Design, Synthesis and Evaluation of the Antibacterial Enhancement Activities of Amino Dihydroartemisinin Derivatives
Abstract
:1. Introduction
2. Results and Discussion
2.1. Computer-Predicted Ligand Binding to AcrB
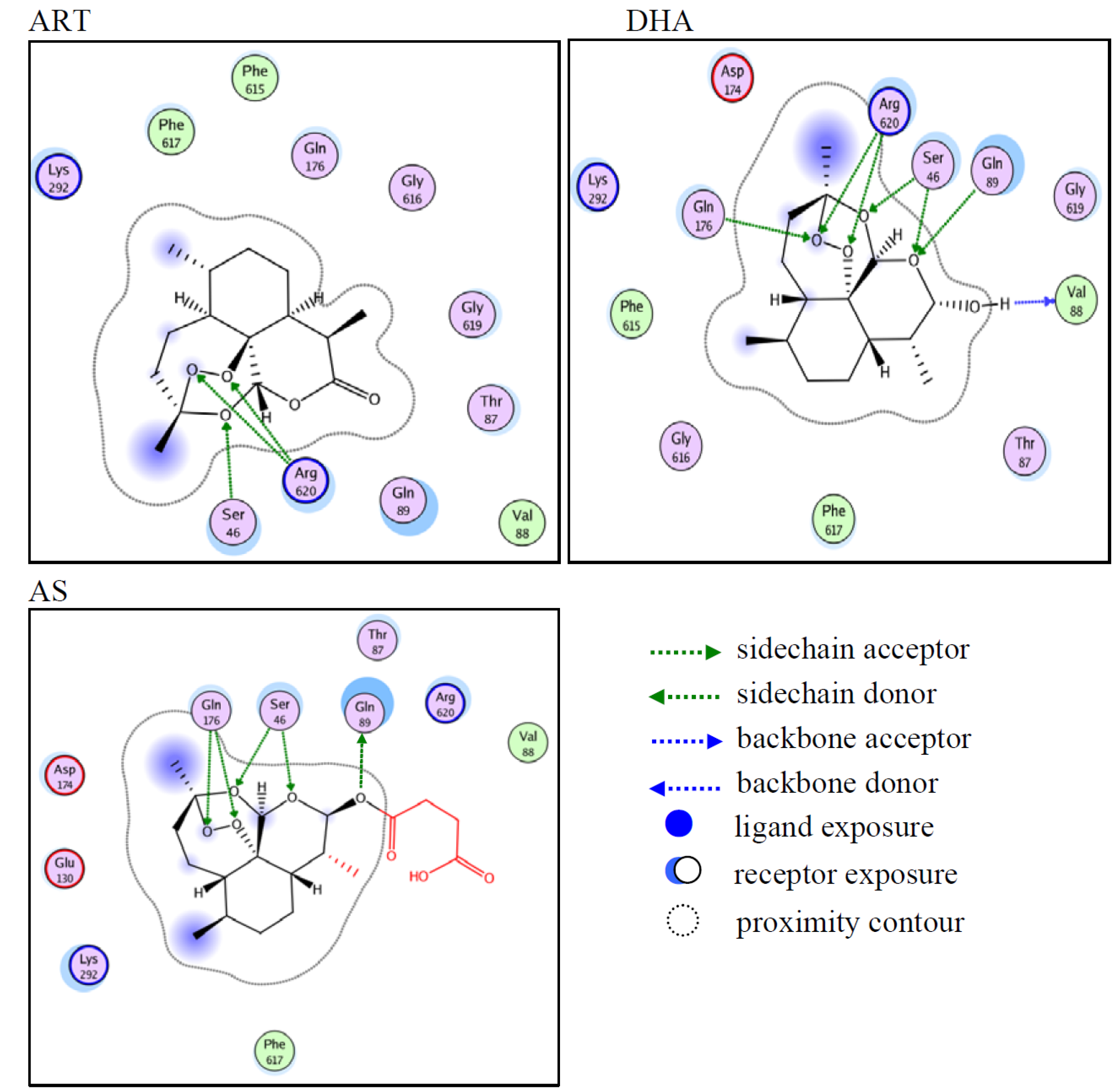
2.2. Synthesis of DHA Derivatives
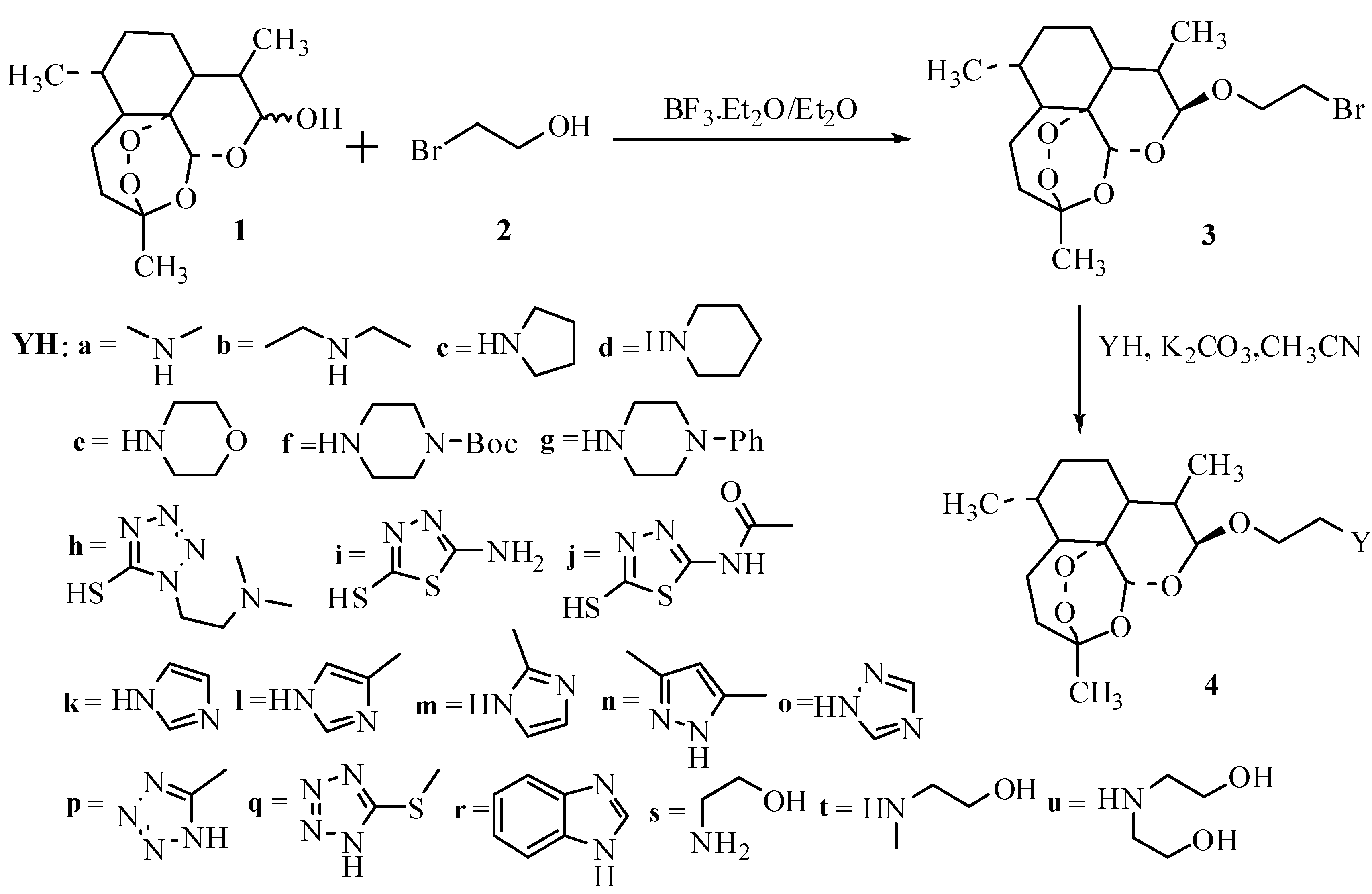
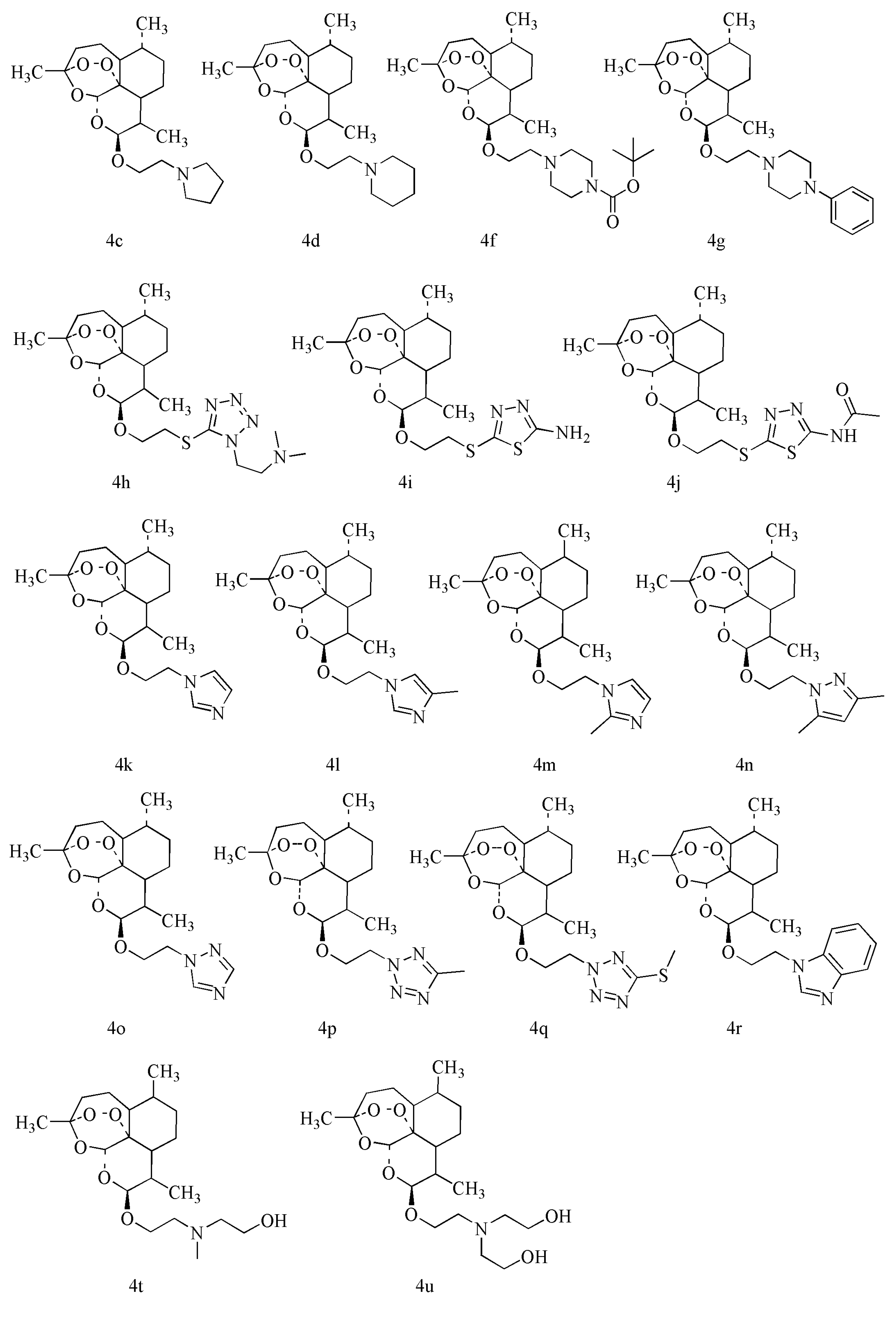
2.3. Antibacterial Enhancement Activity of DHA Derivatives
2.3.1. DHA Derivatives Have no Directly Antibacterial Activity
| Agents | MIC (μg/mL) | Agents | MIC (μg/mL) | |
|---|---|---|---|---|
| AS | >1024 | 4m | 512 | |
| 4a | 2048 | 4n | >2048 | |
| 4b | >2048 | 4o | 1024 | |
| 4c | >2048 | 4p | >2048 | |
| 4d | >2048 | 4r | 1024 | |
| 4e | 1024 | 4s | 1024 | |
| 4h | >2048 | 4t | 2048 | |
| 4i | 2048 | 4u | 1024 | |
| 4k | 512 | AMP | 32 | |
| 4l | 512 | CFX | 512 |
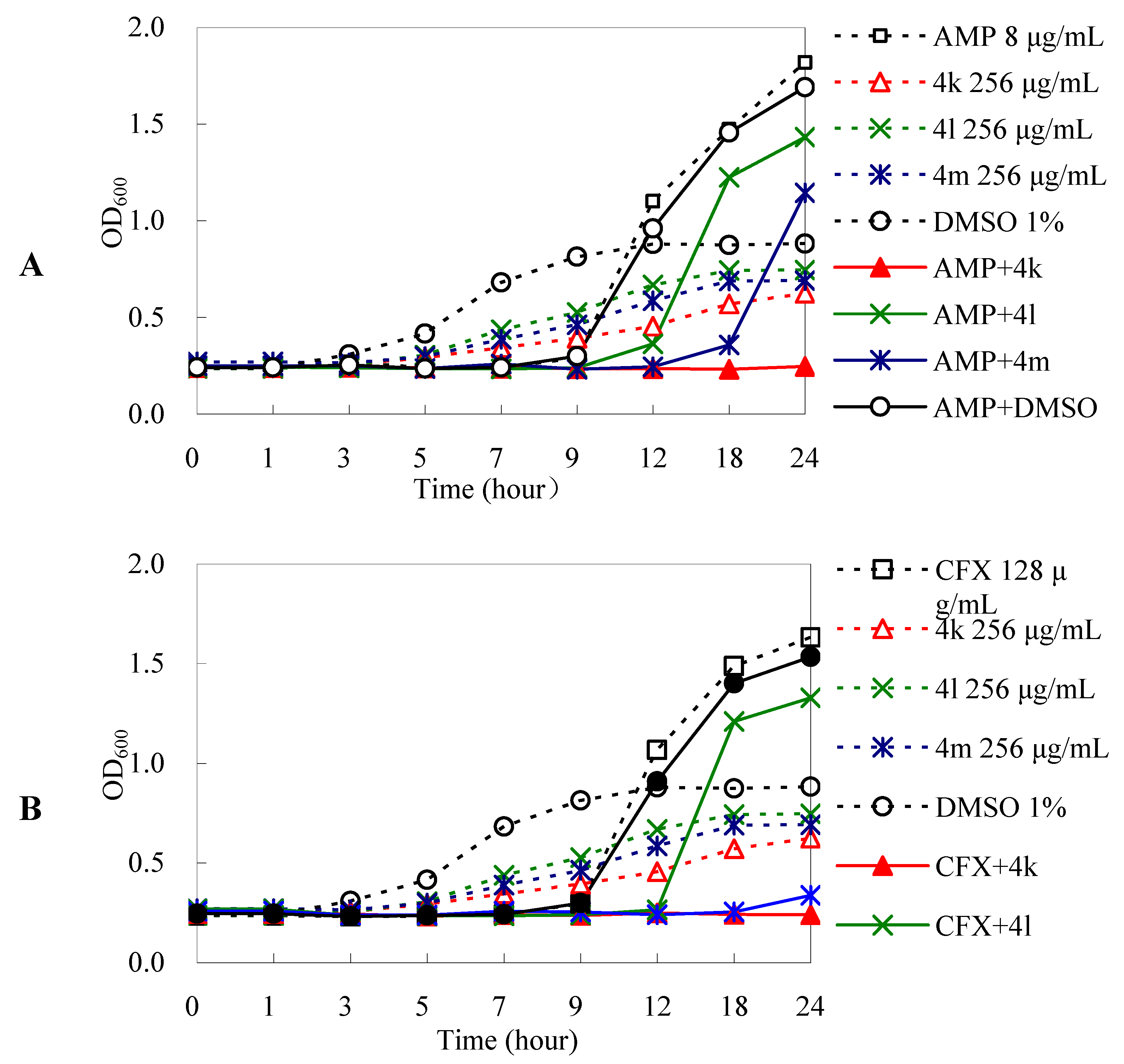
2.3.2. DHA Derivatives Have Antibacterial Enhancement Activities
| Drug concentrations | FICI |
|---|---|
| 1/4 MIC AS + 1/4 MIC AMP | 0.50 |
| 1/4 MIC AS + 1/16 MIC CFX | 0.31 |
| 1/32 MIC 4k + 1/16 MIC AMP | 0.09 |
| 1/64 MIC 4k + 1/64 MIC CFX | 0.03 |
| 1/16 MIC 4l + 1/16 MIC AMP | 0.14 |
| 1/64 MIC 4l + 1/4 MIC CFX | 0.08 |
| 1/16 MIC 4m + 1/16 MIC AMP | 0.13 |
| 1/32 MIC 4m + 1/64 MIC CFX | 0.05 |
| 1/8 MIC 4n + 1/8 MIC AMP | 0.25 |
| 1/8 MIC 4n + 1/64 MIC CFX | 0.14 |
| 1/16 MIC 4r + 1/4 MIC AMP | 0.31 |
| 1/16 MIC 4r + 1/64 MIC CFX | 0.08 |
3. Experimental
3.1. Materials
3.1.1. Reagents and Apparatus
3.1.2. Bacterial Strains
3.2. Methods
3.2.1. Molecular docking
3.2.2. Chemistry
3.2.2.1. Preparation of 10β-(2-Bromoethoxy) Dihydroartemisinin (3)
 + 138.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.94 (3H, d, J = 6.0 Hz, H-13), 0.96 (3H, d, J = 4.8 Hz, H-14), 1.44 (3H, s, H-15), 1.19–2.07 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42(1H, m, H-1), 2.61–2.71 (1H, m, H-11), 3.52 (2H, t, J = 5.7 Hz, H-17), 3.76–3.83 (1H, m, H-16), 4.09–4.17 (1H, m, H-16), 4.85 (1H, d, J = 3.6 Hz, H-12), 5.50 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.1 (C-4), 102.1 (C-12), 88.1 (C-5), 81.1 (C-6), 68.1 (C-16), 52.6 (C-1), 44.3 (C-7), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 31.4 (C-9), 30.9 (C-17), 26.1 (C-8), 24.6 (C-15), 24.3 (C-2), 20.3 (C-14), 13.0 (C-13).
+ 138.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.94 (3H, d, J = 6.0 Hz, H-13), 0.96 (3H, d, J = 4.8 Hz, H-14), 1.44 (3H, s, H-15), 1.19–2.07 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42(1H, m, H-1), 2.61–2.71 (1H, m, H-11), 3.52 (2H, t, J = 5.7 Hz, H-17), 3.76–3.83 (1H, m, H-16), 4.09–4.17 (1H, m, H-16), 4.85 (1H, d, J = 3.6 Hz, H-12), 5.50 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.1 (C-4), 102.1 (C-12), 88.1 (C-5), 81.1 (C-6), 68.1 (C-16), 52.6 (C-1), 44.3 (C-7), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 31.4 (C-9), 30.9 (C-17), 26.1 (C-8), 24.6 (C-15), 24.3 (C-2), 20.3 (C-14), 13.0 (C-13).3.2.2.2. Preparation of Dihydroartemisinin Amino Derivatives 4a–u
| Synthetic Compound | YH | M1/mmol | YH/mmol | K2CO3/mmol | Temp./°C | Time/h | Product/mmol | Yield/% |
|---|---|---|---|---|---|---|---|---|
| 4a | 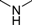 | 1 | 6 | 3 | 11 | 32 | 0.982 | 98.2 |
| 4b | 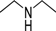 | 1 | 6 | 3 | 45 | 7 | 0.942 | 94.2 |
| 4c | 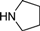 | 1.5 | 9 | 4.5 | 45 | 12 | 1.376 | 91.7 |
| 4d | 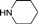 | 1.5 | 9 | 4.5 | 45 | 12 | 1.407 | 93.8 |
| 4e | 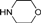 | 1 | 6 | 2 | 10 | 16 | 0.864 | 86.4 |
| 4f | 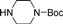 | 1.5 | 3 | 4.5 | 45 | 23 | 1.224 | 81.6 |
| 4g | 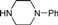 | 1.5 | 3 | 4.5 | 45 | 24 | 1.389 | 92.6 |
| 4h |  | 2 | 3 | 6 | 45 | 6 | 1.424 | 71.2 |
| 4i | 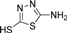 | 1.5 | 3 | 4.5 | 45 | 3 | 1.301 | 86.7 |
| 4j |  | 1.5 | 3 | 4.5 | 45 | 15 | 1.170 | 78.0 |
| 4k | 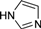 | 1 | 4 | 3 | 45 | 34 | 0.891 | 89.1 |
| 4l | 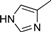 | 1.5 | 3 | 4.5 | 45 | 42 | 1.146 | 76.4 |
| 4m | 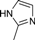 | 1.5 | 3 | 3 | 45 | 23 | 1.146 | 76.4 |
| 4n | 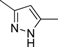 | 2 | 3 | 4 | 45 | 6 | 1.728 | 86.4 |
| 4o |  | 1.5 | 3 | 4.5 | 45 | 22 | 1.431 | 95.4 |
| 4p | 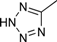 | 1.5 | 3 | 4.5 | 45 | 34 | 0.468 | 31.2 |
| 4q | 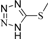 | 1.5 | 3 | 4.5 | 45 | 10.5 | 1.257 | 83.8 |
| 4r | 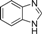 | 2 | 3 | 4 | 45 | 12 | 1.748 | 87.4 |
| 4s |  | 1 | 6 | 2 | 50 | 13 | 0.411 | 41.1 |
| 4t |  | 1 | 6 | 3 | 14 | 24 | 0.925 | 92.5 |
| 4u | 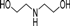 | 1 | 6 | 2 | 50 | 21 | 0.852 | 85.2 |
 + 139.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.44 (3H, s, H-15), 1.22–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.28 (6H, s, H-18), 2.32–2.38 (1H, m, H-1), 2.51–2.54 (2H, m, H-17), 2.60–2.65 (1H, m, H-11), 3.50–3.57 (1H, m, H-16), 3.91–3.98 (1H, m, H-16), 4.80 (1H, d, J = 2.7 Hz, H-12), 5.42 (1H, s, H-5).
+ 139.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.44 (3H, s, H-15), 1.22–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.28 (6H, s, H-18), 2.32–2.38 (1H, m, H-1), 2.51–2.54 (2H, m, H-17), 2.60–2.65 (1H, m, H-11), 3.50–3.57 (1H, m, H-16), 3.91–3.98 (1H, m, H-16), 4.80 (1H, d, J = 2.7 Hz, H-12), 5.42 (1H, s, H-5). + 116.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.05 (6H, t, J = 7.5 Hz, H-19), 1.44 (3H, s, H-15), 1.18–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.32–2.42 (1H, m, H-1), 2.57–2.64 (5H, m, H-18 and H-11), 2.69–2.73 (1H, m, H-17), 3.47–3.55 (1H, m, H-16), 3.91–3.99 (1H, m, H-16), 4.79 (1H, s, H-12), 5.42 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.0 (C-4), 102.1 (C-12), 87.8 (C-5), 81.1 (C-6), 66.7 (C-16), 52.5 (C-1), 52.1 (C-17), 47.4 (C-18), 44.4 (C-7), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 26.2 (C-8), 24.7 (C-15), 24.3 (C-2), 20.4 (C-14), 13.0 (C-13), 11.8 (C-19).
+ 116.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.05 (6H, t, J = 7.5 Hz, H-19), 1.44 (3H, s, H-15), 1.18–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.32–2.42 (1H, m, H-1), 2.57–2.64 (5H, m, H-18 and H-11), 2.69–2.73 (1H, m, H-17), 3.47–3.55 (1H, m, H-16), 3.91–3.99 (1H, m, H-16), 4.79 (1H, s, H-12), 5.42 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.0 (C-4), 102.1 (C-12), 87.8 (C-5), 81.1 (C-6), 66.7 (C-16), 52.5 (C-1), 52.1 (C-17), 47.4 (C-18), 44.4 (C-7), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 26.2 (C-8), 24.7 (C-15), 24.3 (C-2), 20.4 (C-14), 13.0 (C-13), 11.8 (C-19). + 140.0 (c 1.1 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.5 Hz, H-13), 0.95 (3H, d, J = 6.3 Hz, H-14), 1.44 (3H, s, H-15), 1.22–2.06 (14H, m, H-2, H-3, H-7, H-8, H-9, H-10, H-18 and H-19), 2.32–2.42 (1H, m, H-1), 2.59–2.80 (7H, m, H-11, H-17 and H-18), 3.54–3.61 (1H, m, H-16), 3.95–4.02 (1H, m, H-16), 4.61 (1H, s, H-12), 5.44 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.0 (C-4), 102.0 (C-12), 87.8 (C-5), 81.0 (C-6), 67.1 (C-16), 55.3 (C-17) 54.5 (C-18), 52.5 (C-1), 44.4 (C-7), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 23.4 (C-19), 20.3 (C-14), 13.0 (C-13).
+ 140.0 (c 1.1 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.5 Hz, H-13), 0.95 (3H, d, J = 6.3 Hz, H-14), 1.44 (3H, s, H-15), 1.22–2.06 (14H, m, H-2, H-3, H-7, H-8, H-9, H-10, H-18 and H-19), 2.32–2.42 (1H, m, H-1), 2.59–2.80 (7H, m, H-11, H-17 and H-18), 3.54–3.61 (1H, m, H-16), 3.95–4.02 (1H, m, H-16), 4.61 (1H, s, H-12), 5.44 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.0 (C-4), 102.0 (C-12), 87.8 (C-5), 81.0 (C-6), 67.1 (C-16), 55.3 (C-17) 54.5 (C-18), 52.5 (C-1), 44.4 (C-7), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 23.4 (C-19), 20.3 (C-14), 13.0 (C-13). + 135.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-13), 0.96 (3H, d, J = 6.0 Hz, H-14), 1.44 (3H, s, H-15), 1.22–2.06 (16H, m, H-2, H-3, H-7, H-8, H-9, H-10, H-19 and H-20), 2.32–2.60 (8H, m, H-1, H-11, H-17 and H-18), 3.55–3.59 (1H, m, H-16), 3.92–4.00 (1H, m, H-16), 4.79 (1H, s, H-12), 5.44 (1H, s, H-5).
+ 135.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-13), 0.96 (3H, d, J = 6.0 Hz, H-14), 1.44 (3H, s, H-15), 1.22–2.06 (16H, m, H-2, H-3, H-7, H-8, H-9, H-10, H-19 and H-20), 2.32–2.60 (8H, m, H-1, H-11, H-17 and H-18), 3.55–3.59 (1H, m, H-16), 3.92–4.00 (1H, m, H-16), 4.79 (1H, s, H-12), 5.44 (1H, s, H-5). + 59.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.5 Hz, H-13), 0.96 (3H, d, J = 6.3 Hz, H-14), 1.44 (3H, s, H-15), 1.26–2.05 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42 (1H, m, H-1), 2.49–2.63 (7H, m, H-11, H-17 and H-18), 3.54–3.62 (1H, m, H-16), 3.72 (3H, t, J = 4.5 Hz, H-19), 3.94–4.01(1H, m, H-16), 4.81 (1H, d, J = 3.3 Hz, H-12), 5.47 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.0 (C-4), 102.0 (C-12), 87.9 (C-5), 81.1 (C-6), 66.9 (C-19), 65.6 (C-16), 58.2 (C-17) 53.8 (C-18), 52.5 (C-1), 44.4 (C-7), 37.5 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 26.2 (C-8), 24.7 (C-15), 24.4 (C-2), 20.4 (C-14), 13.0 (C-13).
+ 59.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.5 Hz, H-13), 0.96 (3H, d, J = 6.3 Hz, H-14), 1.44 (3H, s, H-15), 1.26–2.05 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42 (1H, m, H-1), 2.49–2.63 (7H, m, H-11, H-17 and H-18), 3.54–3.62 (1H, m, H-16), 3.72 (3H, t, J = 4.5 Hz, H-19), 3.94–4.01(1H, m, H-16), 4.81 (1H, d, J = 3.3 Hz, H-12), 5.47 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.0 (C-4), 102.0 (C-12), 87.9 (C-5), 81.1 (C-6), 66.9 (C-19), 65.6 (C-16), 58.2 (C-17) 53.8 (C-18), 52.5 (C-1), 44.4 (C-7), 37.5 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 26.2 (C-8), 24.7 (C-15), 24.4 (C-2), 20.4 (C-14), 13.0 (C-13). +122.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-13), 0.96 (3H, d, J = 6.0 Hz, H-14), 1.43 (3H, s, H-15), 1.46 (9H, s, H-22), 1.22–2.06 (16H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.65 (8H, m, H-1, H-11, H-17 and H-18), 3.40–3.44 (4H, m, H-19), 3.52–3.58 (1H, m, H-16), 3.92–4.00 (1H, m, H-16), 4.80 (1H, s, H-12), 5.46 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 154.6 (C-21), 104.0 (C-4), 101.9 (C-12), 87.8 (C-5), 81.0 (C-6), 79.7 (C-22), 66.0 (C-16), 57.9 (C-17), 53.5 (C-19), 52.0 (C-1), 49.2 (C-18), 44.4 (C-7), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 28.4 (C-23), 26.4 (C-17), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 20.4 (C-14), 13.0 (C-13).
+122.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-13), 0.96 (3H, d, J = 6.0 Hz, H-14), 1.43 (3H, s, H-15), 1.46 (9H, s, H-22), 1.22–2.06 (16H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.65 (8H, m, H-1, H-11, H-17 and H-18), 3.40–3.44 (4H, m, H-19), 3.52–3.58 (1H, m, H-16), 3.92–4.00 (1H, m, H-16), 4.80 (1H, s, H-12), 5.46 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 154.6 (C-21), 104.0 (C-4), 101.9 (C-12), 87.8 (C-5), 81.0 (C-6), 79.7 (C-22), 66.0 (C-16), 57.9 (C-17), 53.5 (C-19), 52.0 (C-1), 49.2 (C-18), 44.4 (C-7), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 28.4 (C-23), 26.4 (C-17), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 20.4 (C-14), 13.0 (C-13). +74.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-13), 0.95 (3H, d, J = 6.0 Hz, H-14), 1.44 (3H, s, H-15), 1.23–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.32–2.42 (1H, m, H-1), 2.59–2.70 (7H, m, H-11, H-17 and H-18), 3.21 (4H, t, J = 7.5 Hz, H-19), 3.58–3.65 (1H, m, H-16), 3.98–4.05 (1H, m, H-16), 4.83 (1H, d, J = 2.7 Hz, H-12), 5.48 (1H, s, H-5), 6.89 (1H, t, J = 7.2 Hz, H-23), 6.94 (2H, d, J = 7.8 Hz, H-21), 7.25–7.30 (2H, m, H-22); 13C-NMR (CDCl3) δ ppm: 151.3 (C-20), 129.1 (C-22), 120.0 (C-23), 116.0 (C-21), 104.0 (C-4), 102.0 (C-12), 87.9 (C-5), 81.1 (C-6), 66.0 (C-16), 57.9 (C-17), 53.5 (C-19), 52.6 (C-1), 49.2 (C-18), 44.4 (C-7), 37.5 (C-11), 36.4 (C-10), 34.7 (C-3), 30.9 (C-9), 26.2 (C-8), 24.7 (C-15), 24.4 (C-2), 20.3 (C-14), 13.1 (C-13); HRMS calcd for C27H40N2O5 (M+H)+ 473.3010, found 473.3014.
+74.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-13), 0.95 (3H, d, J = 6.0 Hz, H-14), 1.44 (3H, s, H-15), 1.23–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.32–2.42 (1H, m, H-1), 2.59–2.70 (7H, m, H-11, H-17 and H-18), 3.21 (4H, t, J = 7.5 Hz, H-19), 3.58–3.65 (1H, m, H-16), 3.98–4.05 (1H, m, H-16), 4.83 (1H, d, J = 2.7 Hz, H-12), 5.48 (1H, s, H-5), 6.89 (1H, t, J = 7.2 Hz, H-23), 6.94 (2H, d, J = 7.8 Hz, H-21), 7.25–7.30 (2H, m, H-22); 13C-NMR (CDCl3) δ ppm: 151.3 (C-20), 129.1 (C-22), 120.0 (C-23), 116.0 (C-21), 104.0 (C-4), 102.0 (C-12), 87.9 (C-5), 81.1 (C-6), 66.0 (C-16), 57.9 (C-17), 53.5 (C-19), 52.6 (C-1), 49.2 (C-18), 44.4 (C-7), 37.5 (C-11), 36.4 (C-10), 34.7 (C-3), 30.9 (C-9), 26.2 (C-8), 24.7 (C-15), 24.4 (C-2), 20.3 (C-14), 13.1 (C-13); HRMS calcd for C27H40N2O5 (M+H)+ 473.3010, found 473.3014. + 72.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-13), 0.96 (3H, d, J = 5.7 Hz, H-14), 1.44 (3H, s, H-15), 1.20–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.29 (6H, s, H-21), 2.36–2.42(1H, m, H-1), 2.59–2.65(1H, m, H-11), 2.79 (2H, t, J = 6.6 Hz, H-20), 3.60 (2H, t, J = 5.7 Hz, H-17), 3.74–3.81 (1H, m, H-16), 4.11–4.18 (1H, m, H-16), 4.32 (2H, t, J = 6.6 Hz, H-19), 4.82 (1H, s, H-12), 5.42 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 154.1 (C-18), 104.1 (C-4), 102.2 (C-12), 88.0 (C-5), 80.9 (C-6), 66.5 (C-16), 57.2 (C-20), 52.4 (C-1), 45.1 (C-21), 44.2 (C-7), 37.4 (C-11), 36.3 (C-10), 34.5 (C-3), 33.7 (C-17), 30.7 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 20.3 (C-14), 12.9 (C-13).
+ 72.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J = 7.2 Hz, H-13), 0.96 (3H, d, J = 5.7 Hz, H-14), 1.44 (3H, s, H-15), 1.20–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.29 (6H, s, H-21), 2.36–2.42(1H, m, H-1), 2.59–2.65(1H, m, H-11), 2.79 (2H, t, J = 6.6 Hz, H-20), 3.60 (2H, t, J = 5.7 Hz, H-17), 3.74–3.81 (1H, m, H-16), 4.11–4.18 (1H, m, H-16), 4.32 (2H, t, J = 6.6 Hz, H-19), 4.82 (1H, s, H-12), 5.42 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 154.1 (C-18), 104.1 (C-4), 102.2 (C-12), 88.0 (C-5), 80.9 (C-6), 66.5 (C-16), 57.2 (C-20), 52.4 (C-1), 45.1 (C-21), 44.2 (C-7), 37.4 (C-11), 36.3 (C-10), 34.5 (C-3), 33.7 (C-17), 30.7 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 20.3 (C-14), 12.9 (C-13). + 59.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.2 Hz, H-13), 0.96 (3H, d, J =6.0 Hz, H-14), 1.43 (3H, s, H-15), 1.22–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42 (1H, m, H-1), 2.56–2.67 (1H, m, H-11), 3.41 (2H, t, J = 5.7 Hz, H-17), 3.70–3.77 (1H, m, H-16), 4.06–4.14 (1H, m, H-16), 4.82 (1H, s, H-12), 5.44 (1H, s, H-5); 13C-NMR (DMSO and CDCl3) δ ppm: 103.3 (C-4), 101.3 (C-12), 87.2 (C-5), 80.4 (C-6), 66.1 (C-16), 52.0 (C-1), 43.8 (C-7), 36.7 (C-11), 35.9 (C-10), 34.6 (C-3), 34.1 (C-17), 30.3 (C-9), 25.6 (C-8), 24.1 (C-15), 23.8 (C-2), 20.0 (C-14), 12.6 (C-13); HRMS calcd for C19H29N3O5S2 (M+Na)+ 466.1441, found 466.1440.
+ 59.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.2 Hz, H-13), 0.96 (3H, d, J =6.0 Hz, H-14), 1.43 (3H, s, H-15), 1.22–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42 (1H, m, H-1), 2.56–2.67 (1H, m, H-11), 3.41 (2H, t, J = 5.7 Hz, H-17), 3.70–3.77 (1H, m, H-16), 4.06–4.14 (1H, m, H-16), 4.82 (1H, s, H-12), 5.44 (1H, s, H-5); 13C-NMR (DMSO and CDCl3) δ ppm: 103.3 (C-4), 101.3 (C-12), 87.2 (C-5), 80.4 (C-6), 66.1 (C-16), 52.0 (C-1), 43.8 (C-7), 36.7 (C-11), 35.9 (C-10), 34.6 (C-3), 34.1 (C-17), 30.3 (C-9), 25.6 (C-8), 24.1 (C-15), 23.8 (C-2), 20.0 (C-14), 12.6 (C-13); HRMS calcd for C19H29N3O5S2 (M+Na)+ 466.1441, found 466.1440. + 83.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.89 (3H, d, J = 7.2 Hz, H-13), 0.94 (3H, d, J =6.0 Hz, H-14), 1.42 (3H, s, H-15), 1.22–2.05 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42 (1H, m, H-1), 2.49 (3H, s, H-21), 2.58–2.64 (1H, m, H-11), 3.44–3.57 (2H, m, H-17), 3.75–3.81 (1H, m, H-16), 4.13–4.20 (1H, m, H-16), 4.84 (1H, s, H-12), 5.42 (1H, s, H-5), 13.08 (1H, s, H-20); 13C-NMR (CDCl3) δ ppm: 168.5 (C-20), 104.1 (C-4), 102.2 (C-12), 88.0 (C-5), 81.0 (C-6), 66.5 (C-16), 52.5 (C-1), 44.3 (C-7), 37.4 (C-11), 36.3 (C-10), 34.6 (C-3), 34.1 (C-17), 30.8 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 23.0 (C-21), 20.3 (C-14), 12.9 (C-13); HRMS calcd for C21H31N3O6S2 (M+Na)+ 508.1546, found 508.1540.
+ 83.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.89 (3H, d, J = 7.2 Hz, H-13), 0.94 (3H, d, J =6.0 Hz, H-14), 1.42 (3H, s, H-15), 1.22–2.05 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42 (1H, m, H-1), 2.49 (3H, s, H-21), 2.58–2.64 (1H, m, H-11), 3.44–3.57 (2H, m, H-17), 3.75–3.81 (1H, m, H-16), 4.13–4.20 (1H, m, H-16), 4.84 (1H, s, H-12), 5.42 (1H, s, H-5), 13.08 (1H, s, H-20); 13C-NMR (CDCl3) δ ppm: 168.5 (C-20), 104.1 (C-4), 102.2 (C-12), 88.0 (C-5), 81.0 (C-6), 66.5 (C-16), 52.5 (C-1), 44.3 (C-7), 37.4 (C-11), 36.3 (C-10), 34.6 (C-3), 34.1 (C-17), 30.8 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 23.0 (C-21), 20.3 (C-14), 12.9 (C-13); HRMS calcd for C21H31N3O6S2 (M+Na)+ 508.1546, found 508.1540. + 68.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.85 (3H, d, J =7.2 Hz, H-14), 0.94 (3H, d, J = 6.1 Hz, H-13), 1.44 (3H, s, H-15), 1.22–2.07 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.32–2.38 (1H, m, H-1), 2.61–2.63 (1H, m, H-11), 3.62–3.68 (1H, m, H-16), 4.15–4.21 (3H, m, H-16 and H-17), 4.76 (1H, s, H-12), 5.11 (1H, s, H-5). 7.00 (1H, s, H-19), 7.09 (1H, s, H-18), 7.62 (1H, s, H-20); 13C-NMR (CDCl3) δ ppm: 128.7 (C-20), 123.1 (C-19), 118.9 (C-18), 104.1 (C-4), 102.0 (C-12), 87.8 (C-5), 80.8 (C-6), 67.0 (C-16), 52.3 (C-1), 47.2 (C-17), 44.0 (C-7), 37.2 (C-11), 36.3 (C-10), 34.4 (C-3), 30.6 (C-9), 26.1 (C-8), 24.6 (C-15), 24.3 (C-2), 20.3 (C-14), 13.0 (C-13); HRMS calcd for C20H30N2O5 (M+H)+ 379.2228, found 379.2221.
+ 68.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.85 (3H, d, J =7.2 Hz, H-14), 0.94 (3H, d, J = 6.1 Hz, H-13), 1.44 (3H, s, H-15), 1.22–2.07 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.32–2.38 (1H, m, H-1), 2.61–2.63 (1H, m, H-11), 3.62–3.68 (1H, m, H-16), 4.15–4.21 (3H, m, H-16 and H-17), 4.76 (1H, s, H-12), 5.11 (1H, s, H-5). 7.00 (1H, s, H-19), 7.09 (1H, s, H-18), 7.62 (1H, s, H-20); 13C-NMR (CDCl3) δ ppm: 128.7 (C-20), 123.1 (C-19), 118.9 (C-18), 104.1 (C-4), 102.0 (C-12), 87.8 (C-5), 80.8 (C-6), 67.0 (C-16), 52.3 (C-1), 47.2 (C-17), 44.0 (C-7), 37.2 (C-11), 36.3 (C-10), 34.4 (C-3), 30.6 (C-9), 26.1 (C-8), 24.6 (C-15), 24.3 (C-2), 20.3 (C-14), 13.0 (C-13); HRMS calcd for C20H30N2O5 (M+H)+ 379.2228, found 379.2221. +58.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91(3H, d, J = 6.9 Hz, H-14), 0.95 (3H, d, J = 6.3 Hz, H-13), 1.45 (3H, s, H-15), 1.19–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.12 (3H, s, H-21), 2.32–2.43 (1H, m, H-1), 2.59–2.66 (1H, m, H-11), 3.40–3.52 (4H, m, H-16 and H-17), 4.69 (1H, d, J = 2.7 Hz, H-12), 5.39 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 127.8 (C-18), 127.2 (C-20), 118.8 (C-19), 104.2 (C-4), 102.0 (C-12), 87.8 (C-5), 80.9 (C-6), 66.9 (C-16), 52.4 (C-1), 47.8 (C-17), 44.0 (C-7), 37.2 (C-11), 36.3 (C-10), 34.4 (C-3), 30.6 (C-9), 26.1 (C-8), 24.6 (C-15), 24.2 (C-2), 20.3 (C-14), 13.0 (C-13), 12.7 (C-21); HRMS calcd for C21H32N2O5 (M+H)+ 393.2384, found 393.2382.
+58.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91(3H, d, J = 6.9 Hz, H-14), 0.95 (3H, d, J = 6.3 Hz, H-13), 1.45 (3H, s, H-15), 1.19–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.12 (3H, s, H-21), 2.32–2.43 (1H, m, H-1), 2.59–2.66 (1H, m, H-11), 3.40–3.52 (4H, m, H-16 and H-17), 4.69 (1H, d, J = 2.7 Hz, H-12), 5.39 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 127.8 (C-18), 127.2 (C-20), 118.8 (C-19), 104.2 (C-4), 102.0 (C-12), 87.8 (C-5), 80.9 (C-6), 66.9 (C-16), 52.4 (C-1), 47.8 (C-17), 44.0 (C-7), 37.2 (C-11), 36.3 (C-10), 34.4 (C-3), 30.6 (C-9), 26.1 (C-8), 24.6 (C-15), 24.2 (C-2), 20.3 (C-14), 13.0 (C-13), 12.7 (C-21); HRMS calcd for C21H32N2O5 (M+H)+ 393.2384, found 393.2382. + 52.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.85 (3H, d, J =7.2 Hz, H-14), 0.94 (3H, d, J = 4.8 Hz, H-13), 1.43 (3H, s, H-15), 1.22–2.07 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.29–2.34 (1H, m, H-1), 2.41 (3H, s, H-21), 2.61–2.63(1H, m, H-11), 3.58–3.63 (1H, m, H-16), 3.99-4.16 (3H, m, H-16 and H-17), 4.76 (1H, d, J = 3.3 Hz, H-12), 5.08 (1H, s, H-5). 6.88 (1H, s, H-19), 6.92 (1H, s, H-18); 13C-NMR (CDCl3) δ ppm: 126.7 (C-20 and C-19), 118.9 (C-18), 104.2 (C-4), 102.0 (C-12), 87.8 (C-5), 80.9 (C-6), 66.8 (C-16), 52.4 (C-1), 45.8 (C-17), 44.0 (C-7), 37.2 (C-11), 36.3 (C-10), 34.4 (C-3), 30.6 (C-9), 26.1 (C-8), 24.6 (C-15), 24.2 (C-2), 20.3 (C-14), 13.0 (C-13), 12.9 (C-21); HRMS calcd for C21H32N2O5 (M+H)+ 393.2384, found 393.2386.
+ 52.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.85 (3H, d, J =7.2 Hz, H-14), 0.94 (3H, d, J = 4.8 Hz, H-13), 1.43 (3H, s, H-15), 1.22–2.07 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.29–2.34 (1H, m, H-1), 2.41 (3H, s, H-21), 2.61–2.63(1H, m, H-11), 3.58–3.63 (1H, m, H-16), 3.99-4.16 (3H, m, H-16 and H-17), 4.76 (1H, d, J = 3.3 Hz, H-12), 5.08 (1H, s, H-5). 6.88 (1H, s, H-19), 6.92 (1H, s, H-18); 13C-NMR (CDCl3) δ ppm: 126.7 (C-20 and C-19), 118.9 (C-18), 104.2 (C-4), 102.0 (C-12), 87.8 (C-5), 80.9 (C-6), 66.8 (C-16), 52.4 (C-1), 45.8 (C-17), 44.0 (C-7), 37.2 (C-11), 36.3 (C-10), 34.4 (C-3), 30.6 (C-9), 26.1 (C-8), 24.6 (C-15), 24.2 (C-2), 20.3 (C-14), 13.0 (C-13), 12.9 (C-21); HRMS calcd for C21H32N2O5 (M+H)+ 393.2384, found 393.2386. + 49.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.81 (3H, d, J = 7.2 Hz, H-14), 0.87 (3H, d, J = 6.3Hz, H-13), 1.42 (3H, s, H-15), 1.26–2.08 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.29–2.37 (1H, m, H-1), 2.21 (3H, s, H-21), 2.28 (3H, s, H-22), 2.51–2.59 (1H, m, H-11), 3.64–3.69 (1H, m, H-16), 4.12–4.28 (3H, m, H-16 and H-17), 4.75 (1H, d, J = 2.7 Hz, H-12), 5.07 (1H, s, H-5), 5.86 (1H, s, H-19); 13C-NMR (CDCl3) δ ppm: 147.4 (C-20), 139.4 (C-20), 104.6 (C-4), 103.9 (C-19), 101.7 (C-12), 87.6 (C-5), 80.9 (C-6), 66.4 (C-16), 52.3 (C-1), 47.9 (C-17), 44.2 (C-7), 37.0 (C-11), 36.3 (C-10), 34.6 (C-3), 30.7 (C-9), 26.1 (C-8), 24.5 (C-15), 24.0 (C-2), 20.2 (C-14), 13.0 (C-13), 12.8 (C-21), 11.1 (C-22); HRMS calcd for C22H34N2O5 (M+Na)+ 429.2360, found 429.2356.
+ 49.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.81 (3H, d, J = 7.2 Hz, H-14), 0.87 (3H, d, J = 6.3Hz, H-13), 1.42 (3H, s, H-15), 1.26–2.08 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.29–2.37 (1H, m, H-1), 2.21 (3H, s, H-21), 2.28 (3H, s, H-22), 2.51–2.59 (1H, m, H-11), 3.64–3.69 (1H, m, H-16), 4.12–4.28 (3H, m, H-16 and H-17), 4.75 (1H, d, J = 2.7 Hz, H-12), 5.07 (1H, s, H-5), 5.86 (1H, s, H-19); 13C-NMR (CDCl3) δ ppm: 147.4 (C-20), 139.4 (C-20), 104.6 (C-4), 103.9 (C-19), 101.7 (C-12), 87.6 (C-5), 80.9 (C-6), 66.4 (C-16), 52.3 (C-1), 47.9 (C-17), 44.2 (C-7), 37.0 (C-11), 36.3 (C-10), 34.6 (C-3), 30.7 (C-9), 26.1 (C-8), 24.5 (C-15), 24.0 (C-2), 20.2 (C-14), 13.0 (C-13), 12.8 (C-21), 11.1 (C-22); HRMS calcd for C22H34N2O5 (M+Na)+ 429.2360, found 429.2356. + 96.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.78 (3H, d, J =7.2 Hz, H-14), 0.94 (3H, d, J = 4.5 Hz, H-13), 1.42 (3H, s, H-15), 1.19–2.08 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.31–2.39 (1H, m, H-1), 2.58–2.62 (1H, m, H-11), 3.74–3.81 (1H, m, H-16), 4.09–4.17 (1H, m, H-16), 4.36–4.38 (2H, m, H-17), 4.78 (1H, s, H-12), 5.17 (1H, s, H-5), 7.96 (1H, s, H-19), 8.12 (1H, s, H-18).
+ 96.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.78 (3H, d, J =7.2 Hz, H-14), 0.94 (3H, d, J = 4.5 Hz, H-13), 1.42 (3H, s, H-15), 1.19–2.08 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.31–2.39 (1H, m, H-1), 2.58–2.62 (1H, m, H-11), 3.74–3.81 (1H, m, H-16), 4.09–4.17 (1H, m, H-16), 4.36–4.38 (2H, m, H-17), 4.78 (1H, s, H-12), 5.17 (1H, s, H-5), 7.96 (1H, s, H-19), 8.12 (1H, s, H-18). + 93.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.75 (3H, d, J = 7.2 Hz, H-14), 0.94 (3H, d, J = 4.5 Hz, H-13), 1.42 (3H, s, H-15), 1.21–2.04 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.29–2.38 (H, m, H-1), 2.57 (3H, s, H-19), 2.58–2.61 (H, m, H-11), 3.80–3.87 (1H, m, H-16), 4.34–4.37 (1H, m, H-16), 4.46–4.48 (2H, m, H-17), 4.76 (1H, s, H-12), 5.11 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 152.1 (C-18), 104.2 (C-4), 102.3 (C-12), 87.7 (C-5), 80.7 (C-6), 65.8 (C-16), 52.3 (C-1), 47.0 (C-17), 43.9 (C-7), 37.2 (C-11), 36.2 (C-10), 34.3 (C-3), 30.5 (C-9), 26.0 (C-8), 24.5 (C-15), 24.2 (C-2), 20.3 (C-14), 13.0 (C-13), 8.9 (C-19); HRMS calcd for C19H30N4O5 (M+Na)+ 417.2108, found 417.2106.
+ 93.0 (c 1.1 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.75 (3H, d, J = 7.2 Hz, H-14), 0.94 (3H, d, J = 4.5 Hz, H-13), 1.42 (3H, s, H-15), 1.21–2.04 (10H, m, H-2, H-3, H-7, H-8, H-9, and H-10), 2.29–2.38 (H, m, H-1), 2.57 (3H, s, H-19), 2.58–2.61 (H, m, H-11), 3.80–3.87 (1H, m, H-16), 4.34–4.37 (1H, m, H-16), 4.46–4.48 (2H, m, H-17), 4.76 (1H, s, H-12), 5.11 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 152.1 (C-18), 104.2 (C-4), 102.3 (C-12), 87.7 (C-5), 80.7 (C-6), 65.8 (C-16), 52.3 (C-1), 47.0 (C-17), 43.9 (C-7), 37.2 (C-11), 36.2 (C-10), 34.3 (C-3), 30.5 (C-9), 26.0 (C-8), 24.5 (C-15), 24.2 (C-2), 20.3 (C-14), 13.0 (C-13), 8.9 (C-19); HRMS calcd for C19H30N4O5 (M+Na)+ 417.2108, found 417.2106. + 126.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J =7.5 Hz, H-14), 0.96 (3H, d, J = 6.3 Hz, H-13), 1.43 (3H, s, H-15), 1.21–2.04 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.43 (H, m, H-1), 2.61–2.66 (H, m, H-11), 3.63 (2H, t, J =5.7 Hz, H-17), 3.75–3.83 (1H, m, H-16), 3.93 (3H, s, H-19), 4.12–4.19 (H, m, H-16), 4.83 (1H, d, J =2.7 Hz, H-12), 5.43 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 154.0 (C-18), 104.1 (C-4), 102.2 (C-12), 87.9 (C-5), 80.9 (C-6), 66.5 (C-16), 52.4 (C-1), 44.2 (C-7), 37.4 (C-11), 36.3 (C-10), 34.5 (C-3), 33.6 (C-17), 33.4 (C-19), 30.7 (C-9), 26.1 (C-8), 24.6 (C-15), 24.3 (C-2), 20.3 (C-14), 12.9 (C-13); HRMS calcd for C19H30N4O5S (M+Na)+ 449.1829, found 449.1821.
+ 126.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.90 (3H, d, J =7.5 Hz, H-14), 0.96 (3H, d, J = 6.3 Hz, H-13), 1.43 (3H, s, H-15), 1.21–2.04 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.43 (H, m, H-1), 2.61–2.66 (H, m, H-11), 3.63 (2H, t, J =5.7 Hz, H-17), 3.75–3.83 (1H, m, H-16), 3.93 (3H, s, H-19), 4.12–4.19 (H, m, H-16), 4.83 (1H, d, J =2.7 Hz, H-12), 5.43 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 154.0 (C-18), 104.1 (C-4), 102.2 (C-12), 87.9 (C-5), 80.9 (C-6), 66.5 (C-16), 52.4 (C-1), 44.2 (C-7), 37.4 (C-11), 36.3 (C-10), 34.5 (C-3), 33.6 (C-17), 33.4 (C-19), 30.7 (C-9), 26.1 (C-8), 24.6 (C-15), 24.3 (C-2), 20.3 (C-14), 12.9 (C-13); HRMS calcd for C19H30N4O5S (M+Na)+ 449.1829, found 449.1821. + 94.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.76 (3H, d, J = 7.5 Hz, H-14), 0.83 (3H, d, J = 6.0 Hz, H-13), 1.39 (3H, s, H-15), 1.15–1.99 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.24–2.35 (H, m, H-1), 2.52–2.59 (H, m, H-11), 3.70–3.76 (1H, m, H-16), 4.28–4.43 (3H, m, H-16 and H-17), 4.73 (1H, d, J =2.7 Hz, H-12), 4.91 (1H, s, H-5), 7.30 (2H, t, J =2.4 Hz, H-21 and H-22), 7.42 (1H, d, J =7.5 Hz, H-23), 7.80 (1H, d, J =7.5 Hz, H-20), 7.95 (1H, s, H-18); 13C-NMR (CDCl3) δ ppm: 143.5 (C-18), 143.3 (C-19), 133.6 (C-24), 122.9 (C-20), 122.1 (C-21), 120.2 (C-22), 109.6 (C-23), 104.0 (C-4), 102.0 (C-12), 87.6 (C-5), 80.7 (C-6), 65.7 (C-16), 52.2 (C-1), 44.8 (C-17), 43.9 (C-7), 37.0 (C-11), 36.2 (C-10), 34.2 (C-3), 30.5 (C-9), 26.0 (C-8), 24.4 (C-15), 24.2 (C-2), 20.1 (C-14), 12.8 (C-13).
+ 94.0 (c 1.0 mg/mL, CH2Cl2). 1H-NMR (CDCl3) δ ppm: 0.76 (3H, d, J = 7.5 Hz, H-14), 0.83 (3H, d, J = 6.0 Hz, H-13), 1.39 (3H, s, H-15), 1.15–1.99 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.24–2.35 (H, m, H-1), 2.52–2.59 (H, m, H-11), 3.70–3.76 (1H, m, H-16), 4.28–4.43 (3H, m, H-16 and H-17), 4.73 (1H, d, J =2.7 Hz, H-12), 4.91 (1H, s, H-5), 7.30 (2H, t, J =2.4 Hz, H-21 and H-22), 7.42 (1H, d, J =7.5 Hz, H-23), 7.80 (1H, d, J =7.5 Hz, H-20), 7.95 (1H, s, H-18); 13C-NMR (CDCl3) δ ppm: 143.5 (C-18), 143.3 (C-19), 133.6 (C-24), 122.9 (C-20), 122.1 (C-21), 120.2 (C-22), 109.6 (C-23), 104.0 (C-4), 102.0 (C-12), 87.6 (C-5), 80.7 (C-6), 65.7 (C-16), 52.2 (C-1), 44.8 (C-17), 43.9 (C-7), 37.0 (C-11), 36.2 (C-10), 34.2 (C-3), 30.5 (C-9), 26.0 (C-8), 24.4 (C-15), 24.2 (C-2), 20.1 (C-14), 12.8 (C-13). + 87.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91(3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.44 (3H, s, H-15), 1.22–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.38–2.47(1H, m, H-1), 2.58–2.73 (5H, m, H-11, H-17 and H-18), 3.49–3.59(3H, m, H-16 and H-19), 3.90–3.98 (1H, m, H-16), 4.80 (1H, s, H-12), 5.42 (1H, s, H-5).
+ 87.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91(3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.44 (3H, s, H-15), 1.22–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.38–2.47(1H, m, H-1), 2.58–2.73 (5H, m, H-11, H-17 and H-18), 3.49–3.59(3H, m, H-16 and H-19), 3.90–3.98 (1H, m, H-16), 4.80 (1H, s, H-12), 5.42 (1H, s, H-5). + 182.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.44 (3H, s, H-15), 1.22–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32 (3H, s, H-21), 2.37–2.47 (1H, m, H-1), 2.58–2.73 (5H, m, H-11, H-17 and H-18), 3.49–3.59 (3H, m, H-16 and H-19), 3.90–3.98 (1H, m, H-16), 4.80 (1H, s, H-12), 5.42 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.1(C-4), 102.2 (C-12), 87.9 (C-5), 81.0 (C-6), 66.1 (C-16), 58.9 (C-19), 58.2 (C-17), 56.9 (C-18), 52.5 (C-1), 44.3 (C-7), 42.0 (C-21), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 20.4 (C-14), 13.0 (C-13); HRMS calcd for C20H35NO6 (M+H)+ 386.2537, found 386.2545.
+ 182.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.44 (3H, s, H-15), 1.22–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32 (3H, s, H-21), 2.37–2.47 (1H, m, H-1), 2.58–2.73 (5H, m, H-11, H-17 and H-18), 3.49–3.59 (3H, m, H-16 and H-19), 3.90–3.98 (1H, m, H-16), 4.80 (1H, s, H-12), 5.42 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.1(C-4), 102.2 (C-12), 87.9 (C-5), 81.0 (C-6), 66.1 (C-16), 58.9 (C-19), 58.2 (C-17), 56.9 (C-18), 52.5 (C-1), 44.3 (C-7), 42.0 (C-21), 37.4 (C-11), 36.4 (C-10), 34.6 (C-3), 30.8 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 20.4 (C-14), 13.0 (C-13); HRMS calcd for C20H35NO6 (M+H)+ 386.2537, found 386.2545. + 88.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.44 (3H, s, H-15), 1.27–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42 (1H, m, H-11), 2.65–2.69 (1H, m, H-1), 2.89–2.97 (6H, m, H-17 and H-18), 3.58–3.62 (1H, m, H-16), 3.67–3.73 (4H, m, H-19), 3.99–4.03 (1H, m, H-16), 4.83 (1H, s, H-12), 5.44 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.2 (C-4), 102.4 (C-12), 87.9 (C-5), 80.9 (C-6), 65.6 (C-16), 58.8 (C-19), 54.3 (C-17), 56.4 (C-18), 52.5 (C-1), 44.2 (C-7), 37.3 (C-11), 36.3 (C-10), 34.5 (C-3), 30.7 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 20.3 (C-14), 13.0 (C-13); HRMS calcd for C21H37NO7 (M+Na)+ 438.2462, found 438.2456.
+ 88.0 (c 1.0 mg/mL, CHCl3). 1H-NMR (CDCl3) δ ppm: 0.91 (3H, d, J = 7.2 Hz, H-14), 0.95 (3H, d, J = 6.0 Hz, H-13), 1.44 (3H, s, H-15), 1.27–2.06 (10H, m, H-2, H-3, H-7, H-8, H-9 and H-10), 2.32–2.42 (1H, m, H-11), 2.65–2.69 (1H, m, H-1), 2.89–2.97 (6H, m, H-17 and H-18), 3.58–3.62 (1H, m, H-16), 3.67–3.73 (4H, m, H-19), 3.99–4.03 (1H, m, H-16), 4.83 (1H, s, H-12), 5.44 (1H, s, H-5); 13C-NMR (CDCl3) δ ppm: 104.2 (C-4), 102.4 (C-12), 87.9 (C-5), 80.9 (C-6), 65.6 (C-16), 58.8 (C-19), 54.3 (C-17), 56.4 (C-18), 52.5 (C-1), 44.2 (C-7), 37.3 (C-11), 36.3 (C-10), 34.5 (C-3), 30.7 (C-9), 26.1 (C-8), 24.6 (C-15), 24.4 (C-2), 20.3 (C-14), 13.0 (C-13); HRMS calcd for C21H37NO7 (M+Na)+ 438.2462, found 438.2456.3.2.3. Antibacterial Enhancement Effect
3.2.3.1. Bacterial Growth
3.2.3.2. Drug Susceptibility Assay
3.2.3.3. Dynamic Bacterial Growth Assay
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Chen, S.; Li, Z.; Lu, J.; Song, X.; Liu, W.; Luo, W. Distribution and drug resistance of nosocomial infection bacteria in intensive care Unit. J. Microbiol. 2011, 32, 111–105. [Google Scholar]
- Tu, C.; Fang, X.; Shen, L.; Liu, X.; Wu, Y.; Chen, W. The epidemiological and Drug-resistant study of pathogenic bacteria in Intensive Care Unit. Chongqing Med. 2011, 40, 1843–1844. [Google Scholar]
- Tu, C.L.; Fang, C.X.; Shen, L.; Liu, X.B.; Wu, Y.; Chen, W.; He, Y.H.; Qin, X.G. The epidemiological and Drug-resistant study of pathogenic bacteria in Intensive Care Unit. Chongqing Med. 2011, 40, 1843–1844. [Google Scholar]
- Blair, J.M.; Piddock, L.J. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin. Microbiol. 2009, 12, 512–519. [Google Scholar] [CrossRef]
- Nikaido, H.; Takatsuka, Y. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 2009, 1794, 769–781. [Google Scholar]
- Eicher, T.; Cha, H.J.; Seeger, M.A.; Brandstatter, L.; El-Delik, J.; Bohnert, J.A. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. USA 2012, 109, 5687–5692. [Google Scholar]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 2002, 419, 587–593. [Google Scholar]
- Lu, W.; Chai, Q.; Zhong, M.; Yu, L.; Fang, J.; Wang, T. Assembling of AcrB trimer in cell membrane. J. Mol. Biol. 2012, 423, 123–134. [Google Scholar]
- Husain, F.; Nikaido, H. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol. Microbiol. 2010, 78, 320–330. [Google Scholar] [CrossRef]
- Fujihira, E.; Tamura, N.; Yamaguchi, A. Membrane topology of a multidrug efflux transporter, AcrB, in Escherichia col. J. Biochem. 2002, 131, 145–151. [Google Scholar]
- Pos, K.M. Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta 2009, 1794, 782–793. [Google Scholar]
- Li, B.; Yao, Q.; Pan, X.C.; Wang, N.; Zhang, R.; Li, J. Artesunate enhances the antibacterial effect of β-lactam antibiotics against Escherichia coli by increasing antibiotic accumulation via inhibition of the multidrug efflux pump system AcrAB-TolC. J. Antimicrob. Chemother. 2011, 66, 769–777. [Google Scholar]
- Nakashima, R.; Sakurai, K.; Yamasaki, S.; Nishino, K.; Yamaguchi, A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 2011, 480, 565–569. [Google Scholar]
- Takatsuka, Y.; Chen, C.; Nikaido, H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. USA 2010, 107, 6559–6565. [Google Scholar] [CrossRef]
- Sennhauser, G.; Amstutz, P.; Briand, C.; Storchenegger, O.; Grutter, M.G. Drug export pathway of multidrug exporter AcrB revealed by DARP in inhibitors. PLoS Biol. 2007, 5, e7. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wu, C.; Liu, J.; Pan, X.; Xian, W.; Li, B.; Peng, W.; Wang, J.; Yang, D.; Zhou, H. Design, Synthesis and Evaluation of the Antibacterial Enhancement Activities of Amino Dihydroartemisinin Derivatives. Molecules 2013, 18, 6866-6882. https://doi.org/10.3390/molecules18066866
Wu C, Liu J, Pan X, Xian W, Li B, Peng W, Wang J, Yang D, Zhou H. Design, Synthesis and Evaluation of the Antibacterial Enhancement Activities of Amino Dihydroartemisinin Derivatives. Molecules. 2013; 18(6):6866-6882. https://doi.org/10.3390/molecules18066866
Chicago/Turabian StyleWu, Chong, Jian Liu, Xichun Pan, Wenying Xian, Bin Li, Wei Peng, Jingfang Wang, Dacheng Yang, and Hong Zhou. 2013. "Design, Synthesis and Evaluation of the Antibacterial Enhancement Activities of Amino Dihydroartemisinin Derivatives" Molecules 18, no. 6: 6866-6882. https://doi.org/10.3390/molecules18066866
APA StyleWu, C., Liu, J., Pan, X., Xian, W., Li, B., Peng, W., Wang, J., Yang, D., & Zhou, H. (2013). Design, Synthesis and Evaluation of the Antibacterial Enhancement Activities of Amino Dihydroartemisinin Derivatives. Molecules, 18(6), 6866-6882. https://doi.org/10.3390/molecules18066866




