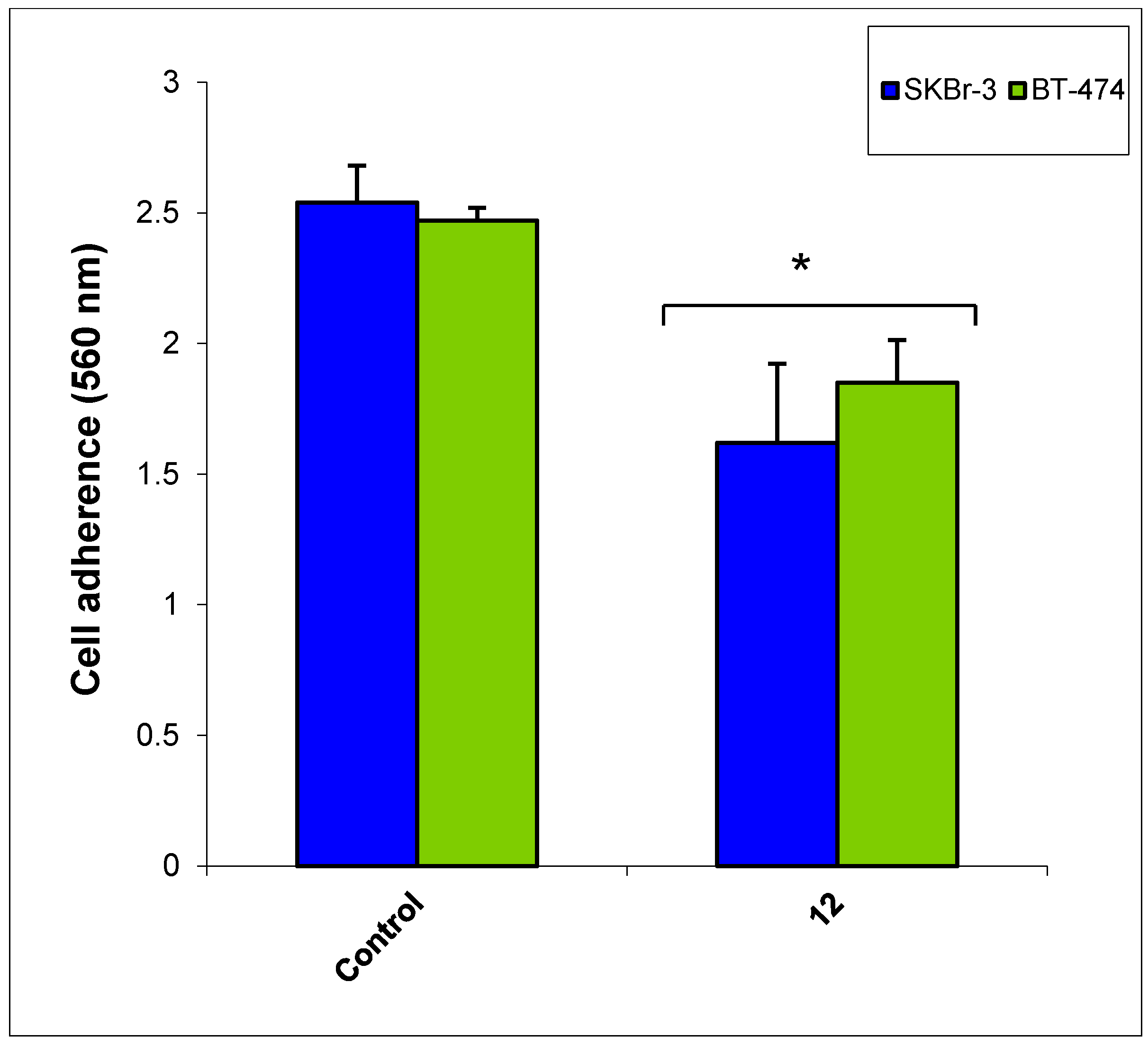
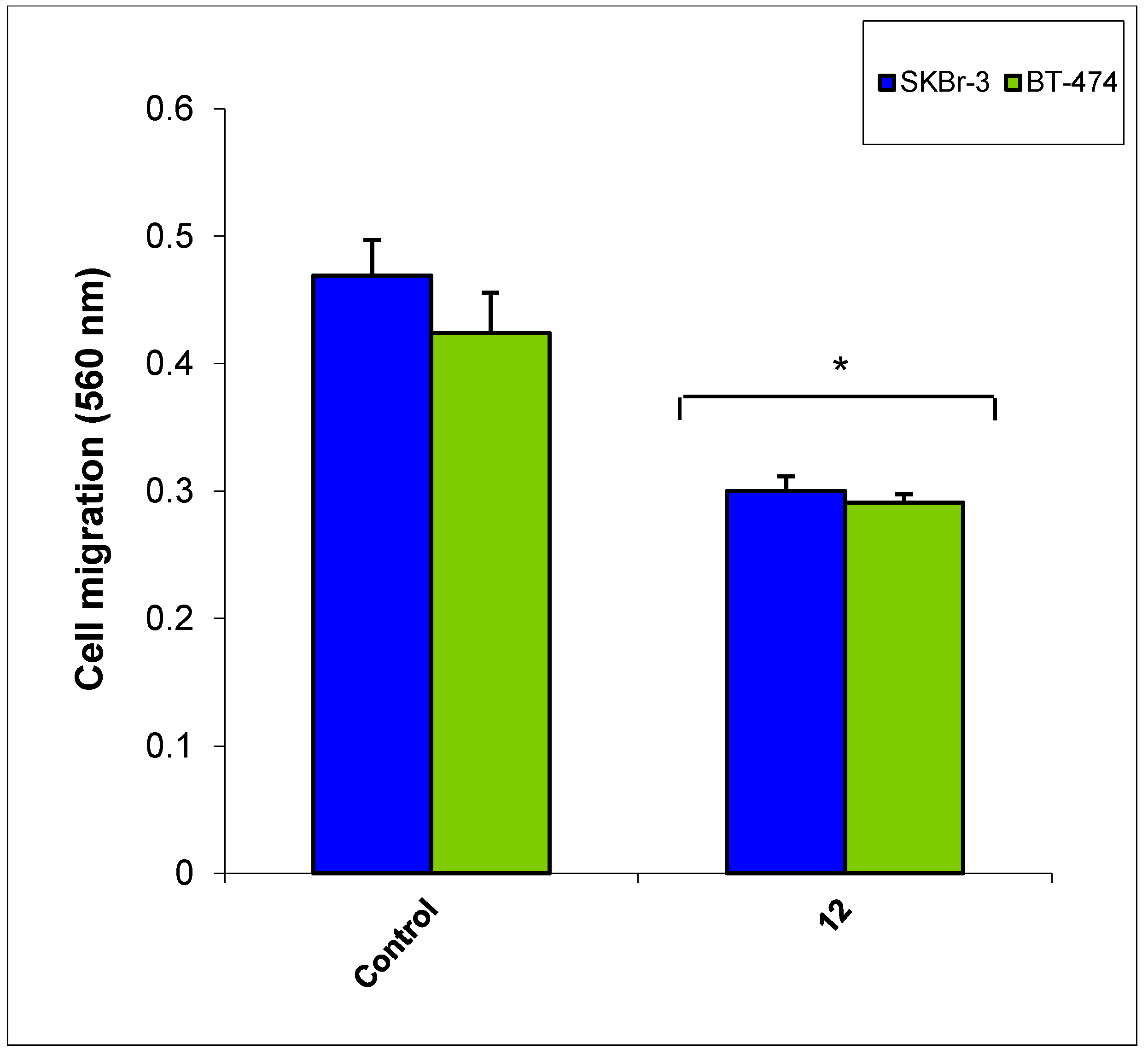
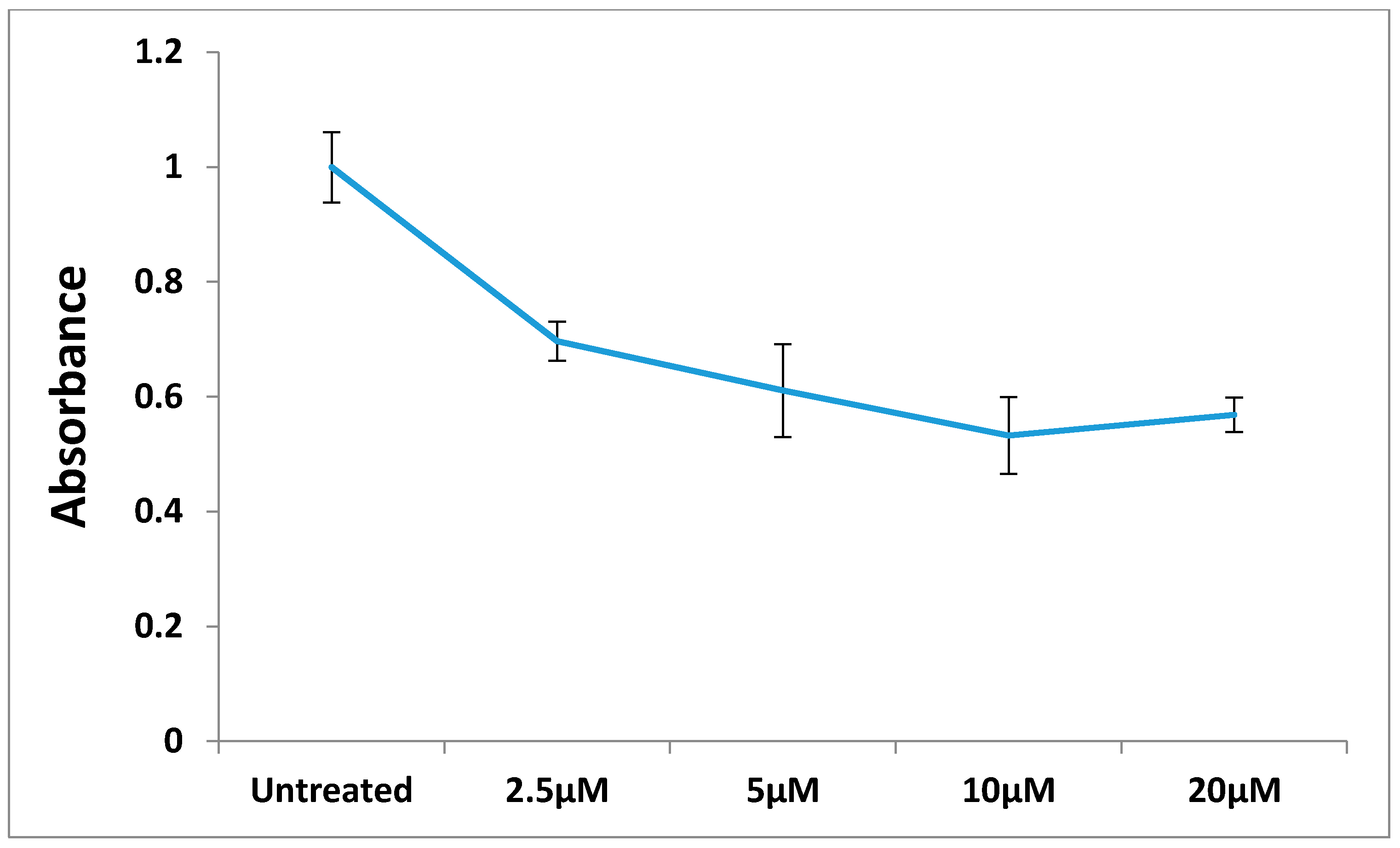
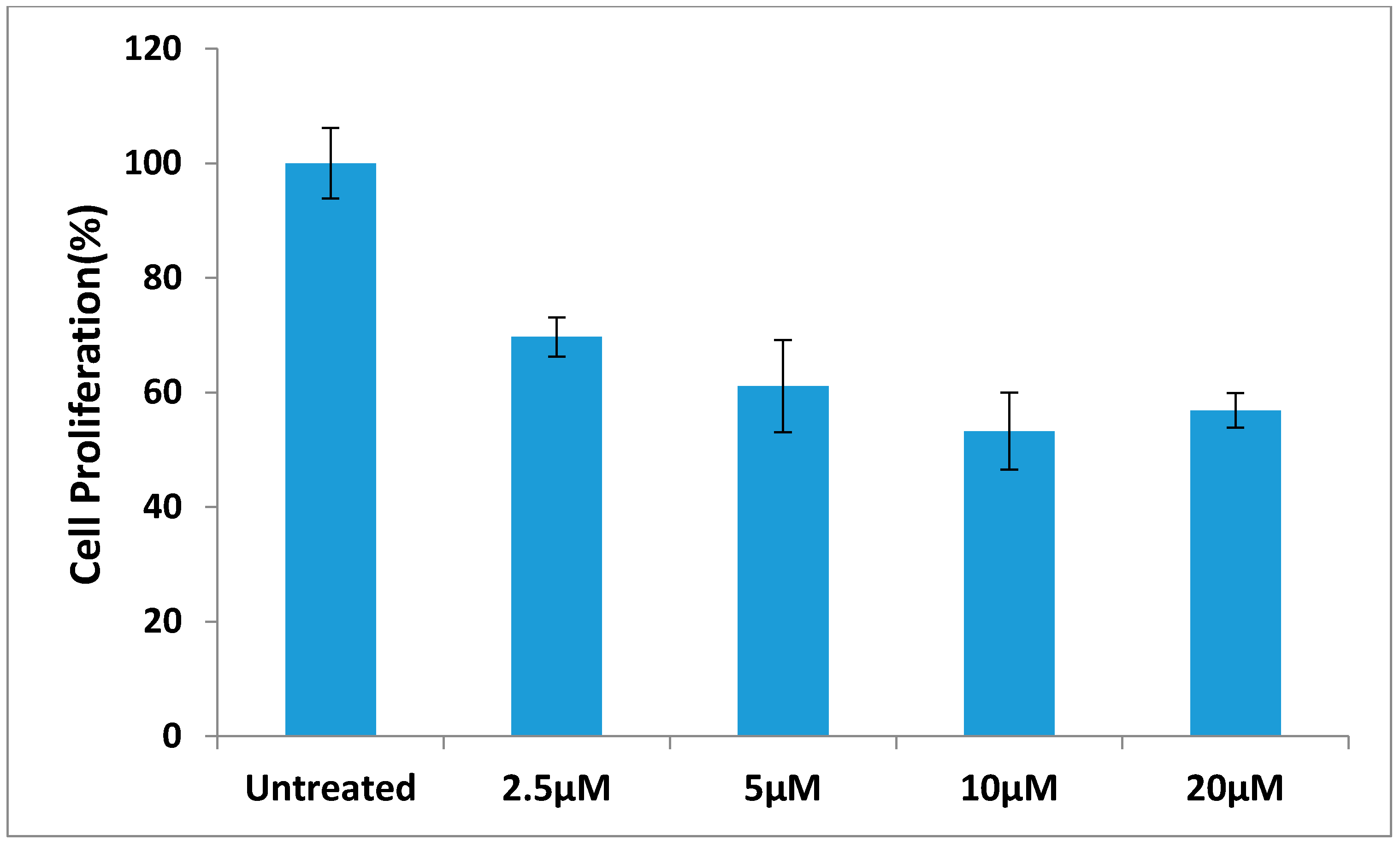
3. Experimental Section
3.1. General Information
3.1.1. Representative Procedure for Synthesis of 1–18
3.2. Cell Lines
3.2.1. WST-1 Cell Proliferation Assay
3.2.2. Measurement of IC50
3.2.3. Flow Cytometric Analysis of Cellular DNA Content
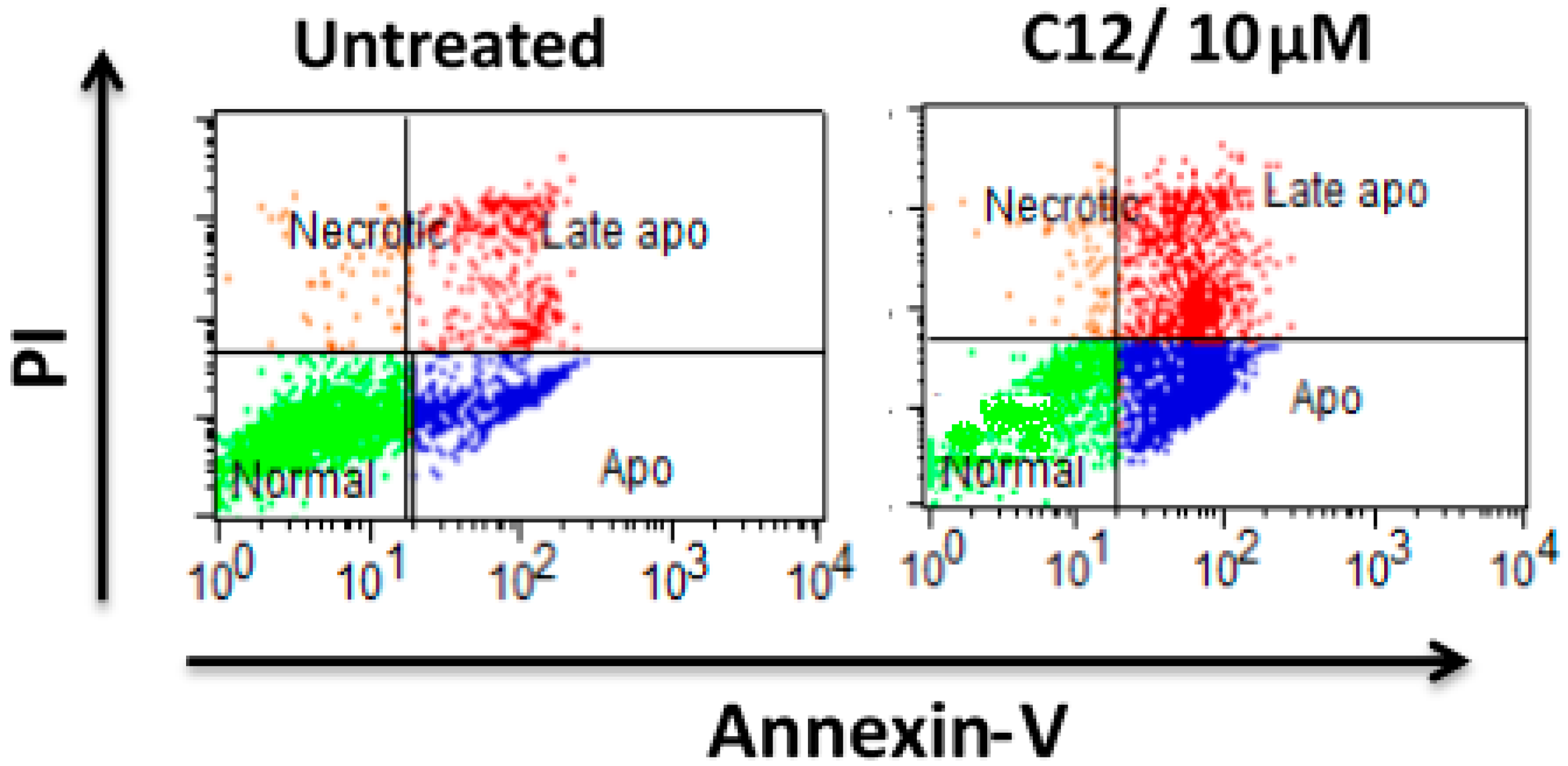
3.2.4. Measurement of Annexin-V Binding by Flow Cytometry
3.2.5. Cancer Cell Migration Assay
3.2.6. Cancer Cell Adhesion Assay
3.2.7. The Effect of Different Concentration of Compound 12 on BT-474 Cells Proliferation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pelosi, G. Thiosemicarbazone metal complexes: from structure to activity. Open. Crystallogr. J. 2010, 3, 16–28. [Google Scholar] [CrossRef]
- Dilović, I.; Rubcić, M.; Vrdoljak, V.; Pavelić, S.K.; Kralj, M.; Piantanida, I.; Cindrić, M. Novel thiosemicarbazone derivatives as potential antitumor agents: Synthesis, physicochemical and structural properties, DNA interactions and antiproliferative activity. Bioorg. Med. Chem. 2008, 16, 5189–5198. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, Z.; Chikhani, S.; Lui, G.Y.; Sivagurunathan, S.; Richardson, D.R. The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. Antioxid. Redox Signal. 2013, 10, 874–887. [Google Scholar] [CrossRef] [PubMed]
- Heiner, G.G.; Fatima, N.; Russell, P.K.; Haase, A.T.; Ahmad, N.; Mohammed, N.; Thomas, D.B.; Mack, T.M.; Khan, M.M.; Knatterud, G.L.; et al. Field trials of methisazone as a prophylactic agent against smallpox. Am. J. Epidemiol. 1971, 94, 435–449. [Google Scholar] [PubMed]
- Jutten, P.; Schumann, W.; Hartl, A.; Dahse, H.M.; Grafe, U. Thiosemicarbazones of formyl benzoic acids as novel potent inhibitors of estrone sulfatase. J. Med. Chem. 2007, 50, 3661–3666. [Google Scholar] [CrossRef] [PubMed]
- Yogeeswari, P.; Sriram, D.; Thirumurugan, R.; Raghavendran, J.V.; Sudhan, K.; Pavana, R.K.; Stables, J. Discovery of N-(2,6-dimethylphenyl)-substituted semicarbazones as anticonvulsants: Hybrid pharmacophore-based design. J. Med. Chem. 2005, 48, 6202–6211. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, D.C.; Mackey, Z.; Hansell, E.; Doyle, P.; Gut, J.; Caffrey, C.R.; Lehrman, J.; Rosenthal, P.J.; McKerrow, J.H.; Chibale, K. Synthesis and structure-activity relationships of parasiticidal thiosemicarbazone cysteine protease inhibitors against Plasmodium falciparum, Trypanosoma brucei, and Trypanosoma cruzi. J. Med. Chem. 2004, 47, 3212–3219. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Finch, R.A.; Liu, M.; Grill, S.P.; Rose, W.C.; Loomis, R.; Vasquez, K.M.; Cheng, Y.; Sartorelli, A.C. Triapine (3-aminopyridine-2-carboxaldehyde- thiosemicarbazone): A potent inhibitor of ribonucleotide reductase activity with broad spectrum antitumor activity. Biochem. Pharmacol. 2000, 59, 983–991. [Google Scholar] [CrossRef]
- Winquist, R.J.; Furey, B.F.; Boucher, D.M. Cancer stem cells as the relevant biomass for drug discovery. Curr. Opin. Pharmacol. 2010, 10, 385–390. [Google Scholar] [CrossRef] [PubMed]
- McDermott, S.P.; Wicha, M.S. Targeting breast cancer stem cells. Mol. Oncol. 2010, 4, 404–419. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Ho, M.M.; Ng, A.V.; Lam, S.; Hung, J.Y. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007, 67, 4827–4833. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; de Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Bomken, S.; Fiser, K.; Heidenreich, O.; Vormoor, J. Understanding the cancer stem cell. Br. J. Cancer 2010, 103, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Kim, S.J.; Tanei, T.; Shimazu, K.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Terada, N.; Noguchi, S. Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci. 2009, 100, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Tarpin, C.; Diebel, M.; Esterni, B.; Houvenaeghel, G.; Extra, J.M.; Bertucci, F.; Jacquemier, J.; et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin. Cancer Res. 2010, 16, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Kim, S.J.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef] [PubMed]
- Magnifico, A.; Albano, L.; Campaner, S.; Delia, D.; Castiglioni, F.; Gasparini, P.; Sozzi, G.; Fontanella, E.; Menard, S.; Tagliabue, E. Tumor-initiating cells of HER2-positive carcinoma cell lines express the highest oncoprotein levels and are sensitive to trastuzumab. Clin. Cancer Res. 2009, 15, 2010–2021. [Google Scholar] [CrossRef] [PubMed]
- Knuefermann, C.; Lu, Y.; Liu, B.; Jin, W.; Liang, K.; Wu, L.; Schmidt, M.; Mills, G.B.; Mendelsohn, J.; Fan, Z. HER2/PI-3K/Akt activation leads to a multidrug resistance in human breast adenocarcinoma cells. Oncogene 2003, 22, 3205–3212. [Google Scholar] [CrossRef] [PubMed]
- Compton, C.C. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod. Pathol. 2003, 16, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.A.; Al-Dhfyan, A.; Khan, A.A.; Al-Harbi, N.; Manogaran, P.S.; Alanazi, A.M.; Fun, H.K.; Al-Omar, M.A. Targeting HER-2 over expressed breast cancer cells with 2-cyclohexyl-N-[(Z)-(substituted phenyl/furan-2-yl/thiophene-2-yl)methylidene]hydrazinecarbothioamide. Bioorg. Med. Chem. Lett. 2015, 25, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds (1–18) are available from the authors. The compounds are solid, crystalline in nature with ≥ 99% purity.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhat, M.A.; Al-Dhfyan, A.; Naglah, A.M.; Khan, A.A.; Al-Omar, M.A. Lead Optimization of 2-Cyclohexyl-N-[(Z)-(3-methoxyphenyl/3-hydroxyphenyl) methylidene]hydrazinecarbothioamides for Targeting the HER-2 Overexpressed Breast Cancer Cell Line SKBr-3. Molecules 2015, 20, 18246-18263. https://doi.org/10.3390/molecules201018246
Bhat MA, Al-Dhfyan A, Naglah AM, Khan AA, Al-Omar MA. Lead Optimization of 2-Cyclohexyl-N-[(Z)-(3-methoxyphenyl/3-hydroxyphenyl) methylidene]hydrazinecarbothioamides for Targeting the HER-2 Overexpressed Breast Cancer Cell Line SKBr-3. Molecules. 2015; 20(10):18246-18263. https://doi.org/10.3390/molecules201018246
Chicago/Turabian StyleBhat, Mashooq A., Abdullah Al-Dhfyan, Ahmed M. Naglah, Azmat Ali Khan, and Mohamed A. Al-Omar. 2015. "Lead Optimization of 2-Cyclohexyl-N-[(Z)-(3-methoxyphenyl/3-hydroxyphenyl) methylidene]hydrazinecarbothioamides for Targeting the HER-2 Overexpressed Breast Cancer Cell Line SKBr-3" Molecules 20, no. 10: 18246-18263. https://doi.org/10.3390/molecules201018246
APA StyleBhat, M. A., Al-Dhfyan, A., Naglah, A. M., Khan, A. A., & Al-Omar, M. A. (2015). Lead Optimization of 2-Cyclohexyl-N-[(Z)-(3-methoxyphenyl/3-hydroxyphenyl) methylidene]hydrazinecarbothioamides for Targeting the HER-2 Overexpressed Breast Cancer Cell Line SKBr-3. Molecules, 20(10), 18246-18263. https://doi.org/10.3390/molecules201018246