Fatty Acids as a Tool to Understand Microbial Diversity and Their Role in Food Webs of Mediterranean Temporary Ponds
Abstract
:1. Introduction
| Fatty Acid | Category | Reference |
|---|---|---|
| Mono-Unsaturated Fatty Acids (MUFA) | ||
| 16:1ω7c | Bacteria | [24,26,27,38] |
| Bacillariophyceae (diatoms) | ||
| Cyanophyceae (cyanobacteria) | ||
| Prymnesiophyceae | ||
| 16:1ω5c | mycorrhizal fungi | [30,39] |
| 16:1ω8 | Type I methanotrophs (gamma-proteobacteria) | [40] |
| 18:1ω9c | Chlorophyceae (green algae) | [24,26,27,35,38] |
| Cyanophyceae | ||
| Dinophyceae | ||
| Prymnesiophyceae | ||
| Gram-positive bacteria | ||
| 18:1ω7c | Bacillariophyceae (up to 10-fold more 18:1ω7c than 18:1ω9c) | [24,26,38] |
| Cryptophyceae | ||
| Cyanophyceae (less amount than 18:1ω9c) | ||
| Prymnesiophyceae | ||
| 18:1ω7t | Gram-negative bacteria | [35] |
| 18:1ω8 | Type II methanotrophs (alpha-proteobacteria) | [39,40] |
| Hydroxy substituted Fatty Acids (OH FA) | ||
| (e.g., 3-OH 10:0) | Gram-negative bacteria | [41] |
| Cyclopropyl saturated Fatty Acids (cyFA) | ||
| (e.g., cy17:0, cy19:0) | Gram-negative bacteria, anaerobic bacteria | [29,31] |
| Iso- and anteiso-branched Fatty Acids | ||
| (e.g., i-15:0, a-17:0) | Gram-positive bacteria | [29,31] |
| Methyl-branched Fatty Acids (10-Me FA) | ||
| e.g., 10-Me 16:0 | Actinomycetales (Actinobacteria) | [42] |
| Polyunsaturated Fatty Acids (PUFA) | ||
| 16:2ω7 | Bacillariophyceae | [24] |
| 16:2ω6 | Chlorophyta | [24] |
| 16:2ω4 | Bacillariophyceae | [24,43] |
| Prasinophyceae | ||
| 16:3ω4 | Bacillariophyceae | [24,43] |
| 16:3ω3 | Chlorophyta | [24] |
| 16:4ω3 | Chlorophyceae | [24] |
| Prasinophyceae | ||
| 16:4ω1 | Bacillariophyceae (diatoms) | [24,43] |
| 18:2ω6 | Chlorophyta | [24,26,27,29,31,38] |
| Cyanophyceae (freshwater) | ||
| Dinophyceae | ||
| Prymnesiophyceae | ||
| Fungi | ||
| 18:3ω6 | Cyanophyceae (freshwater) | [26,31,44] |
| Saprophytic fungi | ||
| 18:3ω3 | Chlorophyceae | [24,26,38] |
| Crypophyceae | ||
| Cyanophyceae | ||
| Dinophyceae | ||
| Prasinophyceae | ||
| Prymnesiophyceae | ||
| 18:4ω3 | Various algal groups (both marine and freshwater) | [24,26,38] |
| 18:5ω3 | Dynophyceae | [24] |
| 20:4ω6 | Bacillariophyceae | [24] |
| Rhodophyceae | ||
| 20:5ω3 | Bacillariophyceae | [24,38] |
| Cryptophyceae | ||
| Dinophyceae | ||
| Pavlovophyceae | ||
| Rhodophyceae | ||
| 22:5ω3 | Bacillariophyceae | [24] |
| Cryptophyceae | ||
| Prasinophyceae | ||
| 22:6ω3 | Bacillariophyceae | [24,26,38] |
| Cryptophyceae | ||
| Dinophyceae | ||
| Haptophyta (Prymnesiophyceae and Pavlovophyceae) | ||
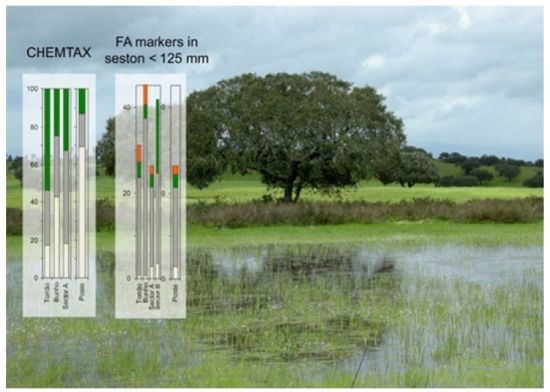
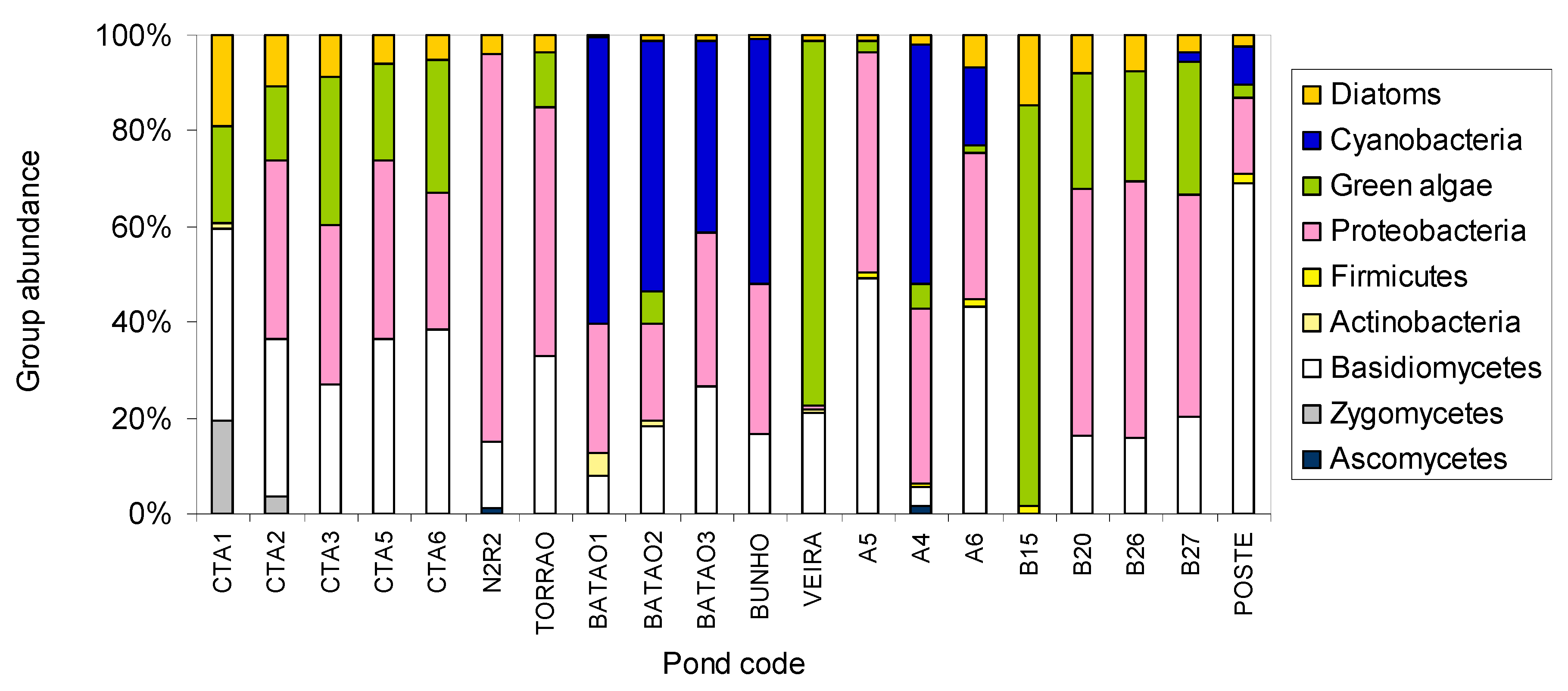
2. Results and Discussion
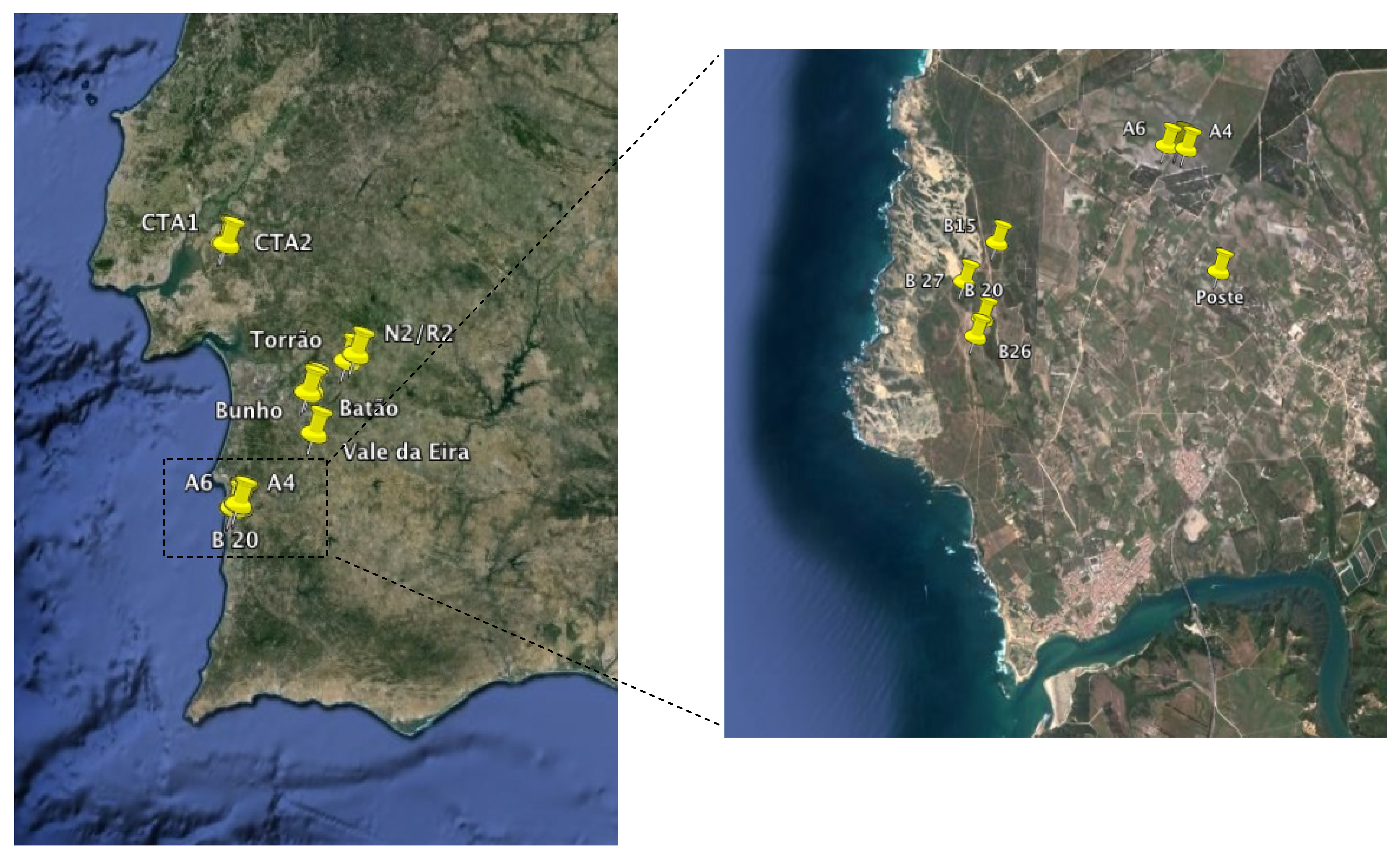
| Code | Location | GPS coordinates | Depth | Temperature | Conductivity | Salinity | Chlorophyll-a |
|---|---|---|---|---|---|---|---|
| m | °C | μS | ppt | μg L−1 | |||
| CTA1 | FFR of Alcochete | N 38° 46.745' W 8° 51.621' | 0.4 | 17.4 | 177.7 | 0.1 | 40.8 |
| CTA2 | FFR of Alcochete | N 38° 46.687' W 8° 50.979' | 0.4 | 17.8 | 112 | 0.1 | 47.7 |
| CTA3 | FFR of Alcochete | N 38° 46.634' W 8° 50.238' | 0.4 | 15.8 | 265.1 | 0.1 | 38.2 |
| CTA5 | FFR of Alcochete | N 38° 46.363' W 8° 47.374' | 0.6 | 16.6 | 153 | 0.1 | 4.4 |
| CTA6 | FFR of Alcochete | N 38° 46.363' W 8° 47.374' | 1.1 | 16.6 | 135.3 | 0.1 | 14.6 |
| N2/R2 | Torrão | N 38° 21.234' W 8° 11.959' | 0.5 | 13.7 | 116.6 | 0.1 | 3.9 |
| TORRAO | Torrão | N 38° 19.670' W 8° 14.587' | 1.2 | 13 | 54.4 | 0 | 4.1 |
| BATAO 1 | Batão | N 38° 12.863' W 8° 23.675’ | 0.6 | 12.4 | 360 | 0.1 | 78.3 |
| BATAO 2 | Batão | N 38° 12.562' W 8° 24.791' | 0.6 | 12.3 | 297 | 0.1 | 28.6 |
| BATAO 3 | Batão | N 38° 12.870' W 8° 25.270' | 0.6 | 12.4 | 281 | 0.1 | 7.1 |
| BUNHO | Bunho | N 38° 12.287' W 8° 26.168' | 0.6 | 17.4 | 111 | 0.1 | 27.1 |
| V EIRA | V. da Eira | N 38° 2.718' W 8° 24.193' | 0.6 | 14.6 | 596 | 0.1 | 23.9 |
| A5 | PNSACV, Sector A | N 37° 46.147' W 8° 46.335' | 0.5 | 11.6 | 158.1 | 0.1 | 1.3 |
| A4 | PNSACV, Sector A | N 37° 46.128' W 8° 46.277' | 0.7 | 13.9 | 119.2 | 0.1 | 0.7 |
| A6 | PNSACV, Sector A | N 37° 46.147' W 8° 46.410' | 0.7 | 12.1 | 167.2 | 0.1 | 2.8 |
| B15 | PNSACV, Sector B | N 37° 45.570' W 8° 47.666' | 0.9 | 14.2 | 175 | 0.1 | 1.4 |
| B20 | PNSACV, Sector B | N 37° 45.124' W 8° 47.769' | 0.7 | 13.8 | 542 | 0.1 | 2.2 |
| B26 | PNSACV, Sector B | N 37° 45.018' W 8° 47.820' | 0.2 | 13.9 | 715 | 0.1 | 3.3 |
| B27 | PNSACV, Sector B | N 37° 44.997' W 8° 47.903' | 0.8 | 13.3 | 621 | 0.1 | 0.5 |
| POSTE | PNSACV, Sector A | N 37° 45.411' W 8° 46.017' | 0.2 | 14 | 139.2 | 0.1 | 0.9 |

| Class/ | Ascomycetes | Zygomycetes | Basidiomycetes | Actinobacteria | Firmicutes | Proteobacteria | Green algae | Cyanobacteria | Diatoms |
|---|---|---|---|---|---|---|---|---|---|
| Fatty acid | |||||||||
| Input matrix ratio | |||||||||
| 12:00 | 0.06 | 0.03 | 0.04 | 0.01 | 0.05 | 0.03 | |||
| 14:00 | 0.09 | 0.06 | 0.05 | 0.28 | 0.31 | 0.07 | 0.06 | 0.03 | 0.99 |
| 15:00 | 0.11 | 0.1 | 0.02 | ||||||
| 15:0 anteiso | 1.13 | 3.88 | 0.16 | ||||||
| 15:0 iso | 0.78 | 2.25 | 0.1 | ||||||
| 16:00 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 16:1ω7c | 1.66 | 0.16 | 0.1 | 0.01 | 0.05 | 0.02 | 0.21 | 0.2 | 1.93 |
| 16:1ω9c | 0.29 | 0.02 | |||||||
| 16:2ω4 | 0.39 | ||||||||
| 16:3ω4 | 0.69 | ||||||||
| 17:00 | 0.04 | 0.03 | 0.03 | 0.04 | 0.02 | 0.02 | |||
| 17:0 iso | 0.13 | 0.3 | 0.03 | ||||||
| 18:00 | 0.24 | 0.2 | 0.17 | 0.07 | 0.06 | 0.05 | 0.09 | 0.05 | 0.07 |
| 18:1ω9c | 2.87 | 1.98 | 1.1 | 0.64 | 0.04 | 0.01 | 0.3 | 0.38 | 0.07 |
| 18:1ω7c | 0.13 | 0.09 | 0.11 | 0.09 | 0.15 | 0.16 | |||
| 18:2ω6c | 0.94 | 0.66 | 3.28 | 0.38 | 0.45 | 0.08 | |||
| 18:3ω3 | 0.07 | 0.49 | 0.6 | 2.27 | 0.83 | 0.03 | |||
| 20:5ω3 | 0.01 | 1.76 | |||||||
| 22:6ω3 | 0.19 | ||||||||
| Output matrix ratio | |||||||||
| 12:00 | 0.06 | 0.03 | 0.04 | 0.01 | 0.05 | 0.03 | |||
| 14:00 | 0.09 | 0.06 | 0.05 | 0.28 | 0.31 | 0.07 | 0.06 | 0.03 | 0.99 |
| 15:00 | 0.11 | 0.1 | 0.02 | ||||||
| 15:0 anteiso | 1.13 | 3.88 | 0.09 | ||||||
| 15:0 iso | 0.78 | 2.25 | 0.07 | ||||||
| 16:0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 16:1ω7c | 1.66 | 0.16 | 0.05 | 0.01 | 0.05 | 0.02 | 0.21 | 0.2 | 1.93 |
| 16:1ω9c | 0.29 | 0.02 | |||||||
| 16:2ω4 | 0.39 | ||||||||
| 16:3ω4 | 0.69 | ||||||||
| 17:00 | 0.04 | 0.03 | 0.03 | 0.04 | 0.02 | 0.02 | |||
| 17:0 iso | 0.13 | 0.3 | 0.03 | ||||||
| 18:00 | 0.24 | 0.2 | 0.17 | 0.07 | 0.06 | 0.05 | 0.09 | 0.05 | 0.07 |
| 18:1ω9c | 2.87 | 1.98 | 0.88 | 0.64 | 0.04 | 0.01 | 0.3 | 0.38 | 0.07 |
| 18:1ω7c | 0.13 | 0.09 | 0.11 | 0.09 | 0.15 | 0.16 | |||
| 18:2ω6c | 0.94 | 0.66 | 3.28 | 0.38 | 0.45 | 0.08 | |||
| 18:3ω3 | 0.07 | 0.49 | 0.6 | 2.27 | 0.83 | 0.03 | |||
| 20:5ω3 | 0.01 | 1.76 | |||||||
| 22:6ω3 | 0.19 | ||||||||
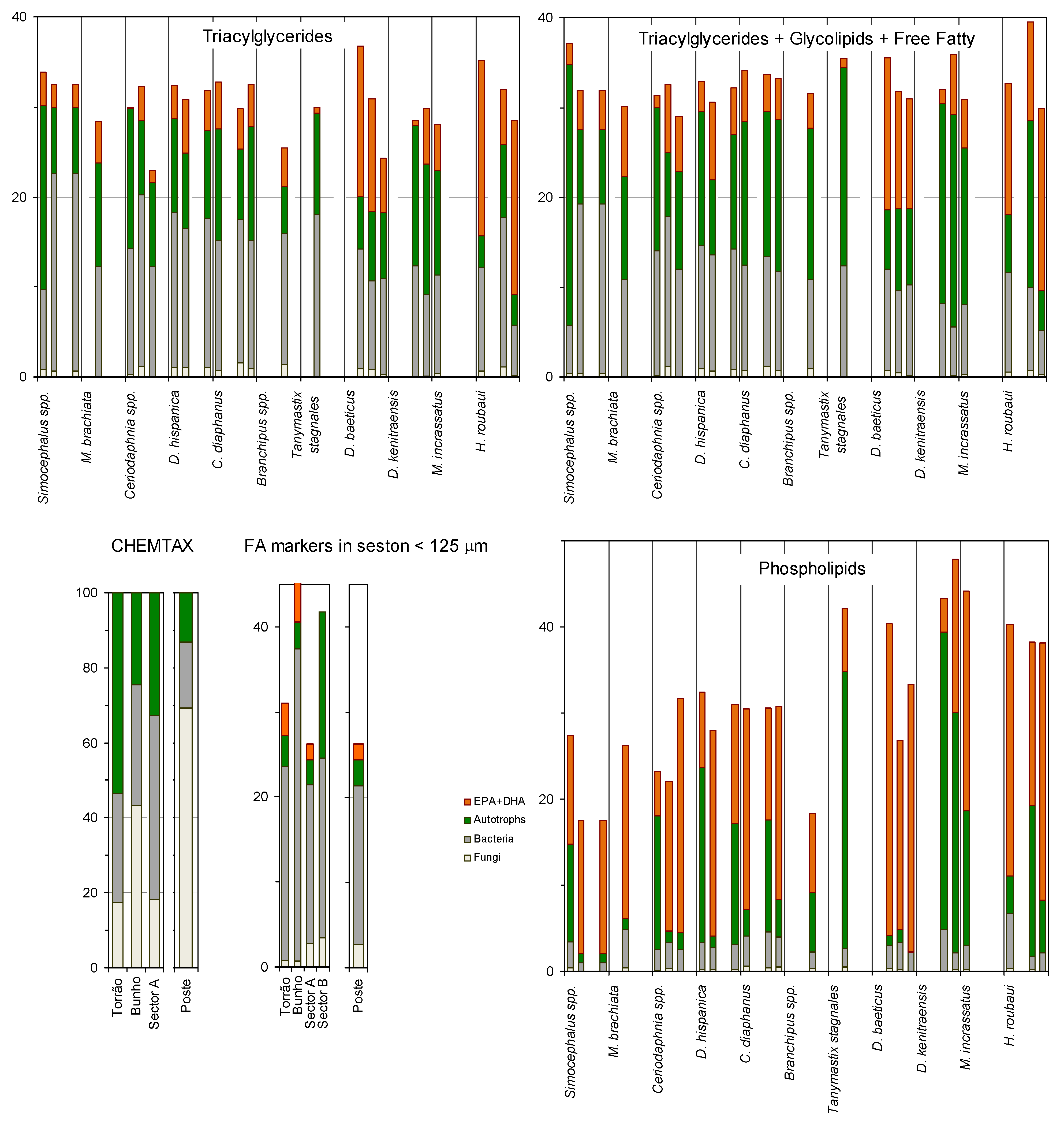
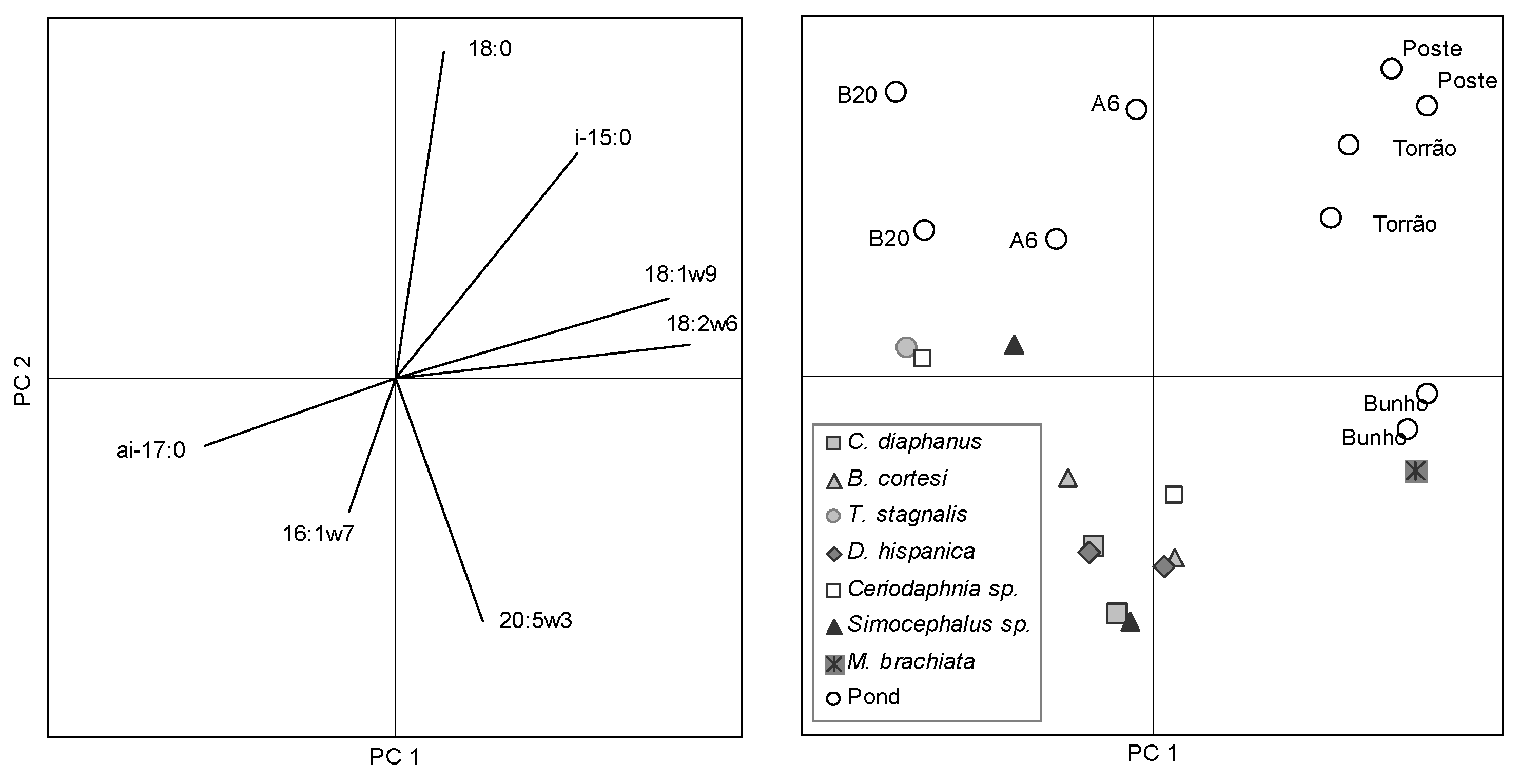
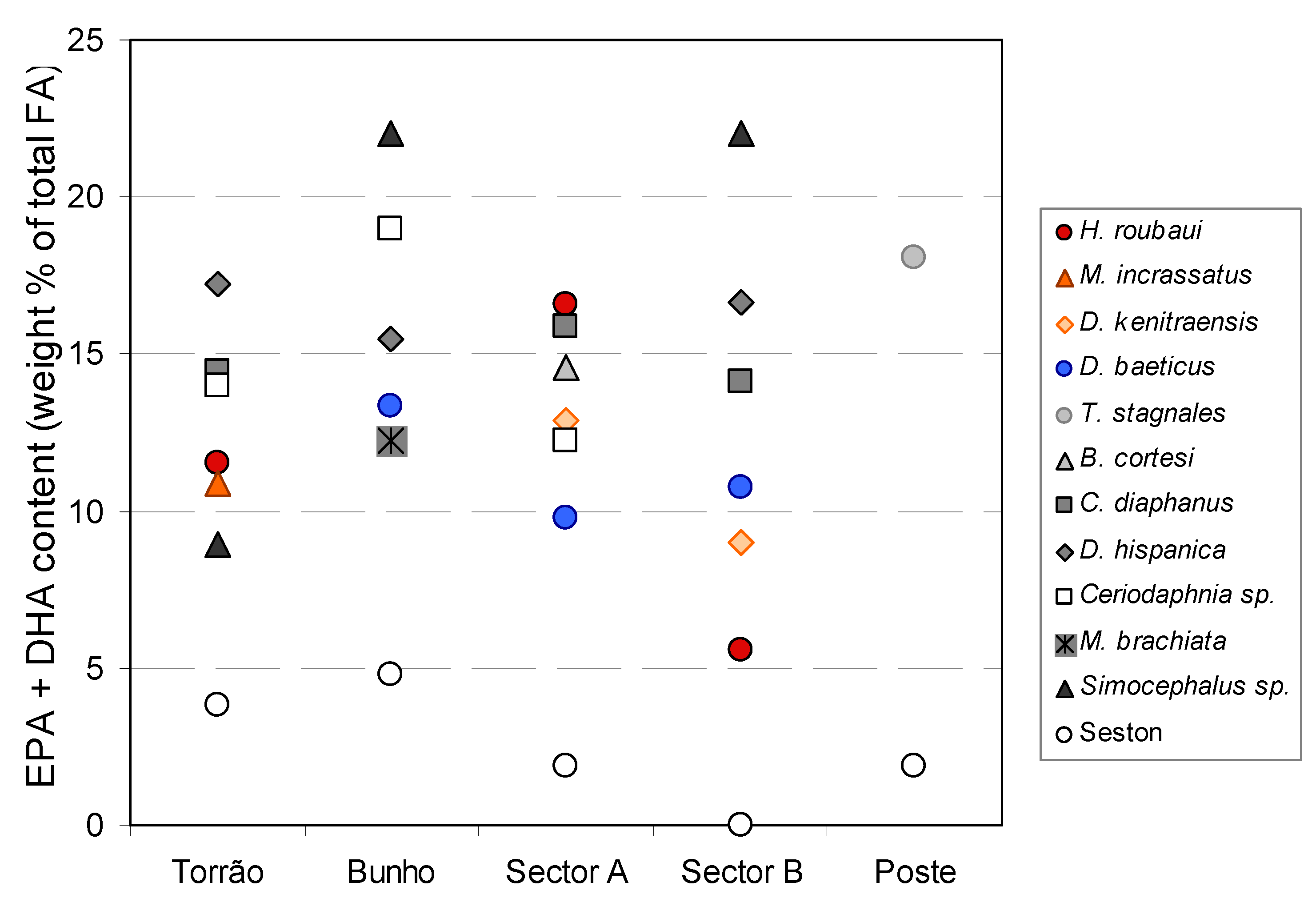
3. Experimental
3.1. Sample Collection and Measurement of Physical Parameters
3.2. Lipid Extraction and Fractioning
3.3. Data Analysis
Microbial Groups
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Desvilettes, C.; Bourdier, G.; Breton, J.-C. Lipid class and fatty acid composition of planktivorous larval pike Esox lucius living in a natural pond. Aquat. Living Resour. 1994, 7, 67–77. [Google Scholar] [CrossRef]
- Nix, M.H.; Jenkins, D.G. Life history comparisons of Daphnia obtusa from temporary ponds, cultured with a low-quality food. Aquat. Ecol. 2000, 34, 19–27. [Google Scholar] [CrossRef]
- Overmann, J.; Garcia-Pichel, F. The phototrophic way of life. In The Prokaryotes; Rosenberg, E., DeLong, E., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer-Verlag: Berlin Heidelberg, Germany, 2013; pp. 203–257. [Google Scholar]
- Howarth, R.W.; Marino, R.; Cole, J.J. Nitrogen fixation in freshwater, estuarine, and marine ecosystem. 2. Biogeochemical controls. Limnol. Oceanogr. 1988, 33, 688–701. [Google Scholar] [CrossRef]
- Hairston, N.G.J.; Hairston, N.G.S. Cause-effect relationships in energy flow, trophic structure, and interspecific interactions. Am. Nat. 1993, 142, 379–411. [Google Scholar]
- Felton, M.; Cooney, J.J.; Moore, W.G. A quantitative study of bacteria of a temporary pond. J. Gen. Microbiol. 1967, 47, 25–31. [Google Scholar] [CrossRef]
- Rubbo, M.J.; Belden, L.K.; Storrs-Mendez, S.I.; Cole, J.J.; Kiesecker, J.M. Species loss in the brown world: Are heterotrophic systems inherently stable? Aquat. Sci. 2012, 74, 397–404. [Google Scholar] [CrossRef]
- Auer, B.; Elzer, U.; Arndt, H. Comparison of pelagic food webs in lakes along a trophic gradient and with seasonal aspects: Influence of resource and predation. J. Plankton Res. 2004, 26, 697–709. [Google Scholar] [CrossRef]
- Butler, H.G. Seasonal dynamics of the planktonic microbial community in a maritime Antarctic lake undergoing eutrophication. J. Plankton Res. 1999, 21, 2393–2419. [Google Scholar] [CrossRef]
- Azam, F.; Fenchel, T.; Field, J.G.; Gray, J.S.; Meyer-Reil, L.A.; Thingstad, F. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 1983, 10, 257–263. [Google Scholar]
- Sherr, E.; Sherr, B. Role of microbes in pelagic food webs—a revised concept. Limnol. Oceanogr. 1988, 33, 1225–1227. [Google Scholar] [CrossRef]
- Ahlgren, G.; Lundstedt, L.; Brett, M.; Forsber, C. Lipid composition and food quality of some freshwater phytoplankton for cladoceran zooplankters. J. Plankton Res. 1990, 12, 809–818. [Google Scholar] [CrossRef]
- Gulati, R.D.; DeMott, W.R. The role of food quality for zooplankton: Remarks on the state-of-the-art, perspectives and priorities. Freshwat. Biol. 1997, 38, 753–768. [Google Scholar] [CrossRef]
- Müller-Navarra, D.C. Evidence that a highly unsaturated fatty-acid limits Daphnia growth in nature. Arch. Hydrobiol. 1995, 132, 297–307. [Google Scholar]
- Müller-Navarra, D.C.; Brett, M.T.; Park, S.; Chandra, S.; Ballantyne, A.P.; Zorita, E.; Goldman, C.R. Unsaturated fatty acid content in seston and tropho-dynamic coupling in lakes. Nature 2004, 427, 69–72. [Google Scholar] [CrossRef]
- Ederington, M.C.; McManus, G.B.; Harvey, H.R. Trophic transfer of fatty acids, sterols, and a triterpenoid alcohol between bacteria, a ciliate, and the copepod Acartia. tonsa. Limnol. Oceanogr. 1995, 40, 860–867. [Google Scholar] [CrossRef]
- Klein Breteler, W.C.M.; Koski, M.; Rampen, S. Role of essential lipids in copepod nutrition: No evidence for trophic upgrading of food quality by a marine ciliate. Mar. Ecol. Prog. Ser. 2004, 274, 199–208. [Google Scholar] [CrossRef]
- Bec, A.; Martin-Creuzburg, D.; Elert, E.V. Trophic upgrading of autotrophic picoplankton by the heterotrophic nanoflagellate Paraphysomonas. sp. Limnol. Oceanogr. 2006, 51, 1699–1707. [Google Scholar] [CrossRef]
- Desvilettes, C.; Bec, A. Formation and transfer of fatty acids in aquatic microbial food webs: Role of heterotrophic protists. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M.T., Arts, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 25–42. [Google Scholar]
- Martin-Creuzburg, D.; Beck, B.; Freese, H.M. Food quality of heterotrophic bacteria for Daphnia magna: Evidence for a limitation by sterols. FEMS Microbiol. Ecol. 2011, 76, 592–601. [Google Scholar] [CrossRef]
- Caramujo, M.J.; Boavida, M.J. Biological diversity of copepods and cladocerans in Mediterranean temporary ponds under periods of contrasting rainfall. J. Limnol. 2010, 69, 64–75. [Google Scholar]
- Piotrowska-Seget, Z.; Mrozik, A. Signature lipid biomarker (SLB) analysis in determining changes in community structure of soil microorganisms. Pol. J. Environ. Stud. 2003, 12, 669–675. [Google Scholar]
- Pinkart, H.G.; Ringelberg, D.B.; Piceno, Y.M.; MacNaughton, S.J.; White, D.C. Biochemical approaches to biomass measurements and community structure. In Manual of Environmental Microbiology; Hurst, C.J., Crawford, R.L., Knudsen, G.R., McInerney, M.J., Stetzenbach, L.D., Eds.; American Society for Microbiology Press: Washington, DC, USA, 2002; pp. 101–113. [Google Scholar]
- Dijkman, N.A.; Kromkamp, J.C. Phospholipid-derived fatty acids as chemotaxonomic markers for phytoplankton: Application for inferring phytoplankton composition. Mar. Ecol. Prog. Ser. 2006, 324, 113–125. [Google Scholar] [CrossRef]
- Napolitano, G.E.; Pollero, R.J.; Gayoso, A.M.; Macdonald, B.A.; Thompson, R.J. Fatty acids as trophic markers of phytoplankton blooms in the Bahía Blanca estuary (Buenos Aires, Argentina) and in Trinity Bay (Newfoundland, Canada). Biochem. Syst. Ecol. 1997, 25, 739–755. [Google Scholar] [CrossRef]
- Ahlgren, G.; Gustafsson, I.-B.; Boberg, M. Fatty acid content and chemical composition of freshwater microalgae. J. Phycol. 1992, 28, 37–50. [Google Scholar]
- Dijkman, N.A.; Boschker, H.T.S.; Stal, L.J.; Kromkamp, J.C. Composition and heterogeneity of the microbial community in a coastal microbial mat as revealed by the analysis of pigments and phospholipid-derived fatty acids. J. Sea Res. 2010, 63, 62–70. [Google Scholar] [CrossRef]
- Findlay, R.H.; King, G.M.; Watling, L. Efficacy of phospholipid analysis in determining microbial biomass in sediments. Appl. Environ. Microbiol. 1989, 55, 2888–2893. [Google Scholar]
- Frostegård, A.; Bååth, E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fertility Soils 1996, 22, 59–65. [Google Scholar] [CrossRef]
- Olsson, P.A. Signature fatty acids provide tools for determination of the distribution and interactions of mycorrhizal fungi in soil. FEMS Microbiol. Ecol. 1999, 29, 303–310. [Google Scholar] [CrossRef]
- Zelles, L. Phospholipid fatty acid profiles in selected members of soil microbial communities. Chemosphere 1997, 35, 275–294. [Google Scholar] [CrossRef]
- Guckert, J.B.; Nold, S.C.; Boston, H.L.; White, D.C. Periphyton response in an industrial receiving stream: Lipid-based physiological stress analysis and pattern recognition of microbial community structure. Can. J. Fish. Aquat. Sci. 1992, 49, 2579–2587. [Google Scholar] [CrossRef]
- Findlay, R. The use of phospholipid fatty acids to determine microbial community structure. In Molecular Microbial Ecology Manual; Akkermans, A.D.L., van Elsas, J.D., de Bruijn, F., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1996; pp. 1–17. [Google Scholar]
- Van den Meersche, K.; Soetaert, K.; Middelburg, J.J. A Bayesian compositional estimator for microbial taxonomy based on biomarkers. Limnol. Oceanogr-Methods 2008, 6, 190–199. [Google Scholar]
- Zelles, L. Identification of single cultured micro-organisms based on their whole-community fatty acid profiles, using an extended extraction procedure. Chemosphere 1999, 39, 665–682. [Google Scholar] [CrossRef]
- White, D.C.; Stair, J.O.; Ringelberg, D.B. Quantitative comparisons of in situ microbial biodiversity by signature biomarker analysis. J. Ind. Microbiol. 1996, 17, 185–196. [Google Scholar] [CrossRef]
- Mackey, M.; Mackey, D.; Higgins, H.; Wright, S. CHEMTAX—a program for estimating class abundances from chemical markers: Application to HPLC measurements of phytoplankton. Mar. Ecol. Prog. Ser. 1996, 144, 265–283. [Google Scholar] [CrossRef]
- Dalsgaard, J.; St John, M.; Kattner, G.; Müller-Navarra, D.; Hagen, W. Fatty acid trophic markers in the pelagic marine environment. Adv. Mar. Biol. 2003, 46, 225–340. [Google Scholar]
- Bowman, J.P.; Skerratt, J.H.; Nichols, P.D.; Sly, L.I. Phospholipid fatty acid and lipopolysaccharide fatty acid signature lipids in methane-utilizing bacteria. FEMS Microbiol. Lett. 1991, 85, 15–21. [Google Scholar] [CrossRef]
- Costello, A.M.; Auman, A.J.; Macalady, J.L.; Scow, K.M.; Lidstrom, M.E. Estimation of methanotroph abundance in a freshwater lake sediment. Environ. Microbiol. 2002, 4, 443–450. [Google Scholar] [CrossRef]
- Lee, A.K.Y.; Chan, C.K.; Fang, M.; Lau, A.P.S. The 3-hydroxy fatty acids as biomarkers for quantification and characterization of endotoxins and Gram-negative bacteria in atmospheric aerosols in Hong Kong. Atmos. Environ. 2004, 38, 6307–6317. [Google Scholar]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl. Environ. Microbiol. 1993, 59, 3605–3617. [Google Scholar]
- Dunstan, G.A.; Volkman, J.K.; Barrett, S.M.; Leroi, J.-M.; Jeffrey, S.W. Essential polyunsaturated fatty acids from 14 species of diatom (Bacillariophyceae). Phytochemistry 1993, 35, 155–161. [Google Scholar] [CrossRef]
- Stahl, P.D.; Klug, M.J. Characterization and differentiation of filamentous fungi based on fatty acid composition. Appl. Environ. Microbiol. 1996, 62, 4136–4146. [Google Scholar]
- Iverson, S. Tracing Aquatic Food Webs Using Fatty Acids: From Qualitative Indicators to Quantitative Determination. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M.T., Arts, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 281–308. [Google Scholar]
- Brett, M.; Müller-Navarra, D.; Persson, J. Crustacean zooplankton fatty acid composition. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M.T., Arts, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 115–146. [Google Scholar]
- Bull, A.T.; Stach, J.E.M. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef]
- Müller-Navarra, D.C.; Brett, M.T.; Liston, A.M.; Goldman, C.R. A highly unsaturated fatty acid predicts carbon transfer between primary producers and consumers. Nature 2000, 403, 74–77. [Google Scholar] [CrossRef]
- Sargent, J.; McEvoy, L.; Estevez, A.; Bell, G.; Bell, M.; Henderson, J.; Tocher, D. Lipid nutrition of marine fish during early development: Current status and future directions. Aquaculture 1999, 179, 217–229. [Google Scholar] [CrossRef]
- Arts, M.; Kohler, C. Health and condition in fish: The influence of lipids on membrane competency and immune response. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M.T., Arts, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 237–256. [Google Scholar]
- Pernthaler, J. Freshwater Microbial Communities. In The Prokaryotes; Rosenberg, E., DeLong, E., Lory, S., Stackebrandt, E., Thompson, F, Eds.; Springer: Heidelberg, Germany, 2013; pp. 97–112. [Google Scholar]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar]
- Haack, S.K.; Garchow, H.; Odelson, D.A.; Forney, L.J.; Klug, M.J. Accuracy, reproducibility, and interpretation of fatty acid methyl ester profiles of model bacterial communities. Appl. Environ. Microbiol. 1994, 60, 2483–2493. [Google Scholar]
- de Carvalho, C.C.C.R.; Caramujo, M.J. Bacterial diversity assessed by cultivation-based techniques shows predominance of Staphylococccus species on coins collected in Lisbon and Casablanca. FEMS Microbiol. Ecol. 2014, 88, 26–37. [Google Scholar] [CrossRef]
- Kunitsky, C.; Osterhout, G.; Sasser, M. Identification of microorganisms using fatty acid methyl ester (FAME) analysis and the MIDI sherlock microbial identification system. In Encyclopedia of Rapid Microbiological Methods; Miller, M.J., Ed.; Davis Healthcare International Publishing, LLC: Baltimore, MD, USA, 2005; Volume III, pp. 1–17. [Google Scholar]
- Carrino-Kyker, S.R.; Swanson, A.K. Temporal and spatial patterns of eukaryotic and bacterial communities found in vernal pools. Appl. Environ. Microbiol. 2008, 74, 2554–2557. [Google Scholar] [CrossRef]
- Romaní, A.; Amalfitano, S.; Artigas, J.; Fazi, S.; Sabater, S.; Timoner, X.; Ylla, I.; Zoppini, A. Microbial biofilm structure and organic matter use in mediterranean streams. Hydrobiologia 2013, 719, 43–58. [Google Scholar] [CrossRef]
- Zoppini, A.; Amalfitano, S.; Fazi, S.; Puddu, A. Dynamics of a benthic microbial community in a riverine environment subject to hydrological fluctuations (Mulargia River, Italy). Hydrobiologia 2010, 657, 37–51. [Google Scholar] [CrossRef]
- Santmire, J.A.; Leff, L.G. The influence of stream sediment particle size on bacterial abundance and community composition. Aquat. Ecol. 2007, 41, 153–160. [Google Scholar] [CrossRef]
- Agogué, H.; Joux, F.; Obernosterer, I.; Lebaron, P. Resistance of marine bacterioneuston to solar radiation. Appl. Environ. Microbiol. 2005, 71, 5282–5289. [Google Scholar] [CrossRef]
- Schiaffino, M.R.; Unrein, F.; Gasol, J.M.; Massana, R.; Balague, V.; Izaguirre, I. Bacterial community structure in a latitudinal gradient of lakes: The roles of spatial versus environmental factors. Freshwat. Biol. 2011, 56, 1973–1991. [Google Scholar] [CrossRef]
- Jürgens, K.; Güde, H. The potential importance of grazing-resistant bacteria in planktonic systems. Mar. Ecol. Prog. Ser. 1994, 112, 169–188. [Google Scholar] [CrossRef]
- Allgaier, M.; Grossart, H.-P. Seasonal dynamics and phylogenetic diversity of free-living and particle-associated bacterial communities in four lakes in northeastern Germany. Aquat. Microb. Ecol. 2006, 45, 115–128. [Google Scholar] [CrossRef]
- Bellinger, E.G.; Sigee, D.C. A key to the more frequently occurring freshwater algae. In Freshwater Algae; John Wiley & Sons, Ltd.: Chichester, UK, 2010; pp. 137–244. [Google Scholar]
- Barnett, J.A.; Payne, R.W.; Yarrow, D. Yeasts: Characteristics and Identification, 3th ed.; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Bessey, E.A. Morphology and Taxonomy of Fungi; Hafner Pub. Co.: New York, NY, USA, 1961; p. 791. [Google Scholar]
- Wong, M.M.; Goh, T.-K.; Hodgkiss, I.J.; Hyde, K.; Ranghoo, V.M.; Tsui, C.M.; Ho, W.-H.; Wong, W.W.; Yuen, T.-K. Role of fungi in freshwater ecosystems. Biodivers. Conserv. 1998, 7, 1187–1206. [Google Scholar] [CrossRef]
- Sigee, D. Freshwater Microbiology: Biodiversity and Dynamic Interactions of Microorganisms in the Aquatic Environment; John Wiley & Sons: Chichester, UK, 2005; p. 544. [Google Scholar]
- Jones, E.B.G.; Choeyklin, R. Ecology of Marine and Freshwater Basidiomycetes. In Ecology of Saprotrophic Basidiomycetes; Boddy, L., Frankland, J., West, P.V., Eds.; Academic Press: London, UK, 2008. [Google Scholar]
- White, D.C.; Ringelberg, D.B.; Macnaughton, S.J.; Alugupalli, S.; Schram, D. Signature lipid biomarker analysis for quantitative assessment in situ of environmental microbial ecology. In Molecular markers in environmental geochemistry; Eganhouse, R.P., Ed.; ACS Symposium Series 671; American Chemical Society: Washington, DC, USA, 1997; Chapter 2; pp. 22–34. [Google Scholar]
- Buyer, J.S.; Sasser, M. High throughput phospholipid fatty acid analysis of soils. Appl. Soil Ecol. 2012, 61, 127–130. [Google Scholar] [CrossRef]
- Mendes, C.R.B.; Tavano, V.M.; Leal, M.C.; de Souza, M.S.; Brotas, V.; Garcia, C.A.E. Shifts in the dominance between diatoms and cryptophytes during three late summers in the Bransfield Strait (Antarctic Peninsula). Polar Biol. 2013, 36, 537–547. [Google Scholar] [CrossRef]
- Wright, S.W.; van den Enden, R.L.; Pearce, I.; Davidson, A.T.; Scott, F.J.; Westwood, K.J. Phytoplankton community structure and stocks in the Southern Ocean (30–80°E) determined by CHEMTAX analysis of HPLC pigment signatures. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 2010, 57, 758–778. [Google Scholar] [CrossRef]
- Wright, S.W.; Jeffrey, S.W. Pigment Markers for Phytoplankton Production. In Marine Organic Matter: Biomarkers, Isotopes and DNA; Volkman, J., Ed.; Springer: Heidelberg, Germany, 2006; Volume 2N, pp. 71–104. [Google Scholar]
- Latasa, M. Improving estimations of phytoplankton class abundances using CHEMTAX. Mar. Ecol. Prog. Ser. 2007, 329, 13–21. [Google Scholar]
- Bardgett, R.D.; Hobbs, P.J.; Frostegård, A. Changes in soil fungal:bacterial biomass ratios following reductions in the intensity of management of an upland grassland. Biol. Fertility Soils 1996, 22, 261–264. [Google Scholar] [CrossRef]
- de Vries, F.T.; Hoffland, E.; van Eekeren, N.; Brussaard, L.; Bloem, J. Fungal/bacterial ratios in grasslands with contrasting nitrogen management. Soil Biol. Biochem. 2006, 38, 2092–2103. [Google Scholar] [CrossRef]
- Spring, S.; Schulze, R.; Overmann, J.; Schleifer, K.-H. Identification and characterization of ecologically significant prokaryotes in the sediment of freshwater lakes: Molecular and cultivation studies. FEMS Microbiol. Rev. 2000, 24, 573–590. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Fuchs, B.M.; Amann, R. Bacterioplankton compositions of lakes and oceans: A first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 1999, 65, 3721–3726. [Google Scholar]
- Roberts, B.J.; Howarth, R.W. Nutrient and light availability regulate the relative contribution of autotrophs and heterotrophs to respiration in freshwater pelagic ecosystems. Limnol. Oceanogr. 2006, 51, 288–298. [Google Scholar] [CrossRef]
- Carlson, R.E. A trophic state index for lakes. Limnol. Oceanogr. 1977, 22, 361–369. [Google Scholar] [CrossRef]
- Beach, D.H.; Harrington, G.W.; Holz, G.G. The polyunsaturated fatty acids of marine and freshwater Cryptomonads. J. Protozool. 1970, 17, 501–510. [Google Scholar] [CrossRef]
- Véra, A.; Desvilettes, C.; Bec, A.; Bourdier, G. Fatty acid composition of freshwater heterotrophic flagellates: An experimental study. Aquat. Microb. Ecol. 2001, 25, 271–279. [Google Scholar] [CrossRef]
- Bec, A.; Desvilettes, C.; Véra, A.; Lemarchand, C.; Fontvieille, D.; Bourdier, G. Nutritional quality of a freshwater heterotrophic flagellate: Trophic upgrading of its microalgal diet for Daphnia hyalina. Aquat. Microb. Ecol. 2003, 32, 203–207. [Google Scholar] [CrossRef]
- Desvilettes, C.H.; Bourdier, G.; Amblard, C.H.; Barth, B. Use of fatty acids for the assessment of zooplankton grazing on bacteria, protozoans and microalgae. Freshwat. Biol. 1997, 38, 629–637. [Google Scholar] [CrossRef]
- Farkas, T. Fats in fresh water crustaceans. 1. Fatty acid composition of lipids obtained from Eudiaptomus gracilis G.O. Sars (Copepoda) and Daphnia cucullata G.O. Sars (Cladocera). Acta Biol. Hung. 1970, 21, 225–233. [Google Scholar]
- Thompson, G.A., Jr. The Regulation of Membrane Lipid Metabolism; CRC Press: Boca Raton, FL, USA, 1992; p. 240. [Google Scholar]
- Hall, J.M.; Parrish, C.C.; Thompson, R.J. Eicosapentaenoic acid regulates scallop (Placopecten magellanicus) membrane fluidity in response to cold. Biol. Bull. 2002, 202, 201–203. [Google Scholar] [CrossRef]
- Persson, J.; Vrede, T. Polyunsaturated fatty acids in zooplankton: Variation due to taxonomy and trophic position. Freshwat. Biol. 2006, 51, 887–900. [Google Scholar] [CrossRef]
- Stockner, J.G.; Shortreed, K.S. Algal picoplankton production and contribution to food-webs in oligotrophic British Columbia lakes. Hydrobiologia 1989, 173, 151–166. [Google Scholar] [CrossRef]
- Parrish, C. Essential Fatty Acids in Aquatic Food Webs. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M.T., Arts, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 309–326. [Google Scholar]
- Lee, R.F.; Nevenzel, J.C.; Paffenhöfer, G.A. Importance of wax esters and other lipids in the marine food chain: Phytoplankton and copepods. Mar. Biol. 1971, 9, 99–108. [Google Scholar] [CrossRef]
- Kainz, M.; Arts, M.T.; Mazumder, A. Essential fatty acids in the planktonic food web and their ecological role for higher trophic levels. Limnol. Oceanogr. 2004, 49, 1784–1793. [Google Scholar] [CrossRef]
- Schlechtriem, C.; Arts, M.T.; Zellmer, I.D. Effect of temperature on the fatty acid composition and temporal trajectories of fatty acids in fasting Daphnia pulex (Crustacea, cladocera). Lipids 2006, 41, 397–400. [Google Scholar] [CrossRef]
- Holm-hansen, O.; Riemann, B. Chlorophyll-a determination: Improvements in methodology. Oikos 1978, 30, 438–447. [Google Scholar] [CrossRef]
- Yentsch, C.S.; Menzel, D.W. A method for the determination of phytoplankton chlorophyll and phaeophytin by fluorescence. Deep Sea Res. Oceanogr. Abstr. 1963, 10, 221–231. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Dussart, B. Les Copépodes des eaux continentales d'Europe occidentale. Tome II: Cyclopoides et Biologie, Editions N ed; Boubée & Cie: Paris, France, 1969. [Google Scholar]
- Dussart, B. Les Copépodes des Eaux continentales d'Europe occidentale. Tome I: Calanoides et Harpacticoides, Editions N ed; Boublée & Cie: Paris, France, 1967. [Google Scholar]
- Kiefer, F.; Fryer, G. Das Zooplankton der Binnengewässer, 2. Teil Freilebende Copepoda. Die Binnengewässer 1978, 26, 1–343. (In German) [Google Scholar]
- Scourfield, D.J.; Harding, J.P. A Key to the British Species of Freshwater Cladocera, With Notes on Their Ecology (no 5); Freshwater Biological Association: Windermere, UK, 1958. [Google Scholar]
- Alonso, M. Crustacea. Branchiopoda. In Fauna Ibérica; Ramos., M.A., Ed.; Museo Nacional de Ciencias Naturales; CSIC: Madrid, Spain, 1996; Volume 7, p. 486. [Google Scholar]
- Miracle, M.R. Biogeography of the freshwater zooplanktonic communities of Spain. J. Biogeogr. 1982, 9, 455–467. [Google Scholar] [CrossRef]
- Marrone, F.; Barone, R.; Flores, L.N. Ecological characterization and cladocerans, calanoid copepods and large branchiopods of temporary ponds in a Mediterranean island (Sicily, southern Italy). Chem. Ecol. 2006, 22, 181–190. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Dijkman, N.A.; Boschker, H.T.S.; Middelburg, J.J.; Kromkamp, J.C. Group-specific primary production based on stable-isotope labeling of phospholipid-derived fatty acids. Limnol. Oceanogr-Methods 2009, 7, 612–625. [Google Scholar] [CrossRef]
- Moss, C.W.; Shinoda, T.; Samuels, J.W. Determination of cellular fatty acid compositions of various yeasts by gas-liquid chromatography. J. Clin. Microbiol. 1982, 16, 1073–1079. [Google Scholar]
- Müller, M.M.; Kantola, R.; Kitunen, V. Combining sterol and fatty acid profiles for the characterization of fungi. Mycol. Res. 1994, 98, 593–603. [Google Scholar] [CrossRef]
- Jongman, R.H.G.; Braak, C.J.F.T.; Tongeren, O.F.R.V. Data Analysis in Community and Landscape Ecology; Cambridge University Press: Cambridge, UK, 1995; p. 324. [Google Scholar]
- Sample Availability: Not available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
De Carvalho, C.C.C.R.; Caramujo, M.-J. Fatty Acids as a Tool to Understand Microbial Diversity and Their Role in Food Webs of Mediterranean Temporary Ponds. Molecules 2014, 19, 5570-5598. https://doi.org/10.3390/molecules19055570
De Carvalho CCCR, Caramujo M-J. Fatty Acids as a Tool to Understand Microbial Diversity and Their Role in Food Webs of Mediterranean Temporary Ponds. Molecules. 2014; 19(5):5570-5598. https://doi.org/10.3390/molecules19055570
Chicago/Turabian StyleDe Carvalho, Carla C. C. R., and Maria-José Caramujo. 2014. "Fatty Acids as a Tool to Understand Microbial Diversity and Their Role in Food Webs of Mediterranean Temporary Ponds" Molecules 19, no. 5: 5570-5598. https://doi.org/10.3390/molecules19055570
APA StyleDe Carvalho, C. C. C. R., & Caramujo, M.-J. (2014). Fatty Acids as a Tool to Understand Microbial Diversity and Their Role in Food Webs of Mediterranean Temporary Ponds. Molecules, 19(5), 5570-5598. https://doi.org/10.3390/molecules19055570