miR-221/222 Promotes S-Phase Entry and Cellular Migration in Control of Basal-Like Breast Cancer
Abstract
:1. Introduction
2. Results and Discussion
2.1. High Expression of miR-221/222 in Highly Invasive BLBC

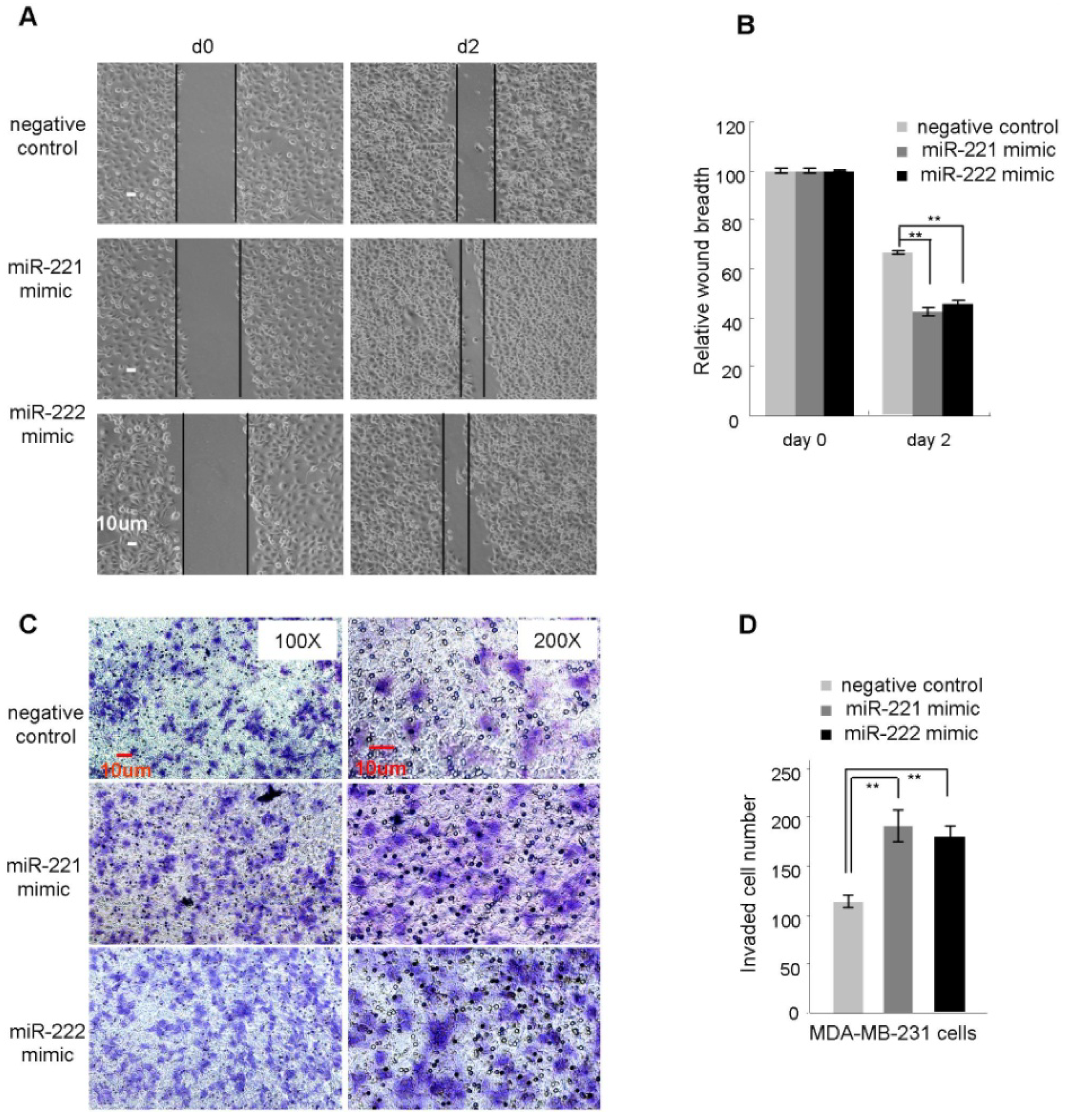
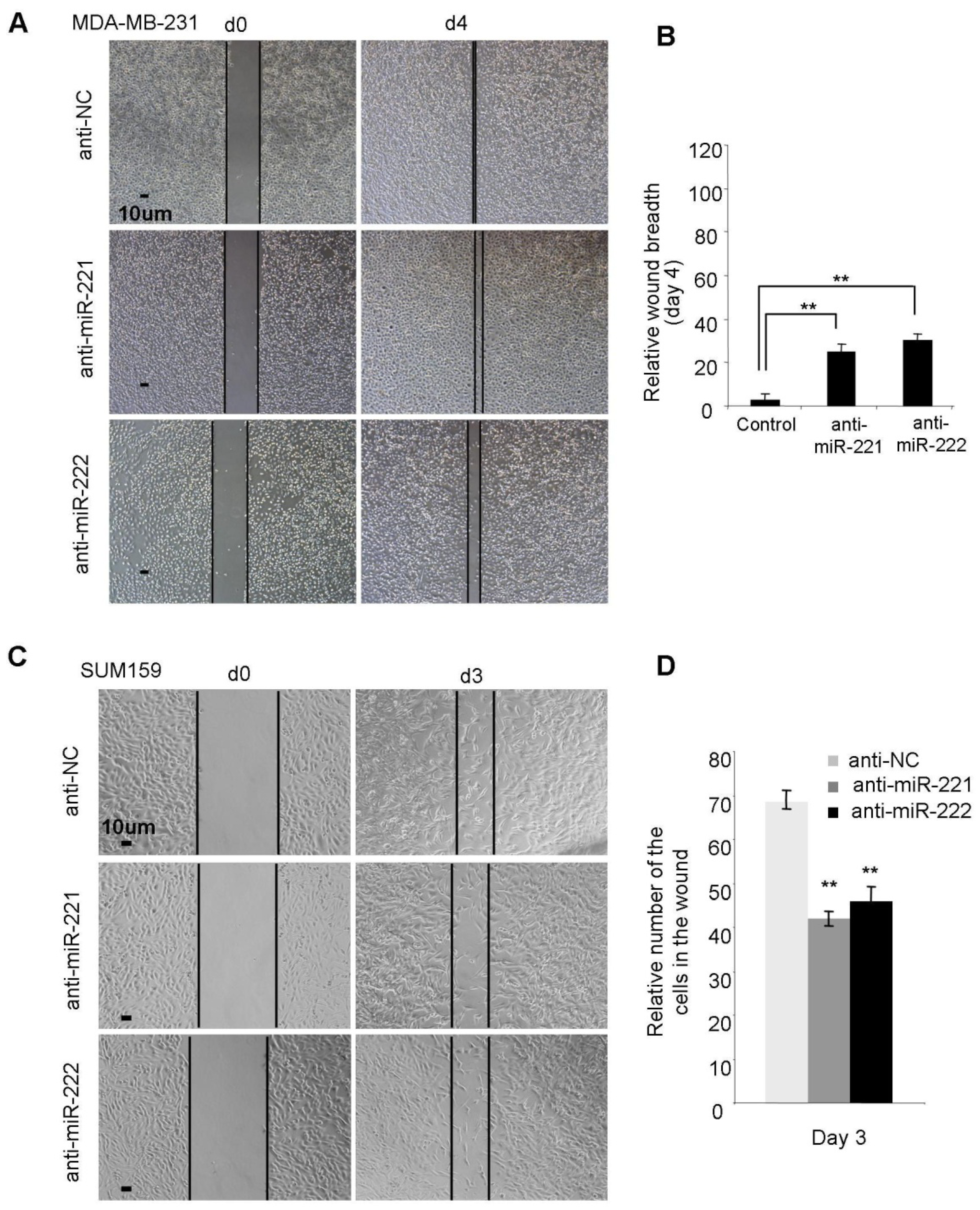
2.2. miR-221/222 Promotes Cellular Migration and Invasion in BLBC
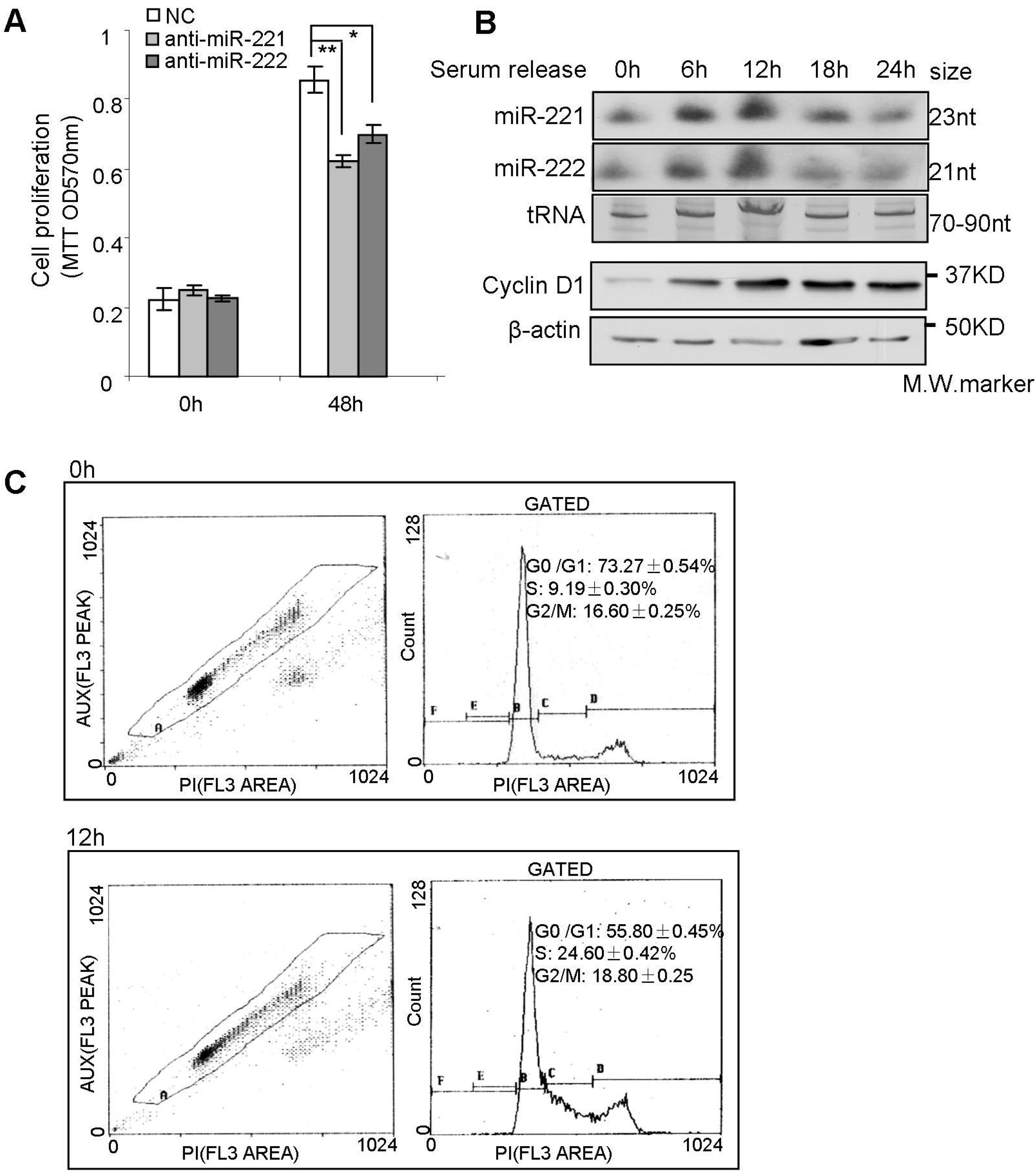
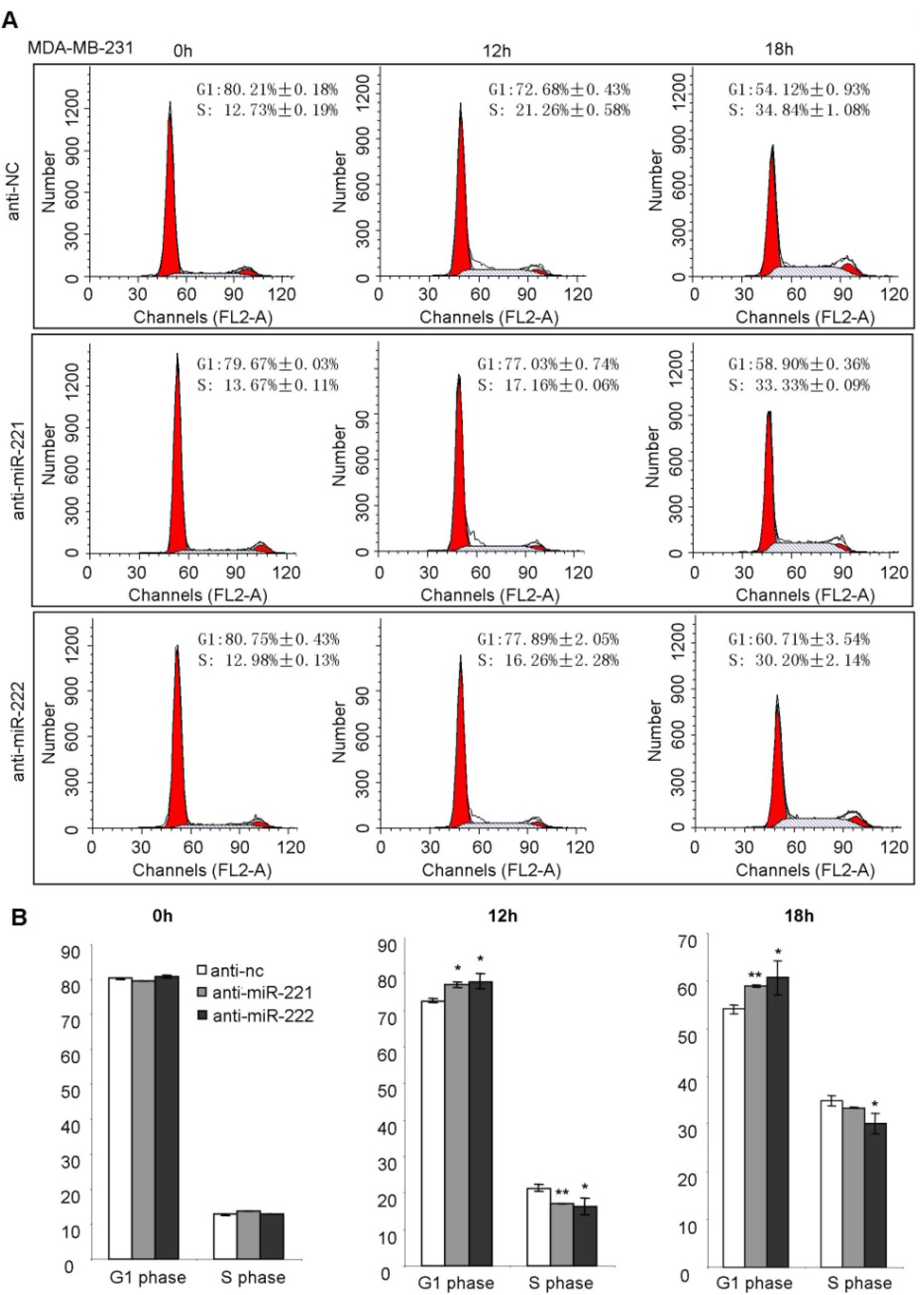
2.3. miR-221/222 Induce G1/S Transition of the Cell Cycle in BLBC
2.4. miR-221/222 Inhibits the Expression of Tumor Suppressor Genes, SOCS1 and CDKN1B

3. Experimental
3.1. Cell Lines and Cell Culture
3.2. Oligos and Transfection
3.3. miRNA Real Time RT-PCR Analysis
3.4. Northern Blot Analysis
3.5. Western Blot Analysis
3.6. Wound Healing Assay
3.7. Cellular Invasion Assay
3.8. Cell Proliferation Assays
3.9. Cell Cycle Analysis
3.10. Plasmid Transfection and Luciferase Reporter Assay
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chang, E.L.; Lo, S. Diagnosis and management of central nervous system metastases from breast cancer. Oncologist 2003, 8, 398–410. [Google Scholar]
- Sanchez-Munoz, A.; Perez-Ruiz, E.; Ribelles, N.; Marquez, A.; Alba, E. Maintenance treatment in metastatic breast cancer. Expert Rev. Anticancer Ther. 2008, 8, 1907–1912. [Google Scholar]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar]
- Yu, Z.; Baserga, R.; Chen, L.; Wang, C.; Lisanti, M.P.; Pestell, R.G. microRNA, cell cycle, and human breast cance. Am. J. Pathol. 2010, 176, 1058–1064. [Google Scholar]
- Cho, W.C. MicroRNAs: Potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int. J. Biochem. Cell. Biol. 2010, 42, 1273–1281. [Google Scholar]
- Cho, W.C. Exploiting the therapeutic potential of microRNAs in human cancer. Expert Opin. Ther. Targets 2012, 16, 345–350. [Google Scholar]
- Yu, Z.; Pestell, R.G. Small non-coding RNAs govern mammary gland tumorigenesis. J Mammary Gland Biol. Neoplasi. 2012, 17, 59–64. [Google Scholar]
- Yu, Z.; Wang, L.; Wang, C.; Ju, X.; Wang, M.; Chen, K.; Loro, E.; Li, Z.; Zhang, Y.; Wu, K.; et al. Cyclin D1 induction of dicer governs microRNA processing and expression in breast cancer. Nat. Commun. 2013, 4, 2812. [Google Scholar]
- Zhang, L.; Huang, J.; Yang, N.; Greshock, J.; Megraw, M.S.; Giannakakis, A.; Liang, S.; Naylor, T.L.; Barchetti, A.; Ward, M.R.; et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 2006, 103, 9136–9141. [Google Scholar]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar]
- Frankel, L.B.; Christoffersen, N.R.; Jacobsen, A.; Lindow, M.; Krogh, A.; Lund, A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008, 283, 1026–1033. [Google Scholar]
- Mertens-Talcott, S.U.; Chintharlapalli, S.; Li, X.; Safe, S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007, 67, 11001–11011. [Google Scholar]
- Yu, Z.; Wang, C.; Wang, M.; Li, Z.; Casimiro, M.C.; Liu, M.; Wu, K.; Whittle, J.; Ju, X.; Hyslop, T.; et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J. Cell Biol. 2008, 182, 509–517. [Google Scholar]
- Shan, S.W.; Lee, D.Y.; Deng, Z.; Shatseva, T.; Jeyapalan, Z.; Du, W.W.; Zhang, Y.; Xuan, J.W.; Yee, S.P.; Siragam, V.; et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Natl. Cell Biol. 2009, 11, 1031–1038. [Google Scholar]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar]
- Tavazoie, S.F.; Alarcón, C.; Oskarsson, T.; Padua, D.; Wang, Q.; Bos, P.D.; Gerald, W.L.; Massagué, J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008, 451, 147–152. [Google Scholar]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 Regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar]
- Richardson, A.L.; Wang, Z.C.; de Nicolo, A.; Lu, X.; Brown, M.; Miron, A.; Liao, X.; Iglehart, J.D.; Livingston, D.M.; Ganesan, S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 2006, 9, 121–132. [Google Scholar]
- Li, Y.; Liu, M,; Zhang, Y.; Han, C.; You, J.; Yang, J.; Cao, C.; Jiao, S. Effects of ARHI on breast cancer cell biological behavior regulated by microRNA-221. Tumour Biol. 2013, 34, 3545–54. [Google Scholar]
- Chen, W.X.; Hu, Q.; Qiu, M.T.; Zhong, S.L.; Xu, J.J.; Tang, J.H.; Zhao, J.H. miR-221/222: Promising biomarkers for breast cancer. Tumour Biol. 2013, 34, 1361–1370. [Google Scholar]
- Zhao, L.; Sun, Y.; Hou, Y.; Peng, Q.; Wang, L.; Luo, H.; Tang, X.; Zeng, Z.; Liu, M. MiRNA expression analysis of cancer-associated fibroblasts and normal fibroblasts in breast cancer. Int. J. Biochem. Cell Biol. 2012, 44, 2051–2059. [Google Scholar]
- Zhao, R.; Wu, J.; Jia, W.; Gong, C.; Yu, F.; Ren, Z.; Chen, K.; He, J.; Su, F. Plasma miR-221 as a predictive biomarker for chemoresistance in breast cancer patients who previously received neoadjuvant chemotherapy. Onkologie 2011, 34, 675–680. [Google Scholar]
- Shah, M.Y.; Calin, G.A. MicroRNAs miR-221 and miR-222: A new level of regulation in aggressive breast cancer. Genome Med. 2011, 3, 56. [Google Scholar]
- Nassirpour, R.; Mehta, P.P.; Baxi, S.M.; Yin, M.J. miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS One 2013, 8, e62170. [Google Scholar]
- Guttilla, I.K.; Phoenix, K.N.; Hong, X.; Tirnauer, J.S.; Claffey, K.P.; White, B.A. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. Breast Cancer Res. Treat. 2012, 132, 75–85. [Google Scholar]
- Stinson, S.; Lackner, M.R.; Adai, A.T.; Yu, N.; Kim, H.J.; O’Brien, C.; Spoerke, J.; Jhunjhunwala, S.; Boyd, Z.; Januario, T.; et al. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci. Signal. 2011, 4, pt5. [Google Scholar]
- Yu, Z.; Willmarth, N.E.; Zhou, J.; Katiyar, S.; Wang, M.; Liu, Y.; McCue, P.A.; Quong, A.A.; Lisanti, M.P.; Pestell, R.G. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc. Natl. Acad. Sci.USA 2010, 107, 8231–8236. [Google Scholar]
- Stinson, S.; Lackner, M.R.; Adai, A.T.; Yu, N.; Kim, H.J.; O’Brien, C.; Spoerke, J.; Jhunjhunwala, S.; Boyd, Z.; Januario, T.; et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci. Signal. 2011, 4, ra41. [Google Scholar]
- Volinia, S.; Galasso, M.; Sana, M.E.; Wise, T.F.; Palatini, J.; Huebner, K.; Croce, C.M. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc. Natl. Acad. Sci. USA 2012, 109, 3024–3029. [Google Scholar]
- Sutherland, K.D.; Lindeman, G.J.; Choong, D.Y.; Wittlin, S.; Brentzell, L.; Phillips, W.; Campbell, I.G.; Visvader, J.E. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene 2004, 23, 7726–7733. [Google Scholar]
- Zou, L.; Ding, Z.; Roy, P. Profilin-1 overexpression inhibits proliferation of MDA-MB-231 breast cancer cells partly through p27kip1 upregulation. J. Cell Physiol. 2010, 223, 623–629. [Google Scholar]
- Slattery, M.L.; Lundgreen, A.; Kadlubar, S.A.; Bondurant, K.L.; Wolff, R.K. JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol. Carcinog. 2013, 52, 155–66. [Google Scholar]
- Xie, T.X.; Wei, D.; Liu, M.; Gao, A.C.; Ali-Osman, F.; Sawaya, R.; Huang, S. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene 2004, 23, 3550–3560. [Google Scholar]
- Chiu, W.T.; Lee, H.T.; Huang, F.J.; Aldape, K.D.; Yao, J.; Steeg, P.S.; Chou, C.Y.; Lu, Z.; Xie, K.; Huang, S. Caveolin-1 upregulation mediates suppression of primary breast tumor growth and brain metastases by stat3 inhibition. Cancer Res. 2011, 71, 4932–4943. [Google Scholar]
- Liu, M.; Casimiro, M.C.; Wang, C.; Shirley, L.A.; Jiao, X.; Katiyar, S.; Ju, X.; Li, Z.; Yu, Z.; Zhou, J.; et al. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc. Natl. Acad. Sci. USA 2009, 106, 19035–19039. [Google Scholar]
- Jackson, R.J.; Adnane, J.; Coppola, D.; Cantor, A.; Sebti, S.M.; Pledger, W.J. Loss of the cell cycle inhibitors p21(Cip1) and p27(Kip1) enhances tumorigenesis in knockout mouse models. Oncogene 2002, 21, 8486–8497. [Google Scholar]
- Sample Availability: Samples of the all compounds are available from the authors.
© 2014 by the authors. licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, Y.; Liang, C.; Ma, H.; Zhao, Q.; Lu, Y.; Xiang, Z.; Li, L.; Qin, J.; Chen, Y.; Cho, W.C.; et al. miR-221/222 Promotes S-Phase Entry and Cellular Migration in Control of Basal-Like Breast Cancer. Molecules 2014, 19, 7122-7137. https://doi.org/10.3390/molecules19067122
Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, Li L, Qin J, Chen Y, Cho WC, et al. miR-221/222 Promotes S-Phase Entry and Cellular Migration in Control of Basal-Like Breast Cancer. Molecules. 2014; 19(6):7122-7137. https://doi.org/10.3390/molecules19067122
Chicago/Turabian StyleLi, Yuan, Chunli Liang, Haizhong Ma, Qian Zhao, Ying Lu, Zhendong Xiang, Li Li, Jie Qin, Yihan Chen, William C. Cho, and et al. 2014. "miR-221/222 Promotes S-Phase Entry and Cellular Migration in Control of Basal-Like Breast Cancer" Molecules 19, no. 6: 7122-7137. https://doi.org/10.3390/molecules19067122
APA StyleLi, Y., Liang, C., Ma, H., Zhao, Q., Lu, Y., Xiang, Z., Li, L., Qin, J., Chen, Y., Cho, W. C., Pestell, R. G., Liang, L., & Yu, Z. (2014). miR-221/222 Promotes S-Phase Entry and Cellular Migration in Control of Basal-Like Breast Cancer. Molecules, 19(6), 7122-7137. https://doi.org/10.3390/molecules19067122





