Abstract
Toward the expansion of the genetic alphabet, an unnatural base pair between 7-(2-thienyl)imidazo[4,5-b]pyridine (Ds) and pyrrole-2-carbaldehyde (Pa) functions as a third base pair in replication and transcription, and provides a useful tool for the site-specific, enzymatic incorporation of functional components into nucleic acids. We have synthesized several modified-Pa substrates, such as alkylamino-, biotin-, TAMRA-, FAM-, and digoxigenin-linked PaTPs, and examined their transcription by T7 RNA polymerase using Ds-containing DNA templates with various sequences. The Pa substrates modified with relatively small functional groups, such as alkylamino and biotin, were efficiently incorporated into RNA transcripts at the internal positions, except for those less than 10 bases from the 3′-terminus. We found that the efficient incorporation into a position close to the 3′-terminus of a transcript depended on the natural base contexts neighboring the unnatural base, and that pyrimidine-Ds-pyrimidine sequences in templates were generally favorable, relative to purine-Ds-purine sequences. The unnatural base pair transcription system provides a method for the site-specific functionalization of large RNA molecules.
1. Introduction
Expansion of the genetic alphabet of DNA by an unnatural base pair system could be a powerful tool for the site-specific incorporation of extra functional components into nucleic acids and proteins. The realization of this genetic expansion system requires the development of an unnatural base pair that functions in biological systems, such as replication, transcription, and translation, with highly exclusive selectivity as a third base pair, along with the natural A–T and G–C pairs. Researchers are attempting to create expanded systems, and many unnatural base pairs have been designed and tested in in vitro biological systems [1,2,3,4,5]. Among them, some unnatural base pairs have exhibited high selectivity as a third base pair in PCR amplification and/or transcription [6,7,8,9,10,11,12,13,14,15,16,17,18,19].
In the course of our research, we developed several unnatural base pairs, such as those between 2-amino-6-thienylpurine (s) and 2-oxopyridine (y) [13,20,21,22], 7-(2-thienyl)-imidazo[4,5-b]pyridine (Ds) and pyrrole-2-carbaldehyde (Pa) [6], Ds and 2-nitropyrrole (Pn) [23], and Ds and 2-nitro-4-propynylpyrrole (Px) [7,8], toward practical use in DNA/RNA-based biotechnologies. The Ds–Px pair exhibits the highest selectivity and efficiency in PCR amplification, and the Ds–Pa pair is suitable for the site-specific incorporation of Pa and its modified bases, as well as Ds, into RNA by transcription. The substrates of modified-Pa bases, in which several functional groups are attached to position 4 of Pa via a propynyl linker, can be site-specifically incorporated into RNA, opposite Ds in templates, by T7 RNA polymerase. For transcription involving the Ds–Pa pair, DNA templates containing Ds are prepared by PCR amplification using the Ds–Px pair. Thus, the combination of the Ds–Px and Ds–Pa pairs would be useful for the direct site-specific modification of long RNA molecules by T7 transcription.
To provide a highly versatile method for generating functional RNA molecules with an increased number of components by the Ds–Pa pair system, we here report the site-specific incorporation of several modified-Pa substrates, linked with alkylamino, biotin, or fluorescent groups, into RNA transcripts (from 17-mer to 48-mer) using DNA templates containing Ds with different sequence contexts and lengths. We previously reported that the PCR amplification efficiency involving the Ds–Px pair depends on the natural base contexts around the unnatural base. For example, we found that the efficiency of pyrimidine-Ds-pyrimidine sequences in both the template and complementary growing strands is higher than that of purine-Ds-purine sequences [8]. In this report, we examined whether the sequence dependency is also observed in transcription involving the Ds–Pa pair, as in the case of PCR amplification involving the Ds–Px pair. We found that Pa substrates modified with relatively small functional groups, such as alkylamino and biotin, were efficiently and selectively incorporated into RNA transcripts at the internal positions, except for those less than 10 bases from the 3′-terminus, without any sequence dependency. Although the incorporation efficiency and selectivity were reduced for Pa substrates modified with relatively large functional groups, such as TAMRA, FAM, and digoxigenin, their incorporation can be practically used for the site-specific labeling of RNA transcripts. In addition, the efficiency of the site-specific incorporation of all modified-Pa substrates into a position close to the 3′-terminus of a transcript significantly depends on the natural base sequence contexts around the unnatural base in templates. In this case, the sequence dependency is similar to that in replication involving the Ds–Px pair. Overall, the specific transcription using the Ds–Pa pair could be a useful tool for the site-specific functionalization of long RNA molecules, based on a fundamental understanding of the characteristics of the Ds–Pa pair in transcription.
2. Results and Discussion
2.1. Chemical Syntheses of Modified-Pa Triphosphates (Modified-PaTPs) and Ds-Containing DNA Templates
To examine the site-specific incorporation efficiency of a variety of modified-Pa triphosphate substrates (modified-PaTPs) into RNA transcripts by the Ds–Pa pair system, we chemically synthesized several modified-PaTPs with a variety of functional groups, such as alkylamino groups, biotin, digoxigenin (Dig), and fluorescent dyes (FAM and TAMRA), which are attached to position 4 of the Pa base via a propynyl linker (Figure 1).
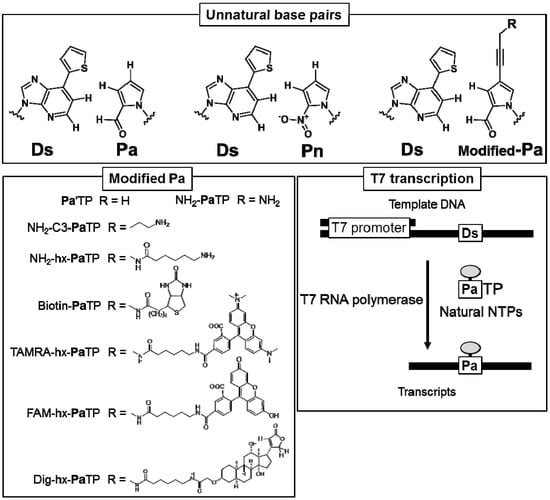
Figure 1.
Chemical structures of unnatural base pairs used for T7 transcription in this study. A variety of substituent groups can be attached to the Pa base moiety. The scheme of T7 transcription mediated by the unnatural Ds–Pa pair is shown (lower right).
The modified-Pa substrates of Pa′TP [6], NH2-PaTP [6], NH2-C3-PaTP, and NH2-hx-PaTP were prepared by palladium-mediated cross-coupling of a 4-iodopyrrole-2-carbaldehyde nucleoside with the functional groups of 1-propyne, 3-(dichloroacetamido)-1-propyne, 5-trifluoroacetamide-1-pentyne, and N-(2-propynyl)-6-trifluoroacetamidohexanamide, respectively (Scheme 1 and Scheme 2).
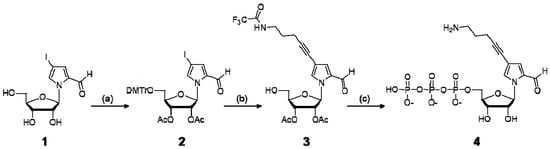
Scheme 1.
Synthesis of the nucleoside 5′-triphosphate of NH2-C3-Pa.
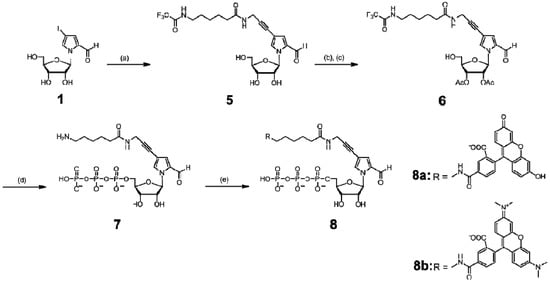
Scheme 2.
Synthesis of the nucleoside 5′-triphosphates of TAMRA- and FAM-hx-Pa.
These nucleoside derivatives were then converted to the triphosphate substrates (bearing free amino groups for alkylamino-PaTPs) by Eckstein’s method. Biotin-PaTP [6] and Dig-hx-PaTP were prepared by the treatment of NH2-PaTP with biotin-N-hydroxysuccinimide and digoxigenin-3-O-methylcarbonyl-ε-aminocaproic acid-N-hydroxysuccinimide ester, respectively (Scheme 3). TAMRA-hx-PaTP and FAM-hx-PaTP were prepared by the treatment of NH2-hx-PaTP with 5-carboxytetramethylrhodamine N-hydroxysuccinimidyl ester and 5-carboxyfluorescein N-hydroxy-succinimidyl ester, respectively (Scheme 2). The relatively large groups, such as TAMRA, FAM, and digoxigenin, were attached to the 4-aminopropynyl Pa base via an aminohexyl linker, to reduce the steric hindrance in the nucleic acid and polymerase complexes during transcription. All modified-Pa triphosphates were purified by DEAE Sephadex column chromatography and C18 HPLC. Their structures were identified by 1H- and 31P-NMR and mass spectrometry.
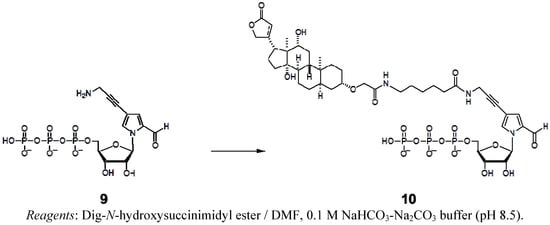
Scheme 3.
Synthesis of the nucleoside 5′-triphosphate of Dig-hx-Pa.
DNA templates containing Ds were prepared by standard phosphoramidite chemistry, using a DNA synthesizer [6]. The synthesized DNA fragments were purified by gel electrophoresis. Full-length template strands (from 35-mer to 65-mer) containing one Ds base were hybridized with a 21-mer non-template strand including the T7 promoter region for transcription (Figure 1).
2.2. T7 Transcription for 48-Mer Transcripts Containing Modified-Pa at Internal Positions
We first examined the incorporation efficiency of each modified-PaTP, as well as PnTP, in transcription by T7 RNA polymerase, using 65-mer DNA templates containing one Ds base, in which Ds was located at the complementary site corresponding to position 21 in the 48-mer transcripts (Figure 2A). To investigate the natural base sequence dependency around the Ds base in templates, we used four templates containing Ds in different sequence contexts, 3′-CDsC-5′, 3′-CDsT-5′, 3′-TDsC-5′, and 3′-GDsG-5′, as well as control templates, 3′-CAT-5′ and 3′-GAG-5′, comprising only the natural bases. Transcription was performed in the presence of 1 mM natural base NTPs, 0 or 1 mM modified-PaTP or PnTP, [γ-32P] GTP, DNA templates, and T7 RNA polymerase, at 37 °C for 3 h. After transcription, the 5′-labeled transcripts were analyzed by gel electrophoresis (Figure 2B), and the relative yields of the full-length transcripts (48-mer) were determined by comparison to the yield of transcription using the control template (Figure 2B and Figure 3A). On the gel, the mobilities of the full-length transcripts differed, depending on the modifications of the Pa base. The incorporation of PaTPs modified with large molecules (TAMRA-hx-PaTP, FAM-hx-PaTP, and Dig-hx-PaTP) reduced the mobility of the transcripts on the gel, and the incorporation of TAMRA-hx-Pa or FAM-hx-Pa into the full-length transcripts was confirmed by monitoring their fluorescence with a bio-imager, FLA-7000 (data not shown). Truncated products (20-mer), which resulted from pausing before the unnatural base position, were also detected. All of the unnatural base substrates, PaTP, PnTP, and modified-PaTPs, except for the PaTPs modified with the large molecules, were efficiently incorporated into RNA by T7 transcription, and the transcription efficiency (86−139%) was independent of both the sequence contexts and the amounts of the truncated products (20-mer). Furthermore, we found that PnTP, with a 2-nitro group instead of the 2-aldehyde group of Pa, is also a favorable substrate as a pairing partner of Ds in transcription, as shown in Figure 2 and Figure 3A.
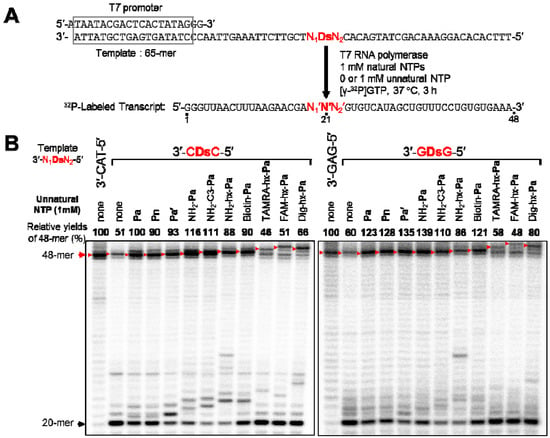
Figure 2.
T7 transcription mediated by the Ds–Pa pair for 48-mer transcripts. (a) Scheme of the experiments. (b) Gel electrophoresis of transcripts generated from a template containing a 3′-CDsC-5′ or 3′-GDsG-5′ sequence with natural NTPs (1 mM) and each modified-PaTP or PnTP (1 mM). Transcripts were labeled at their 5′-termini with [γ-32P]GTP. The positions of the full-length 48-mer products are indicated by red arrows. Due to the differences in the substituent groups attached to the Pa base moiety, the mobilities of the transcripts containing modified-Pa differed. The relative yield of each full-length transcript was determined by comparison to the yield of the native transcript from the template consisting of the natural bases only (3′-CAT-5′). The black arrow indicates the position of 20-mer transcripts, which paused at the previous position of Ds in the template. The experiments were independently repeated three times, and representative data are shown.
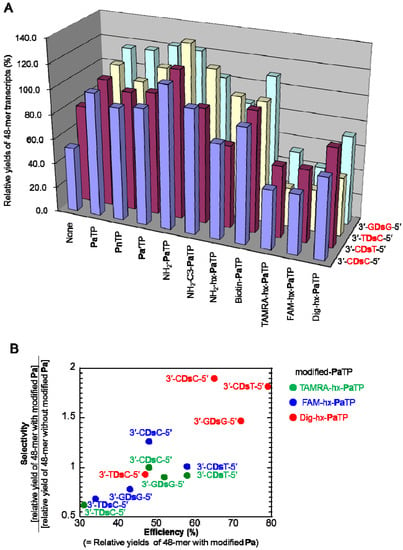
Figure 3.
Efficiency and selectivity analysis of the 48-mer transcripts. (a) Plot of relative yields of the 48-mer transcripts from templates containing 3′-CDsC-5′, 3′-CDsT-5′, 3′-TDsC-5′, or 3′-GDsG-5′, in the presence of each modified-PaTP or PnTP. The relative yield of each full-length transcript was determined by comparison to the yield of the native transcript from the template consisting of the natural bases only (3′-N1DsN2-5′ = 3′-CAT-5′ for 3′-CDsC-5′, 3′-CDsT-5′, 3′-TDsC-5′; 3′-N1DsN2-5′ = 3′-GAG-5′ for 3′-GDsG-5′). The values were averaged from three data sets. (b) Correlation between the incorporation efficiency and selectivity of PaTPs modified with large molecules in T7 transcription. The selectivity was determined as the ratio of the relative yield of the 48-mer transcripts with and without modified-Pa bases (TAMRA-hx-Pa, FAM-hx-Pa, and Dig-hx-Pa) from the gel analysis of transcripts.
The PaTPs modified with the large molecules were less effective, and the transcription yields were reduced by nearly half. In addition, the PaTPs modified with the large molecules exhibited reduced incorporation selectivity opposite Ds in templates, and the full-length transcripts without modified-Pa also appeared on the gel. We determined the incorporation selectivities of TAMRA-hx-PaTP, FAM-hx-PaTP, and Dig-hx-PaTP, from the ratios of the amounts of the full-length transcripts with modified-Pa to those without modified-Pa (Figure 3B).
Since the bands corresponding to the full-length transcripts containing these modified-Pa were shifted on the gel, the incorporation selectivity of each modified-Pa could be determined by comparing the band densities between the full-length transcripts with and without the modified-Pa bases. Although the incorporation selectivities of the PaTPs modified with the large molecules were relatively low, the selectivity of each modified-Pa correlated well with its incorporation efficiency into RNA. Depending on the sequence context of the templates, more efficient incorporation tended to lead to more selective incorporations. Next, we determined the incorporation position of Biotin-Pa in the full-length transcripts. We previously reported the high selectivity of the PaTP and Biotin-PaTP incorporations opposite Ds by transcription, using DNA templates containing 3′-CDsT-5′ and 3′-ADsT-5′ sequences, respectively [6].
Therefore, we confirmed the incorporation positions of Biotin-Pa in the full-length transcripts, obtained by transcription using DNA templates with four different sequence contexts, 3′-CDsC-5′, 3′-CDsT-5′, 3′-TDsC-5′, and 3′-GDsG-5′. The 32P-labeled full-length transcripts were partially digested by either alkali or RNase T1, and the digested products were incubated with streptavidin magnetic beads. This treatment separated the Biotin-Pa-containing fragments from the digested products, and the remaining fragments without Biotin-Pa were analyzed by gel electrophoresis (Figure 4). By comparing the sequencing ladder patterns obtained by the alkali and RNase T1 treatments, the position of Biotin-Pa incorporation was determined from the initial disappearance position of the streptavidin-treated ladders.
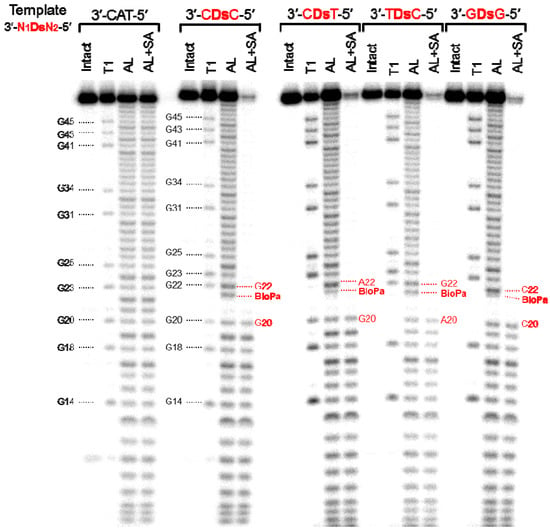
Figure 4.
Sequence analysis of the 48-mer transcripts containing Biotin-Pa or U at position 21. The 5′ 32P-labeled transcripts were partially digested with either RNase T1 (T1) or with alkali (AL). A portion of the partially alkali-digested transcripts was treated with streptavidin magnetic beads, to capture the RNA fragments containing Biotin-Pa (AL+SA). Each digested fragment was analyzed on a 10% polyacrylamide gel containing 7 M urea.
On the patterns obtained by the streptavidin treatment of all four transcripts with different sequences, the ladders corresponding to the fragments larger than 21-mer were almost undetectable. Based on the disappearance ratio of the ladders, more than 90% of Biotin-Pa was incorporated at the desired position in the transcripts. In the alkali-digested products without the streptavidin treatment, the ladders corresponding to the fragments larger than 21-mer were largely shifted on the gel. From these results, we concluded that Biotin-PaTP was site-specifically incorporated into the transcripts at position 21, opposite Ds in the templates. The incorporation selectivity of Biotin-PaTP opposite Ds was more than 90%, and was independent of the sequence context.
2.3. T7 Transcription for Transcripts Containing Pa at Different Positions from the 3′-Terminus
We next examined how the position of Pa incorporation, especially close to the 3′-terminus, in transcripts affects the incorporation efficiency. We previously reported that transcription efficiency involving the Ds–Pa pairing decreased when using short DNA templates (such as 35-mer) [6].
Thus, we prepared a series of DNA templates (42–65-mer), which were shortened stepwise from the terminus, and examined T7 transcription using the templates (Figure 5). For the experiments, we chose the DNA template containing the 3′-GDsG-5′ sequence, which was expected to be an inferior template based on our replication experiments involving related unnatural base pairs [7,8]. Using the DNA templates and PaTP, we compared the yields of transcripts (25-, 28-, 33-, 38-, 43-, and 48-mer).
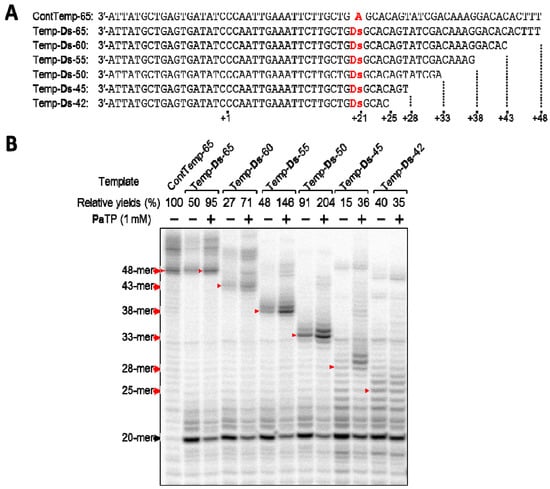
Figure 5.
T7 transcription mediated by the Ds–Pa pair from DNA templates containing Ds, with different lengths after the unnatural base position. (a) Sequences of templates used in this study. (b) Gel electrophoresis of transcripts from different length templates with natural NTPs (1 mM) and in the presence or absence of PaTP (1 mM). Transcripts were labeled at their 5′-termini with [γ-32P]GTP. The positions of the full-length products are indicated by red arrows. The relative yield of each full-length transcript was determined by comparison to the yield of the native transcript from the template consisting of the natural bases only. The experiments were independently repeated three times, and representative data are shown.
The full-length transcripts (48-, 43-, 38-, and 33-mer) obtained from 65-, 60-, 55-, and 50-mer templates were observed as fairly clear bands on the gel, and the relative yields of these transcripts were more than 71%, as compared to that using the control template (ContTemp-65) with natural NTPs. In contrast, the full-length transcripts (28-mer and 25-mer) from the 45- and 42-mer templates were difficult to discern as a single main band on the gel, and the transcription yields were also reduced, to 35–36%. When using the 45-mer and 42-mer templates, several ladders, which were larger than the full-length transcripts, appeared on the gel. Although we still do not know how the ladders were generated, this might be an intrinsic problem caused by transcript-transcript or transcript-template interactions, depending on the natural base sequence contexts [24] or the instability of the complex between the template, resulting transcript, substrate, and T7 RNA polymerase. Even allowing for this problem, the incorporation of Pa at a position less than 12-bases from the 3′-terminus of the transcripts significantly reduces the transcription efficiency involving the Ds–Pa pairing, when using templates with the inferior 3′-GDsG-5′ sequence. These results compelled us to examine how the sequence contexts in templates affect the transcription efficiency, when the Pa incorporation site is close to the 3′-terminus in the transcripts.
2.4. Pa Incorporation into the 3′-Region of Transcripts with Different Sequence Contexts around the Unnatural Base
We then examined the transcription efficiency using short DNA templates (35-mer) containing 20 sequence combinations, including all 3′-CTNDsN-5′ sequences, where N = A, G, C or T, as well as 3′-CACDsT-5′, 3′-CGCDsT-5′, 3′-CCCDsT-5′, and 3′-ATCDsT-5′. RNA fragments (17-mer) containing Pa at position 13, 5-bases from the 3′-terminus, were transcribed as full-length products, using the same method described above. The representative gel data are displayed in Figure 6, and the transcription efficiencies for all 16 templates are summarized in Figure 7.
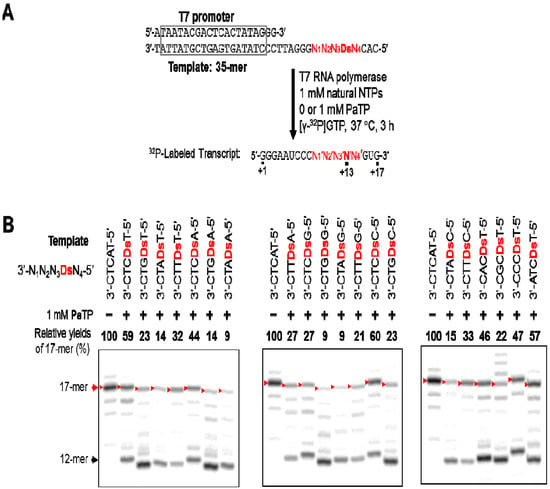
Figure 6.
T7 transcription mediated by the Ds–Pa pair for 17-mer transcripts. (a) Scheme of the experiments. (b) Gel electrophoresis of transcripts generated from templates with different sequences around Ds, with natural NTPs (1 mM), in the presence or absence of PaTP (1 mM). Transcripts were labeled at their 5′-termini with [γ-32P]GTP. The positions of the full-length 17-mer products are indicated by red arrows. Due to the different sequence contexts, the mobilities of the transcripts differed. The relative yield of each full-length transcript was determined by comparison to the yield of the native transcript from the template consisting of the natural bases only. The black arrow indicates the approximate position of the 12-mer transcripts, which paused at the previous position of Ds in the template. The experiments were independently repeated three times, and representative data are shown.
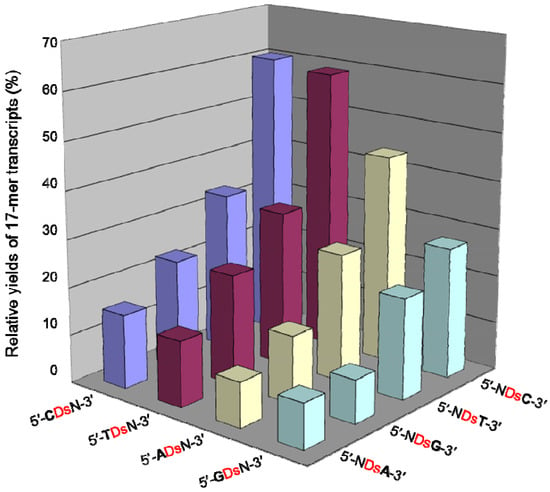
Figure 7.
Plot of the relative yields of the 17-mer transcripts from the template containing a 3′-N1DsN2-5′ sequence (N1, N2 = A, G, C, or T), in the presence of PaTP. The relative yield of each full-length transcript was determined by comparison to the yield of the native transcript from the template consisting of the natural bases only. The values were averaged from three data sets.
We found that the adjacent natural base contexts around the unnatural base significantly affected the transcription efficiency involving the Ds–Pa pairing, when the Pa incorporation site is close to the 3′-terminus of the transcript. The order of the effective sequences was 3′-CDsC-5′ ≈ 3′-CDsT-5′ > 3′-CDsA-5′ > 3′-TDsC-5′ ≈ 3′-TDsT-5′ > 3′-TDsA-5′ ≈ 3′-CDsG-5′ ≈ 3′-GDsC-5′ ≈ 3′-GDsT-5′ > 3′-TDsG-5′ > 3′-ADsC-5′ ≈ 3′-ADsT-5′ ≈ 3′-GDsA-5′ > 3′-ADsA-5′ ≈ 3′-ADsG-5′ ≈ 3′-GDsG-5′ (Figure 7). Thus, 3′-pyrimidine-Ds-pyrimidine-5′ is generally better than 3′-purine-Ds-purine-5′ for efficient transcription. Even using the short templates, the 3′-CDsC-5′ and 3′-CDsT-5′ sequence contexts generated relatively high yields (60%) of the 17-mer transcripts. As shown in Figure 6, templates containing 3′-CTCDsT-5′, 3′-CACDsT-5′, 3′-CCCDsT-5′, and 3′-ATCDsT-5′ sequences provided similar transcription efficiency (relative yields of the 17-mer transcripts = 46–59%). These templates have the same 3′-CDsT-5′ sequence, and thus, the adjacent natural bases around the unnatural base significantly affect the transcription efficiency. The template containing a 3′-CGCDsT-5′ sequence was the only exception, and the relative yield of the 17-mer transcript was 22%. However, in this case, some transcripts (longer than 17-mer) appeared on the gel. These products might be obtained by unusual interactions between the transcripts and templates [24], and thus the low yield of the 17-mer transcript is due to the by-products and is not related to the Ds–Pa pairing in transcription. We also determined the incorporation efficiencies of PaTP, PnTP, and eight modified-PaTPs using short DNA templates (35-mer) containing 3′-CDsC-5′, 3′-CDsT-5′, 3′-TDsC-5′ or 3′-GDsG-5′ sequences (Figure 8). The yields for the pyrimidine-Ds-pyrimidine contexts of 3′-CDsC-5′ and 3′-CDsT-5′ were much higher than those for the purine-Ds-purine context of 3′-GDsG-5′ in all substrates. We previously reported that the incorporation selectivity was also high (>95%) for the PaTP and Pa′TP incorporations, using short DNA templates containing a 3′-CDsT-5′ sequence [6]. However, PaTPs modified with large molecules exhibited very low incorporation efficiencies even with 3′-TDsC-5′, besides 3′-GDsG-5′.
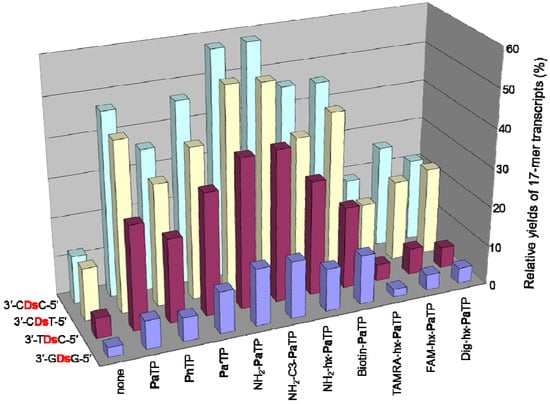
Figure 8.
Plot of the relative yields of the 17-mer transcripts from templates with different sequences around Ds, with natural NTPs (1 mM), in the presence of each modified-PaTP or PnTP (1 mM). The relative yield of each full-length transcript was determined by comparison to the yield of the native transcript from the template consisting of the natural bases only (3′-N1DsN2-5′ = 3′-CAT-5′ for ′-CDsC-5′, 3′-CDsT-5′, 3′-TDsC-5′; 3′-N1DsN2-5′ = 3′-GAG-5′ for ′-GDsG-5′). The values were averaged from three to four data sets.
Taken together, the incorporations of Pa and modified-Pa, as well as those of Pn, close to the 3′-terminus of the transcript exhibited lower efficiencies, relative to those of the internal incorporations, as shown in Figure 3.
2.5. DNA Template Preparation for the Ds–Pa Transcription System
For the practical uses of the Ds–Pa transcription system demonstrated here, the preparation methods for long Ds-containing templates are an important consideration. The general schemes for Ds-containing template preparation and the specific transcription of modified-PaTPs are summarized in Figure 9. For the modified-PaTP incorporation, Ds-containing DNA templates can be prepared by several methods. Templates (less than 80-mer including the T7 promoter) are prepared by a direct chemical method using a DNA synthesizer, as shown here. In this case, partially double stranded DNA templates for the promoter region can be used. Templates (more than 100-mer) can be prepared by several methods, as follows: (1) Chemical synthesis + enzymatic ligation: Chemically synthesized 5′-phosphorylated DNA fragments are annealed as a double stranded form and ligated by T4 DNA ligase [6,15]. As the complementary position opposite Ds, T can be used instead of the complementary unnatural base, Px. Phosphorylation of DNA fragments is performed during DNA chemical synthesis by using commercially available phosphorylation amidite reagents or T4 polynucleotide kinase. (2) Fusion PCR: For the internal introduction of Ds in long templates (more than 200-mer), fusion PCR methods involving the Ds–Px pair can be used with dDsTP and dPxTP (unpublished data, Kimoto and Hirao). (3) PCR: For the terminal incorporation of Ds close to the 3′-terminus for transcripts, PCR amplification with primers containing Ds is useful [15,25]. In these three methods, the position of Ds introduction into each DNA fragment should be more than 10 bases away from the terminus.
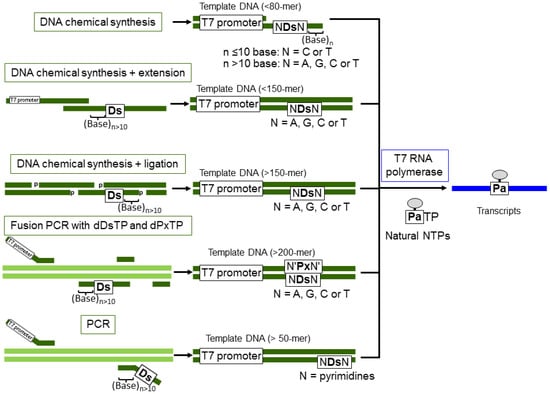
Figure 9.
Schemes for the preparation of Ds-containing templates for T7 transcription, using Ds-containing DNA fragments.
3. Experimental
3.1. General
Reagents and solvents were purchased from standard suppliers without further purification. Reactions were monitored by thin-layer chromatography (TLC), using 0.25 mm silica gel 60 plates impregnated with 254 nm fluorescent indicator (Merck). 1H-NMR, 13C-NMR, and 31P-NMR spectra were recorded on a Bruker (300-AVM) magnetic resonance spectrometer. Nucleoside purification was performed on a Gilson HPLC system with a preparative C18 column (Waters μ-BONDASPHERE, 150 × 19 mm). The triphosphate derivatives were purified by DEAE-Sephadex A-25 column chromatography (300 × 15 mm; eluted by a linear gradient of 50 mM to 1 M triethylammonium bicarbonate) and by HPLC with using a C18 column (CAPCELL PAK, 250 × 4.6 mm, SHISEIDO), eluted with a linear gradient of CH3CN in 100 mM triethylammonium acetate, pH 7.0. High resolution mass spectra (HRMS) and electrospray ionization mass spectra (ESI-MS) were recorded on a JEOL JM 700 or GC mate mass spectrometer and a Waters micromass ZMD 4000 equipped with a Waters 2690 LC system or a Waters UPLC-MS (H class) system, respectively. DNA templates were synthesized with an Applied Biosystems 392 DNA synthesizer, using CE phosphoramidite reagents for the natural and Ds bases (Glen Research), and were purified by gel electrophoresis. The syntheses of PaTP, Pa′TP, NH2-PaTP, and Biotin-PaTP were described previously [6]. Biotin-PaTP is commercially available from Glen Research, and the other modified-PaTPs are available from TagCyx Biotechnologies. T7 RNA polymerase and T4 polynucleotide kinase were purchased from Takara. Antarctic phosphatase and streptavidin magnetic beads were purchased from New England Biolabs. The natural NTP Set (100 mM solutions: ATP, CTP, GTP, and UTP) and RNase T1 were purchased from GE Healthcare. The [γ-32P]GTP and [γ-32P]ATP were purchased from PerkinElmer. The Escherichia coli tRNA mixture was purchased from Sigma. Gel images were analyzed with a bio-imaging analyzer, FLA7000 (Fuji Film).
3.2. Synthesis of NH2-C3-PaTP (4)
1-(2,3-(Di-O-acetyl)-5-O-dimethoxytrityl-β-d-ribofuranosyl)-4-iodopyrrole-2-carbaldehyde (2). 1-(β-d-Ribofuranosyl)-4-iodopyrrole-2-carbaldehyde (1) [6] (945 mg, 2.7 mmol) was dissolved in pyridine and evaporated to dryness in vacuo. The residue was dissolved in pyridine (27 mL), and 4,4′-dimethoxytrityl chloride (998 mg, 2.9 mmol) was added to the solution. The reaction mixture was stirred at room temperature for 1 h. After the reaction, the mixture was diluted with ethyl acetate (EtOAc) and washed with 5% NaHCO3 and brine. The organic phase was dried over Na2SO4, and evaporated in vacuo.
1-(5-O-Dimethoxytrityl-β-d-ribofuranosyl)-4-iodopyrrole-2-carbaldehyde (1.7 g) was purified by silica gel column chromatography (1% MeOH in CH2Cl2), and 2.6 mmol (1.7 g) of the purified product was co-evaporated with dry pyridine three times. The residue was dissolved in pyridine (26 mL), and acetic anhydride (979 μL, 10.4 mmol) was added to the solution. The mixture was stirred at room temperature for 12 h. After the reaction, the solution was diluted with EtOAc and washed with a saturated NaHCO3 solution. The organic layer was washed with brine, dried over MgSO4, and evaporated in vacuo. The product was purified by silica gel column chromatography in CH2Cl2, to give 1.77 g of 2 (89% yield from 1). 1H-NMR (300 MHz, DMSO-d6) δ 9.49 (s, 1H), 7.69 (s, 1H), 7.39–7.24 (m, 10H), 6.89 (d, 4H, J = 7.8 Hz), 6.63 (d, 1H, J = 4.9 Hz), 5.52–5.42 (m, 2H), 4.25 (m, 1H), 3.74 (s, 6H), 3.39–3.25 (m, 2H), 2.05 (s, 3H), 2.04 (s, 3H).
1-(2,3-(Di-O-acetyl)-β-d-ribofuranosyl)-4-(5-trifluoroacetamide-1-pentynyl)pyrrole-2-carbaldehyde (3). The protected nucleoside 2 (148 mg, 0.2 mmol) was dissolved in DMF (1 mL) with CuI (6.1 mg, 0.032 mmol), Pd(PPh3)4 (11.6 mg, 0.01 mmol), and triethylamine (42 μL). To the solution, 5-trifluoroacetamide-1-pentyn (53.7 mg, 0.3 mmol) in DMF (0.6 mL) was added, and the solution was stirred at room temperature for 12 h. After the reaction, the mixture was diluted with EtOAc and washed with 5% NaHCO3 and brine. The organic phase was dried over Na2SO4 and evaporated in vacuo. The product was purified by silica gel column chromatography to give 1-(2,3-(di-O-acetyl)-5-O-dimethoxytrityl-β-d-ribofuranosyl)-4-(5-trifluoroacetamide-1-pentynyl)pyrrole-2-carbaldehyde. To the residue in CH2Cl2 (20 mL), 200 μL of dichloroacetic acid was added, and the solution was stirred at 0 °C for 15 min. The reaction mixture was poured into a saturated NaHCO3 solution, and the product was extracted with CH2Cl2. The combined organic layer was dried over MgSO4, evaporated in vacuo, and purified by silica gel column chromatography (1% MeOH in CH2Cl2), to give 83 mg of 3 (85% yield from 2). 1H-NMR (300 MHz, DMSO-d6) δ 9.50 (s, 1H), 9.46 (bs, 1H), 7.98 (s, 1H), 7.18 (d, 1H, J = 1.7 Hz), 7.66 (d, 1H, J = 4.8 Hz), 5.41 (t, 1H, J = 5.1 Hz), 5.38–5.31 (m, 2H), 4.18 (m, 1H), 3.77–3.60 (m, 2H), 3.33–3.27 (m, 2H), 2.42 (t, 1H, J = 7.1 Hz), 2.08 (s, 3H), 2.02 (s, 3H), 1.76 (m, 2H).
1-(β-d-Ribofuranosyl)-4-(5-amino-1-pentynyl)pyrrole-2-carbaldehyde 5′-triphosphate (4). The protected nucleoside 3 (49 mg, 0.1 mmol) was dissolved in pyridine and evaporated to dryness in vacuo. The residue was dissolved in pyridine (100 μL) and dioxane (300 μL). A 1 M solution of 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one in dioxane (110 μL, 0.11 mmol) was added. After 10 min, tri-n-butylamine (100 μL) and 0.5 M bis(tributylammonium)pyrophosphate in DMF (300 μL, 0.15 mmol) were added to the reaction mixture. The mixture was stirred at room temperature for 10 min. A solution of 1% iodine in pyridine/water (98:2, v/v) (2.0 mL) was then added. After 15 min, a 5% aqueous solution (150 μL) of NaHSO3 was added. The volatile components were removed by evaporation, and then water (5 mL) was added to the residue. The solution was stirred at room temperature for 30 min, and then was treated with concentrated NH4OH (20 mL) at room temperature for 6 h. The solution was evaporated in vacuo, and the product was purified by DEAE-Sephadex A-25 column chromatography and C18 HPLC to give the nucleoside 5′-triphosphate 4 (NH2-C3-PaTP), in a 31% yield from 3. 1H-NMR (300 MHz, D2O) δ 9.36 (s, 1H), 8.04 (s, 1H), 7.16 (s, 1H), 6.48 (d, 1H, J = 2.9 Hz), 4.44 (m, 1H), 4.32–4.24 (m, 4H), 3.16 (m 2H), 3.15 (q, 22H, J = 7.3 Hz, TEA), 2.50 (t, 2H, J = 6.4 Hz), 1.89 (m, 2H), 1.23 (t, 32H, J = 7.3 Hz, TEA). 31P-NMR (121 MHz, D2O) δ −7.42 (bs, 1P), −10.39 (d, 1P, J = 17.2 Hz), −21.73 (t, 1P, J = 18.8 and 18.9 Hz). ESI-MS for C15H23O14N2P3 [M−H]−: calcd., 547.03; found, 546.82. UV-vis spectrum (in 10 mM sodium phosphate buffer, pH 7.0): λmax 258 nm (ε = 12,200), 311 nm (ε = 10,100).
3.3. Synthesis of NH2-hx-PaTP (7), FAM-hx-PaTP (8a), and TAMRA-hx-PaTP (8b)
1-(β-d-Ribofuranosyl)-4-[3-(6-trifluoroacetamidohexanamido)-1-propynyl]pyrrole-2-carbaldehyde (5). The nucleoside 1 (177 mg, 500 μmol, containing an α anomer) was co-evaporated with dry CH3CN twice. The residue was dissolved in DMF (2.5 mL) with CuI (15 mg, 80 μmol), Pd(PPh3)4 (29 mg, 25 μmol), and triethylamine (105 μL, 750 μmol), and the mixture was stirred in the dark at room temperature. To the solution, N-(2-propynyl)-6-trifluoroacetamidohexanamide (198 mg, 750 μmol) in DMF (2 mL) was added, and the solution was stirred at room temperature overnight. The reaction mixture was concentrated in vacuo. The product was purified from the residue by silica gel column chromatography (10% CH3OH in CH2Cl2) and C18 HPLC, to give 126 mg of 5 (51% yield). 1H-NMR (270 MHz, DMSO-d6) δ 9.52 (d, 1H, J = 0.66 Hz), 9.38 (brs, 1H), 8.27 (t, 1H, J = 5.3 Hz), 7.96 (s, 1H), 7.13 (d, 1H, J = 1.6 Hz), 6.32 (d, 1H, J = 3.6 Hz), 5.42–5.32 (brs, 1H), 5.18–5.05 (m, 2H), 4.08–3.95 (m, 4H), 3.90–3.84 (m, 1H), 3.70–3.50 (m, 2H), 3.20–3.10 (m, 2H), 2.08 (t, 2H, J = 7.3 Hz), 1.55–1.40 (m, 4H), 1.30–1.15 (m, 2H). HRMS (FAB, 3-NBA matrix) for C21H27F3N3O7 [M+H]+: calcd, 490.1801; found, 490.1815.
1-(2,3-Di-O-acetyl-β-d-ribofuranosyl)-4-[(3-(6-trifluoroacetamidohexanamido)-1-propynyl]pyrrole-2-carbaldehyde (6). The nucleoside 5 (122 mg, 250 μmol) was co-evaporated with dry pyridine three times. The residue was dissolved in pyridine (4 mL) with 4,4′-dimethoxytrityl chloride (89 mg, 263 μmol), and the solution was stirred at room temperature for 2.5 h. The solution was separated by EtOAc and water. The organic layer was washed with a saturated NaHCO3 solution, dried with MgSO4, and evaporated in vacuo. The residue was purified by silica gel column chromatography (0–0.5% CH3OH in CH2Cl2), to give 1-(5-O-dimethoxytrityl-β-d-ribofuranosyl)-4-[(3-(6-trifluoro- acetamidohexanamido)-1-propynyl]pyrrole-2-carbaldehyde (150 mg). The dimethoxytrityl derivative (149 mg, 188 μmol) was co-evaporated with dry pyridine three times. To the residue in pyridine (2 mL), acetic anhydride (53 μL, 565 μmol) was added. The mixture was stirred at room temperature overnight. The solution was separated by EtOAc and a saturated NaHCO3 solution. The organic layer was washed with brine, dried with MgSO4, and evaporated in vacuo. To the residue in dichloromethane (19 mL), dichloroacetic acid (280 μL) was added, and the solution was stirred at 0 °C for 15 min. The reaction mixture was poured into a saturated NaHCO3 solution and extracted with CH2Cl2. The combined organic layer was dried with MgSO4 and evaporated in vacuo. The product was purified by silica gel column chromatography (1–2.5% CH3OH in CH2Cl2), to give 90 mg of 6 (63% yield from 5). 1H-NMR (270 MHz, DMSO-d6) δ 9.49 (s, 1H), 9.39 (brs, 1H), 8.28 (t, 1H, J = 5.3 Hz), 8.03 (s, 1H), 7.21 (d, 1H, J = 1.6 Hz), 6.63 (d, 1H, J = 5.3 Hz), 5.44 (brs, 1H), 5.40–5.28 (m, 3H), 4.20–4.15 (m, 1H), 4.05 (d, 2H, J = 5.3 Hz), 3.80–3.56 (m, 2H), 3.20–3.10 (m, 2H), 2.14–1.98 (m, 8H), 1.55–1.40 (m, 4H), 1.30–1.15 (m, 2H). HRMS (FAB, 3-NBA matrix) for C25H31F3N3O9 [M+H]+: calcd., 574.2012; found, 574.2061.
1-(β-d-Ribofuranosyl)-4-[3-(6-aminohexanamido)-1-propynyl]pyrrole-2-carbaldehyde 5′-triphosphate (7). The protected nucleoside 6 (57 mg, 100 μmol) was co-evaporated with dry pyridine three times. The residue was dissolved in pyridine (100 μL) and dioxane (300 μL). A 1 M solution of 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one in dioxane (110 μL, 110 μmol) was added to the solution, and then the reaction mixture was stirred at room temperature for 10 min. Aliquots of tri-n-butylamine (100 μL) and 0.5 M bis(tri-n-butylammonium)pyrophosphate in DMF (300 μL) were then added to the reaction mixture. After 10 min, a solution of 1% iodine in pyridine/water (98/2, v/v) (2.0 mL) was added and stirred. After 15 min, a 5% aqueous solution (150 μL) of NaHSO3 was added. The volatile components were removed by evaporation in vacuo, and the residue was dissolved in water (5 mL). The solution was stirred at room temperature for 30 min, and then was treated with concentrated ammonia (20 mL) at room temperature for 8 h. The solution was lyophilized, and the product was purified by DEAE-Sephadex A-25 column chromatography to give the nucleoside 5′-triphosphate 7 (NH2-hx-PaTP), used for modification with a fluorophore, as described below. For further use as a transcription substrate, the triphosphate was purified by C18 HPLC. 1H-NMR (300 MHz, D2O) δ 9.39 (s, 1H), 7.93 (s, 1H), 7.21 (d, 1H, J = 1.1 Hz), 6.49 (d, 1H, J = 4.0 Hz), 4.38 (m, 2H), 4.23 (m, 3H), 4.10 (s, 2H), 3.15 (q, 14H, J = 7.3 Hz, TEA), 2.91 (t, 2H, J = 7.5 Hz), 2.26 (t, 2H, J = 7.1 Hz), 1.63 (m, 4H), 1.35 (m, 2H), 1.23 (t, 21H, J = 7.3 Hz, TEA). 31P-NMR (121 MHz, D2O) δ −9.04 (1P), −11.13 (d, 1P, J = 19.0 Hz), −22.66 (t, 1P, J = 19.8 Hz). ESI-MS for C19H29N3O15P3 [M−H]−: calcd., 632.08; found, 631.72. UV-vis spectrum (in 10 mM sodium phosphate buffer, pH 7.0): λmax 308 nm (ε = 9,200).
1-(β-d-Ribofuranosyl)-4-[3-[6-(fluorescein-5-carboxamido)hexanamido]-1-propynyl]pyrrole-2-carbaldehyde 5′- triphosphate (8a) and 1-(β-d-Ribofuranosyl)-4-[3-[6-(tetramethylrhodamine-5-carboxamido)hexanamido]-1-propynyl]pyrrole-2-carbaldehyde 5′-triphosphate (8b).
(1) FAM-hx-PaTP (8a): A 0.1 M NaHCO3-Na2CO3 buffer solution (pH 8.5, 2.5 mL) of 7 (one-third of the crude product) was reacted with 5-carboxyfluorescein N-hydroxysuccinimidyl ester (FAM-SE, Invitrogen) (9 mg, 19 μmol) in DMF (200 μL), in the dark at room temperature. After 9 h, the reaction was quenched with concentrated ammonia (1 mL). The product was purified by DEAE-Sephadex A-25 column chromatography and C18 HPLC to give 8a (FAM-hx-PaTP). 1H-NMR (300 MHz, D2O): δ 9.07 (s, 1H), 8.20 (s, 1H), 7.93 (d, 1H, J = 8.0 Hz), 7.62 (s, 1H), 7.13 (d, 1H, J = 7.9 Hz), 6.92 (d, 1H, J = 8.9 Hz), 6.89 (d, 1H, J = 9.0 Hz), 6.77 (m, 3H), 6.67 (d, 1H, J = 9.0 Hz), 6.64 (d, 1H, J = 9.2 Hz), 6.10 (d, 1H, J = 3.2 Hz), 4.14–3.93 (m, 5H), 3.87 (s, 2H), 3.32 (m, 2H), 3.04 (q, 21H, J = 7.3 Hz, TEA), 2.16 (t, 2H, J = 6.5 Hz), 1.54 (m, 4H), 1.35–1.09 (m, 35H, TEA). 31P-NMR (121 MHz, D2O) δ −10.90 (1P), −11.38 (1P), −23.25 (1P). ESI-MS for C40H39N3O21P3 [M−H]−: calcd., 990.13; found, 989.78. UV-vis spectrum (in 10 mM sodium phosphate buffer, pH 7.0): λmax 493 nm (ε = 64,500).
(2) TAMRA-hx-PaTP (8b): A 0.1 M NaHCO3-Na2CO3 buffer solution (pH 8.5, 2.5 mL) of 7 (one-third of the crude product) was reacted with 5-carboxytetramethylrhodamine N-hydroxysuccinimidyl ester (TAMRA-SE, Invitrogen) (10 mg, 19 μmol) in DMF (1 mL), in the dark at room temperature. After 8 h, the reaction was quenched with concentrated ammonia (1 mL) for 1 h. The product was purified by DEAE-Sephadex A-25 column chromatography and C18 HPLC to give 8b (TAMRA-hx-PaTP). 1H-NMR (300 MHz, D2O) δ 9.07 (s, 1H), 8.26 (s, 1H), 8.02 (d, 1H, J = 8.0 Hz), 7.69 (s, 1H), 7.35 (d, 1H, J = 7.9 Hz), 7.10 (d, 1H, J = 9.4 Hz), 7.01 (d, 1H, J = 9.4 Hz), 6.83–6.72 (m, 3H), 6.39 (d, 2H, J = 9.2 Hz), 6.06 (d, 1H, J = 3.5 Hz), 4.20 (t, 1H, J = 4.9 Hz), 4.05–3.95 (m, 6H), 3.42 (m, 2H), 3.16–3.07 (m, 33H, TEA), 2.25 (m, 2H), 1.63 (m, 4H), 1.40 (m, 2H), 1.20 (t, 28H, J = 7.3 Hz, TEA). 31P-NMR (121 MHz, D2O) δ −10.84 (d, 1P, J = 18.8 Hz), −11.56 (d, 1P, J = 17.5 Hz), −23.22 (t, 1P, J = 19.0 Hz). ESI-MS for C44H49N5O19P3 [M−H]−: calcd., 1044.22; found, 1044.09. UV-vis spectrum (in 10 mM sodium phosphate buffer, pH 7.0): λmax 553 nm (ε = 107,000).
3.4. Synthesis of Dig-hx-PaTP (10)
1-(β-d-Ribofuranosyl)-4-[3-[6-(digoxigenin-3-O-methylcarbonyl)-aminohexananoyl]-amino-1-propynyl]pyrrole-2-carbaldehyde 5′-triphosphate (10). A 0.1 M NaHCO3-Na2CO3 buffer solution (pH 8.5, 1 mL) of 9 (NH2-PaTP, 8 μmol) was reacted with digoxigenin-3-O-methylcarbonyl-ε-aminocaproic acid-N-hydroxysuccinimide ester (Dig-hx-SE, Fluka) (5 mg, 7.6 μmol) in DMF (500 μL), at room temperature for 20 h. The product was purified by DEAE-Sephadex A-25 column chromatography and C18 HPLC, to give 5.3 μmol of 10 (Dig-hx-PaTP) in a 70% yield. 1H-NMR (300 MHz, D2O) δ 9.39 (s, 1H), 7.88 (s, 1H), 7.21 (s, 1H), 6.47 (d, 1H, J = 4.0 Hz), 5.94 (s, 1H), 4.97 (s, 2H), 4.36 (m, 2H), 4.20 (m, 3H), 4.10 (s, 2H), 3.90 (s, 2H), 3.65 (s, 1H), 3.44 (m, 1H), 3.26–3.11 (m, 23H, TEA), 2.24 (t, 2H, J = 7.0 Hz), 2.19–1.90 (m, 2H), 1.85–1.38 (m, 19H), 1.23 (m, 36H, TEA), 0.89 (s, 3H), 0.71 (s, 3H). 31P-NMR (121 MHz, D2O) δ −10.69 (d, 1P, J = 18.8 Hz), −11.33 (d, 1P, J = 18.5 Hz), −23.13 (t, 1P, J = 19.7 Hz). ESI-MS for C44H63N3O21P3 [M−H]−: calcd., 1062.32; found, 1061.99. UV-vis spectrum (in 10 mM sodium phosphate buffer, pH 7.0): λmax 308 nm (ε = 8,600).
3.5. DNA Templates for T7 Transcription
Each template DNA strand (10 μM; 65-mer, 60-mer, 55-mer, 50-mer, 45-mer, 42-mer, or 35-mer) was annealed with a 21-mer non-template strand (10 μM), in a buffer containing 10 mM Tris-HCl (pH 7.6) and 10 mM NaCl, by heating at 95 °C and slow cooling to 4 °C.
3.6. Transcription
Throughout the experiments, transcription (20 μL) was performed in a buffer containing 40 mM Tris–HCl (pH 8.0), 24 mM MgCl2, 5 mM DTT, 2 mM spermidine, and 0.01% Triton X-100, in the presence of 1 mM each NTP, 0 or 1 mM unnatural NTP, 0.05 μCi/μL [γ-32P]GTP, 2 μM DNA template, and 2.5 U/μL T7 RNA polymerase. After an incubation at 37 °C for 3 h, the reaction was quenched by adding an equivalent volume of a dye solution, containing 10 M urea and 0.05% BPB in 1× TBE. The reaction mixtures were heated at 75 °C for 3 min, and the products were analyzed on a 20% denaturing polyacrylamide gel containing 7 M urea, for the 17-mer transcripts, or a 10% denaturing polyacrylamide gel containing 7 M urea, for the other transcripts.
3.7. Analysis of the Insertion Position of Biotin-Pa in the 48-Mer Transcripts
RNA transcripts (48-mer) with or without Biotin-Pa at position 21 were prepared by T7 transcription using a 65-mer DNA fragment as the template, and were purified by gel electrophoresis. After dephosphorylation by Antarctic phosphatase, each RNA (10 pmol) was labeled with 32P at the 5′-end by [γ-32P]ATP and T4 polynucleotide kinase, and was purified by gel electrophoresis. Each labeled RNA transcript (40,000 to 70,000 cpm) was dissolved in 20 μL of water. The labeled RNA was partially digested under alkaline conditions, by incubating the mixtures of the RNA solution (2 μL) and the H solution (5 μL, 50 mM sodium carbonate (pH 9.1) and 1 mM EDTA) for 10 min and 20 min at 90 °C respectively. The two digested samples (14 μL in total) and 6 μL of 1× PBS were combined and used for further treatment with streptavidin magnetic beads to capture the RNA fragments containing Biotin-Pa. As a sequence marker, we partially digested the labeled RNAs (1 μL) by mixing them with RNase T1 (1 μL, 1 U/μL) and the A solution (9 μL; 20 mM sodium citrate pH 5.0, 7 M urea, 1 mM EDTA, 0.025% BPB, and 0.25 mg/mL tRNA mixture), at 55 °C for 12 min. The partially-digested RNA solution in alkali (10 μL) was incubated with 20 μL of streptavidin magnetic beads (4 mg/mL) for 5 min at room temperature. We analyzed 5 μL aliquots of the supernatants and the other digested samples on a 10% denaturing polyacrylamide gel containing 7 M urea.
4. Conclusions
We have examined the site-specific incorporation of functional components into RNA by an unnatural base pair transcription system, using the Ds–Pa pairing. The Pa substrate can easily be chemically modified with a wide range of functional groups, via the propyne linker. These modified-PaTPs can be site-specifically incorporated into RNA transcripts by conventional T7 transcription, by considering the characteristics of the Ds–Pa pair in transcription.
The PaTPs modified with small molecules (MW less than ~240, such as PaTP, Pa′TP, NH2-PaTP, and Biotin-PaTP) are efficiently incorporated into a specific internal position of a transcript, except for a position less than 10 bases from the 3′-terminus, without any natural base sequence dependency. For incorporation into a position close to the 3′-terminus of a transcript, the pyrimidine-Pa-pyrimidine sequences should be avoided, because of low transcription efficiency. Our previous results revealed that the incorporation selectivity of PaTP opposite Ds in templates is more than 90%, and the misincorporation rate of PaTP opposite the natural bases in templates is around 0.2% per base [6]. The present experiments using Biotin-PaTP also displayed high selectivity (>90%).
The PaTPs modified with large molecules (MW larger than ~480, such as TAMRA-hx-PaTP, FAM-hx-PaTP, and Dig-hx-PaTP) exhibited reduced incorporation efficiency and selectivity in T7 transcription, and their incorporation into a position less than 10 bases from the 3′-terminus of the transcripts should be avoided. However, for their internal incorporation into RNA (less than ~50-mer), the intended transcripts can be isolated by gel electrophoresis, because the mobility of the transcripts containing the modified-Pa bases is significantly shifted on a gel, relative to that of transcripts lacking the modified-Pa bases.
This specific transcription involving the Ds–Pa pair could provide a useful tool for the site-specific introduction of functional groups of interest into RNA molecules with long chains. The incorporation of PaTPs modified with large molecules, such as fluorescent dyes, is unfavorable for specific transcription, and thus, as an alternative method, the efficient incorporation of the NH2-Pa bases can be used as a site for post-transcriptional modifications with large functional molecules.
Acknowledgements
This work was supported by the Targeted Proteins Research Program and the RIKEN Structural Genomics/Proteomics Initiative, the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan, and by Grants-in-Aid for Scientific Research (KAKENHI 19201046 to I.H., 20710176 to M.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References and Notes
- Benner, S.A. Understanding nucleic acids using synthetic chemistry. Acc. Chem. Res. 2004, 37, 784–797. [Google Scholar] [CrossRef] [PubMed]
- Henry, A.A.; Romesberg, F.E. Beyond A, C, G and T: Augmenting nature’s alphabet. Curr. Opin. Chem. Biol. 2003, 7, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Krueger, A.T.; Kool, E.T. Redesigning the architecture of the base pair: Toward biochemical and biological function of new genetic sets. Chem. Biol. 2009, 16, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I. Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 2006, 10, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Cox, R.S., 3rd.; Hirao, I. Unnatural base pair systems for sensing and diagnostic applications. Expert Rev. Mol. Diagn. 2011, 11, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Kimoto, M.; Mitsui, T.; Fujiwara, T.; Kawai, R.; Sato, A.; Harada, Y.; Yokoyama, S. An unnatural hydrophobic base pair system: Site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods 2006, 3, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Kawai, R.; Mitsui, T.; Yokoyama, S.; Hirao, I. An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res. 2009, 37, e14. [Google Scholar] [CrossRef] [PubMed]
- Yamashige, R.; Kimoto, M.; Takezawa, Y.; Sato, A.; Mitsui, T.; Yokoyama, S.; Hirao, I. Highly specific unnatural base pair systems as a third base pair for PCR amplification. Nucleic Acids Res. 2011. [Google Scholar] [CrossRef] [PubMed]
- Malyshev, D.A.; Seo, Y.J.; Ordoukhanian, P.; Romesberg, F.E. PCR with an expanded genetic alphabet. J. Am. Chem. Soc. 2009, 131, 14620–14621. [Google Scholar] [CrossRef] [PubMed]
- Malyshev, D.A.; Pfaff, D.A.; Ippoliti, S.I.; Hwang, G.T.; Dwyer, T.J.; Romesberg, F.E. Solution structure, mechanism of replication, and optimization of an unnatural base pair. Chem. Eur. J. 2010, 16, 12650–12659. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sismour, A.M.; Sheng, P.; Puskar, N.L.; Benner, S.A. Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res. 2007, 35, 4238–4249. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, F.; Alvarado, J.B.; Benner, S.A. Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J. Am. Chem. Soc. 2011, 133, 15105–15112. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Ohtsuki, T.; Fujiwara, T.; Mitsui, T.; Yokogawa, T.; Okuni, T.; Nakayama, H.; Takio, K.; Yabuki, T.; Kigawa, T.; Kodama, K.; Nishikawa, K.; Yokoyama, S. An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol. 2002, 20, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Hikida, Y.; Kimoto, M.; Yokoyama, S.; Hirao, I. Site-specific fluorescent probing of RNA molecules by unnatural base-pair transcription for local structural conformation analysis. Nat. Protoc. 2010, 5, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Hirao, I. Site-specific incorporation of extra components into RNA by transcription using unnatural base pair systems. Methods Mol. Biol. 2010, 634, 355–369. [Google Scholar] [PubMed]
- Seo, Y.J.; Malyshev, D.A.; Lavergne, T.; Ordoukhanian, P.; Romesberg, F.E. Site-specific labeling of DNA and RNA using an efficiently replicated and transcribed class of unnatural base pairs. J. Am. Chem. Soc. 2011, 133, 19878–19888. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.J.; Matsuda, S.; Romesberg, F.E. Transcription of an expanded genetic alphabet. J. Am. Chem. Soc. 2009, 131, 5046–5047. [Google Scholar] [CrossRef] [PubMed]
- Tor, Y.; Dervan, P.B. Site-specific enzymatic incorporation of an unnatural base, N6-(6-aminohexyl)isoguanosine, into RNA. J. Am. Chem. Soc. 1993, 115, 4461–4467. [Google Scholar] [CrossRef]
- Kimoto, M.; Mitsui, T.; Yamashige, R.; Sato, A.; Yokoyama, S.; Hirao, I. A new unnatural base pair system between fluorophore and quencher base analogues for nucleic acid-based imaging technology. J. Am. Chem. Soc. 2010, 132, 15418–15426. [Google Scholar] [CrossRef] [PubMed]
- Kawai, R.; Kimoto, M.; Ikeda, S.; Mitsui, T.; Endo, M.; Yokoyama, S.; Hirao, I. Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J. Am. Chem. Soc. 2005, 127, 17286–17295. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Endo, M.; Mitsui, T.; Okuni, T.; Hirao, I.; Yokoyama, S. Site-specific incorporation of a photo-crosslinking component into RNA by T7 transcription mediated by unnatural base pairs. Chem. Biol. 2004, 11, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, K.; Kimoto, M.; Mitsui, T.; Yokoyama, S.; Hirao, I. Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 2005, 33, e129. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I.; Mitsui, T.; Kimoto, M.; Yokoyama, S. An efficient unnatural base pair for PCR amplification. J. Am. Chem. Soc. 2007, 129, 15549–15555. [Google Scholar] [CrossRef] [PubMed]
- Nacheva, G.A.; Berzal-Herranz, A. Preventing nondesired RNA-primed RNA extension catalyzed by T7 RNA polymerase. Eur. J. Biochem. 2003, 270, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Yamashige, R.; Kimoto, M.; Mitsui, T.; Yokoyama, S.; Hirao, I. Monitoring the site-specific incorporation of dual fluorophore-quencher base analogues for target DNA detection by an unnatural base pair system. Org. Biomol. Chem. 2011, 9, 7504–7509. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of three triphosphate derivatives (NH2-hx-PaTP, Biotin-PaTP, FAM-PaTP) are available from the authors. |
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).