Cleavage of Oligonucleotides Containing a P3’→N5’ Phosphoramidate Linkage Mediated by Single-Stranded Oligonucleotide Templates
Abstract
:1. Introduction
 ) as probes; the TFOs have a P3’→N5’ phosphoramidate (P-N) linkage in the backbone. This linkage was more susceptible to acid-mediated hydrolysis upon triplex formation, and the enhanced susceptibility was due to conformational strain on the P-N linkage induced by triplex formation. Previously, we examined the effects of chemical modifications that alter the microenvironment around the P-N linkage and change the extent of the conformational strain; these chemical modifications had substantial effects on the observed pseudo first-order rate constants (kobss) of the hydrolysis with the dsDNA templates [29]. These findings indicated that when the P-N linkage is subjected to sufficient strain, the linkage promptly breaks upon hybridization to the template. We hypothesized that duplex formation, like triplex formation, could induce conformational strain when oligonucleotides have a certain chemical modification and that such oligonucleotides may be selectively cleaved in the presence of single-stranded templates and, therefore, may be used as probes to detect single-stranded nucleic acids (Figure 2).
) as probes; the TFOs have a P3’→N5’ phosphoramidate (P-N) linkage in the backbone. This linkage was more susceptible to acid-mediated hydrolysis upon triplex formation, and the enhanced susceptibility was due to conformational strain on the P-N linkage induced by triplex formation. Previously, we examined the effects of chemical modifications that alter the microenvironment around the P-N linkage and change the extent of the conformational strain; these chemical modifications had substantial effects on the observed pseudo first-order rate constants (kobss) of the hydrolysis with the dsDNA templates [29]. These findings indicated that when the P-N linkage is subjected to sufficient strain, the linkage promptly breaks upon hybridization to the template. We hypothesized that duplex formation, like triplex formation, could induce conformational strain when oligonucleotides have a certain chemical modification and that such oligonucleotides may be selectively cleaved in the presence of single-stranded templates and, therefore, may be used as probes to detect single-stranded nucleic acids (Figure 2). , Figure 1) in the middle of a sequence with one of two chemical modifications, 2’,4’-BNA/LNA [30,31,32] (designated
, Figure 1) in the middle of a sequence with one of two chemical modifications, 2’,4’-BNA/LNA [30,31,32] (designated  ) or 2’,5’-linked DNA [33,34] (designated
) or 2’,5’-linked DNA [33,34] (designated  ), on adjacent residues (Table 1). The reactivity of these oligonucleotides in the presence of single-stranded DNA (ssDNA) or ssRNA templates was compared with their reactivity in the presence of parallel double-stranded DNA (PDD) templates and in the absence of any template. The parallel (Hoogsteen motif) single-stranded DNA and RNA (PSD and PSR, respectively) and anti-parallel (Watson-Crick motif) single-stranded DNA and RNA (ASD and ASR) were prepared as templates (see Table 1 caption). The formation of different motifs of duplexes was expected to have different effects on the reactivity of the hydrolysis depending on the extent of the strain.
), on adjacent residues (Table 1). The reactivity of these oligonucleotides in the presence of single-stranded DNA (ssDNA) or ssRNA templates was compared with their reactivity in the presence of parallel double-stranded DNA (PDD) templates and in the absence of any template. The parallel (Hoogsteen motif) single-stranded DNA and RNA (PSD and PSR, respectively) and anti-parallel (Watson-Crick motif) single-stranded DNA and RNA (ASD and ASR) were prepared as templates (see Table 1 caption). The formation of different motifs of duplexes was expected to have different effects on the reactivity of the hydrolysis depending on the extent of the strain. (5’-amino-2’,4’-BNA),
(5’-amino-2’,4’-BNA), 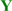 (2’,4’-BNA/LNA), and
(2’,4’-BNA/LNA), and  (2’,5’-linked DNA).
(2’,5’-linked DNA).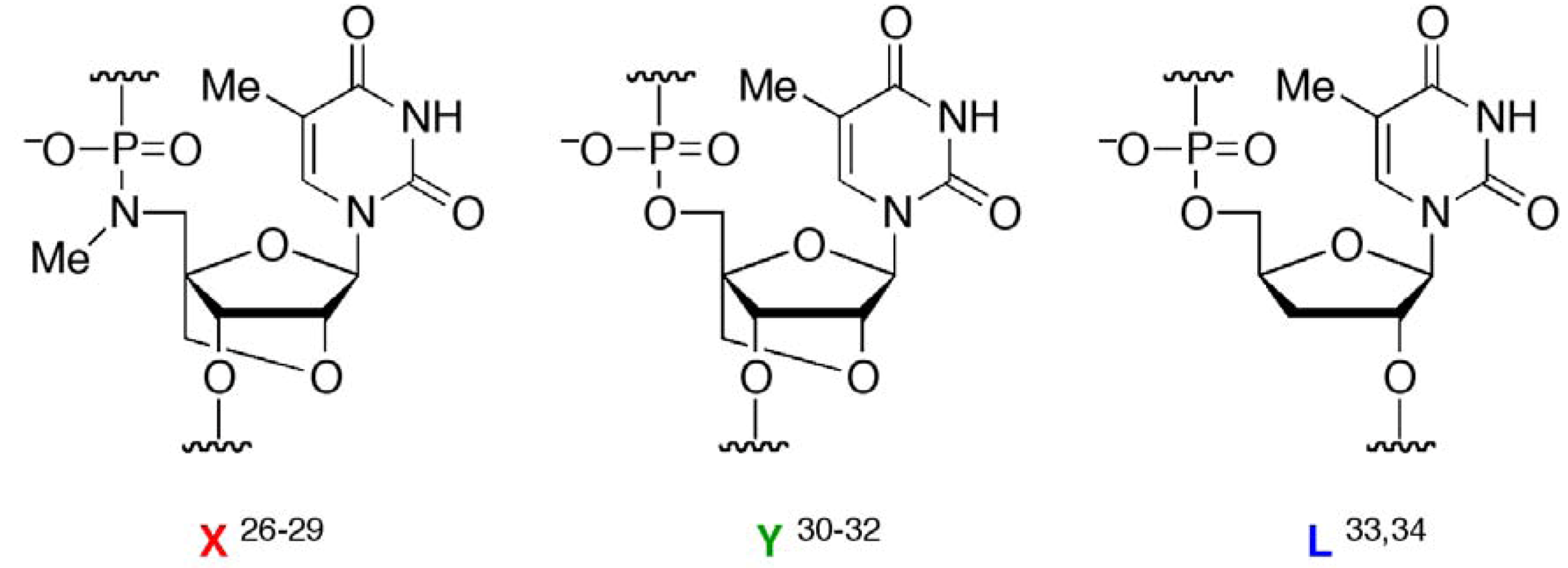
2. Results
2.1. UV Melting Experiments
| ON | Sequence (5’ to 3’) b | Tm (Δ Tm) in °C with | |||
|---|---|---|---|---|---|
| PSD c | PSR c | ASD c | ASR c | ||
| ON-0 | TTTTTmCTTTmCTmCTmCT | 33 (-) | 34 (-) | 52 (-) | 54 (-) |
| ON-0 d | TTTTTmCTTTmCTmCTmCT | 48 (+15) | n.d. e | 48 (−4) | 36 (−18) |
| ON-1 | TTTTTmCT  TmCTmCTmCT TmCTmCTmCT | 38 (+5) | 34 (±0) | 54 (+2) | 57 (+3) |
| ON-2 | TTTTTmC 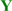  TmCTmCTmCT TmCTmCTmCT | 37 (+4) | 37 (+3) | 53 (+1) | 60 (+6) |
| ON-3 | TTTTTmCT  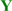 mCTmCTmCT mCTmCTmCT | 41 (+8) | 40 (+6) | 55 (+3) | 62 (+8) |
| ON-4 | TTTTTmC 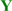  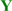 mCTmCTmCT mCTmCTmCT | 44 (+11) | 44 (+10) | 54 (+2) | 65 (+11) |
| ON-5 | TTTT  mCT mCT  TmC TmC  mCTmCT mCTmCT | 45 (+12) | 46 (+12) | 57 (+5) | 69 (+15) |
| ON-6 | TTTTTmC   TmCTmCTmCT TmCTmCTmCT | 33 (±0) | 34 (±0) | 47 (−5) | 55 (+1) |
| ON-7 | TTTTTmCT   mCTmCTmCT mCTmCTmCT | 31 (−2) | 30 (−4) | 49 (−3) | 53 (−1) |
| ON-8 | TTTTTmC    mCTmCTmCT mCTmCTmCT | 31 (−2) | 29 (−5) | 43 (−9) | 51 (−3) |
 , 5’-amino-2’,4’-BNA (NMe);
, 5’-amino-2’,4’-BNA (NMe);  , 2’,4’-BNA/LNA;
, 2’,4’-BNA/LNA;  , 2’,5’-linked DNA; mC, 5-MedC; c PSD (parallel single-stranded DNA), 5’-d(AAAAAGAAAGAGAGA)-3’; PSR (parallel single-stranded RNA), 5’-r(AAAAAGAAAGAGAGA)-3’; ASD (anti-parallel single-stranded DNA), 5’-d(AGAGAGAAAGAAAAA)-3’; ASR (anti-parallel single-stranded RNA), 5’-r(AGAGAGAAAGAAAAA)-3’; d Tm measured at pH 4.0, for detail see experimental section; e: Not determined due to low stability (Tm < 25 °C).
, 2’,5’-linked DNA; mC, 5-MedC; c PSD (parallel single-stranded DNA), 5’-d(AAAAAGAAAGAGAGA)-3’; PSR (parallel single-stranded RNA), 5’-r(AAAAAGAAAGAGAGA)-3’; ASD (anti-parallel single-stranded DNA), 5’-d(AGAGAGAAAGAAAAA)-3’; ASR (anti-parallel single-stranded RNA), 5’-r(AGAGAGAAAGAAAAA)-3’; d Tm measured at pH 4.0, for detail see experimental section; e: Not determined due to low stability (Tm < 25 °C).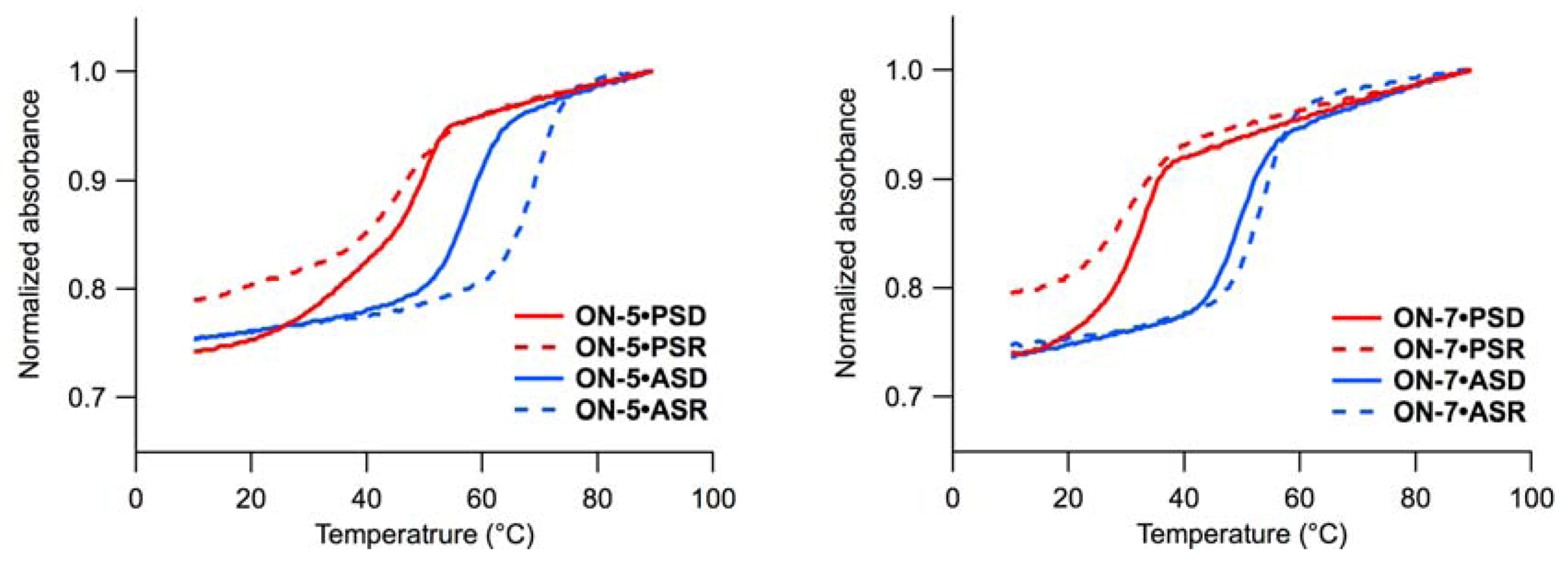
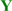 ) (ON-2–ON-5) stabilized the duplexes in most cases [30,31,32], and the stabilizing effects were more apparent for the duplexes with PSD, PSR, and ASR than those with ASD. Comparisons between ON-2 and ON-3 Tms revealed that a
) (ON-2–ON-5) stabilized the duplexes in most cases [30,31,32], and the stabilizing effects were more apparent for the duplexes with PSD, PSR, and ASR than those with ASD. Comparisons between ON-2 and ON-3 Tms revealed that a 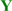 positioned just 3’ of
positioned just 3’ of  (5’-amino-2’,4’-BNA) stabilized all duplexes to a larger extent than a
(5’-amino-2’,4’-BNA) stabilized all duplexes to a larger extent than a 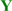 positioned just 5’ of
positioned just 5’ of  . Insertion of two residues between
. Insertion of two residues between  and
and 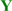 had greater stabilizing effects in all types of duplexes tested here (ON-4 vs. ON-5). Comparison of Tms of duplexes containing 2’,5’-linked DNA (
had greater stabilizing effects in all types of duplexes tested here (ON-4 vs. ON-5). Comparison of Tms of duplexes containing 2’,5’-linked DNA (  ) with those of ON-1 revealed that introduction of
) with those of ON-1 revealed that introduction of  is destabilizing in most cases. The destabilization was less pronounced for the duplexes with ASR, as reported previously [35,36,37]. The melting curves of the duplexes consisting of PSD and ON-3, ON-4, or ON-8 showed some two-transition character.
is destabilizing in most cases. The destabilization was less pronounced for the duplexes with ASR, as reported previously [35,36,37]. The melting curves of the duplexes consisting of PSD and ON-3, ON-4, or ON-8 showed some two-transition character.2.2. Hydrolysis Experiments
| ON | kobs × 103 (s−1) in the presence of | ||||
|---|---|---|---|---|---|
| No template b | PDD b,c | PSD | ASD | ASR | |
| ON-1 | 0.027 ± 0.005 | 0.77 ± 0.03 | 0.57 ± 0.09 | 0.14 ± 0.01 | 0.17 ± <0.01 |
| ON-2 | 0.017 ± 0.006 | 0.051 ± 0.013 | 0.025 ± 0.001 | 0.020 ± 0.002 | 0.032 ± 0.001 |
| ON-3 | 0.038 ± 0.012 | 1.4 ± 0.4 | 0.60 ± 0.05 | 0.10 ± <0.01 | 0.19 ± <0.01 |
| ON-4 | 0.026 ± 0.003 | 0.044 ± 0.025 | 0.032 ± 0.003 | 0.015 ± 0.003 | n.d. d |
| ON-5 | 0.029 ± 0.004 | 1.3 ± 0.4 | 0.86 ± 0.11 | 0.24 ± 0.02 | 0.19 ± <0.01 |
| ON-6 | 0.066 ± 0.024 | n.d. d | 0.021 ± 0.004 | 0.021 ± 0.001 | 0.011 ± 0.001 |
| ON-7 | 0.022 ± 0.002 | 2.1 ± 0.1 | 0.83 ± 0.03 | 0.29 ± <0.01 | 0.18 ± <0.01 |
| ON-8 | 0.058 ± 0.003 | 0.022 ± 0.012 | 0.078 ± 0.003 | 0.035 ± 0.002 | 0.024 ± <0.001 |
| ON | kobs × 103 (s−1) in the presence of | ||
|---|---|---|---|
| No template | PSD | ASR | |
| ON-1 | 0.011 ± <0.001 | 0.27 ± 0.06 | 0.022 ± 0.001 |
| ON-2 | 0.011 ± 0.002 | 0.0067 ± 0.0006 | 0.0059 ± 0.0006 |
| ON-3 | 0.0072 ± 0.0010 | 0.19 ± 0.05 | 0.032 ± 0.002 |
| ON-4 | 0.0062 ± 0.0011 | n.d. b | n.d. b |
| ON-5 | 0.0073 ± 0.0015 | 0.26 ± 0.01 | 0.041 ± <0.001 |
| ON-6 | 0.0067 ± 0.0013 | n.d. b | n.d. b |
| ON-7 | 0.0080 ± 0.0008 | 0.68 ± 0.04 | 0.037 ± 0.005 |
| ON-8 | 0.0070 ± 0.0005 | n.d. b | n.d. b |
2.2.1. Reactivity on Parallel Single-Stranded DNA
 to the 5’-neighboring residue of
to the 5’-neighboring residue of  (the sequence 5’-
(the sequence 5’- 
 -3’ found in ON-2 and ON-4) resulted in inactivation of hydrolysis upon hybridization to PSD (Figure S2).
-3’ found in ON-2 and ON-4) resulted in inactivation of hydrolysis upon hybridization to PSD (Figure S2).  added to the 3’-neighboring residue of
added to the 3’-neighboring residue of  (5’-T
(5’-T 
 -3’) had little effect, resulting in equivalent kobs for ON-1 and ON-3. Accelerated hydrolysis of ON-5 on PSD was observed as non-neighboring residual effects of
-3’) had little effect, resulting in equivalent kobs for ON-1 and ON-3. Accelerated hydrolysis of ON-5 on PSD was observed as non-neighboring residual effects of  . The sequence 5’-
. The sequence 5’- 
 -3’ found in ON-6 and ON-8 eliminated the acceleration associated with hybridization to PSD, while hydrolysis of ON-7 (sequence 5’-T
-3’ found in ON-6 and ON-8 eliminated the acceleration associated with hybridization to PSD, while hydrolysis of ON-7 (sequence 5’-T 
 -3’) was accelerated (Figure 4). At 20 °C, ON-1–ON-8 were less reactive, but the reactivity of ON-7 was least affected, and reactivities of ON-1, ON-3, and ON-5 were equivalent in the presence of PSD (Table 3, Figure S3).
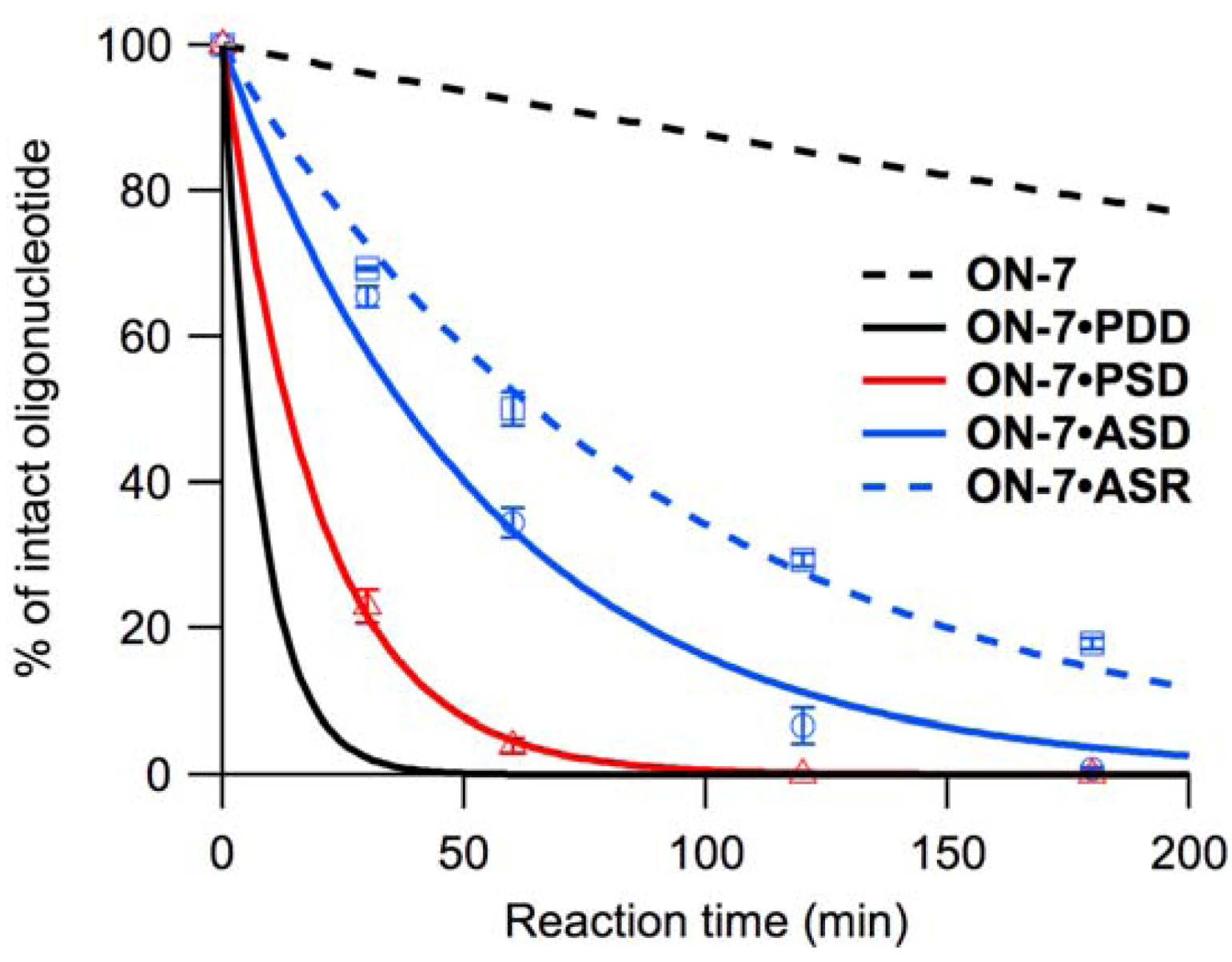
-3’) was accelerated (Figure 4). At 20 °C, ON-1–ON-8 were less reactive, but the reactivity of ON-7 was least affected, and reactivities of ON-1, ON-3, and ON-5 were equivalent in the presence of PSD (Table 3, Figure S3).2.2.2. Reactivity on Anti-Parallel Single-Stranded DNA and RNA
3. Discussion
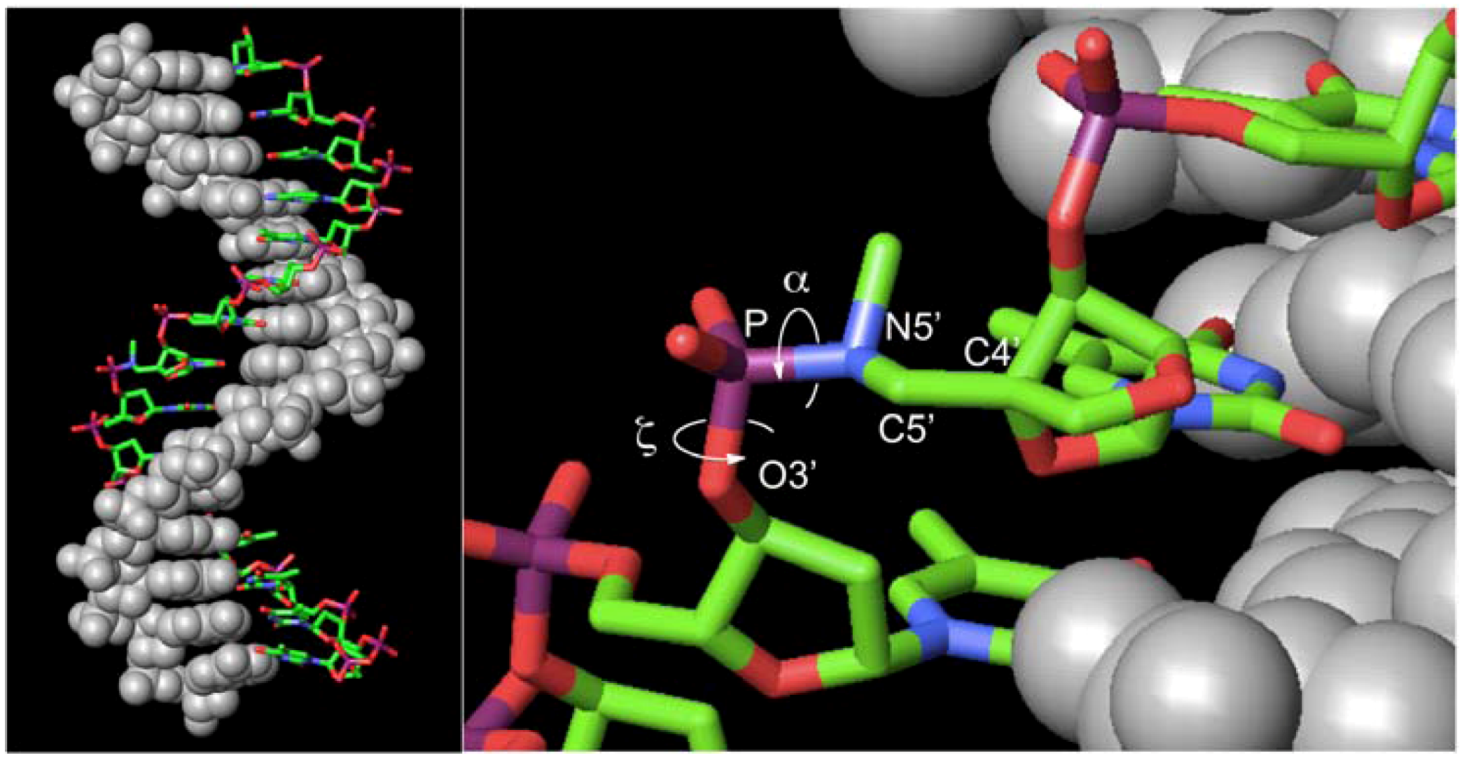
 did not prohibit α and ζ dihedral angles from adopting −sc orientations (Figure 5, Figure S5). However, the high reactivity in triplexes and parallel Hoogsteen duplexes may have been due to a change in the preferred dihedral angles, which would make the phosphoramidates more basic. The quantum chemical calculations using one model compound (N,N,O-trimethylphosphoramidate, R = R’ = Me in Scheme 1) revealed the relative stability between the neutral form (N-protonated) and the anionic form of the phosphoramidate (Scheme 1) as functions of α and ζ.
did not prohibit α and ζ dihedral angles from adopting −sc orientations (Figure 5, Figure S5). However, the high reactivity in triplexes and parallel Hoogsteen duplexes may have been due to a change in the preferred dihedral angles, which would make the phosphoramidates more basic. The quantum chemical calculations using one model compound (N,N,O-trimethylphosphoramidate, R = R’ = Me in Scheme 1) revealed the relative stability between the neutral form (N-protonated) and the anionic form of the phosphoramidate (Scheme 1) as functions of α and ζ. 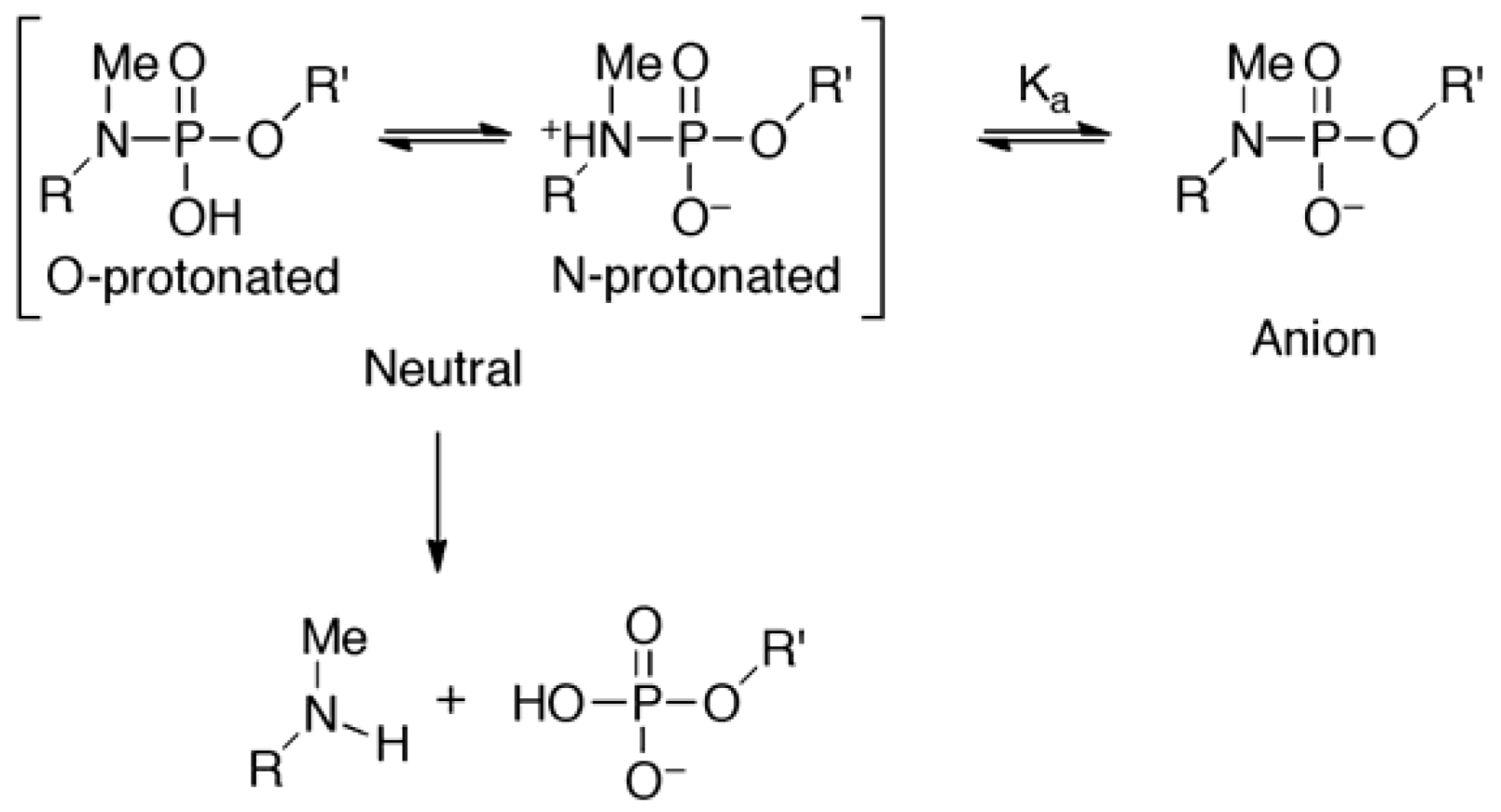
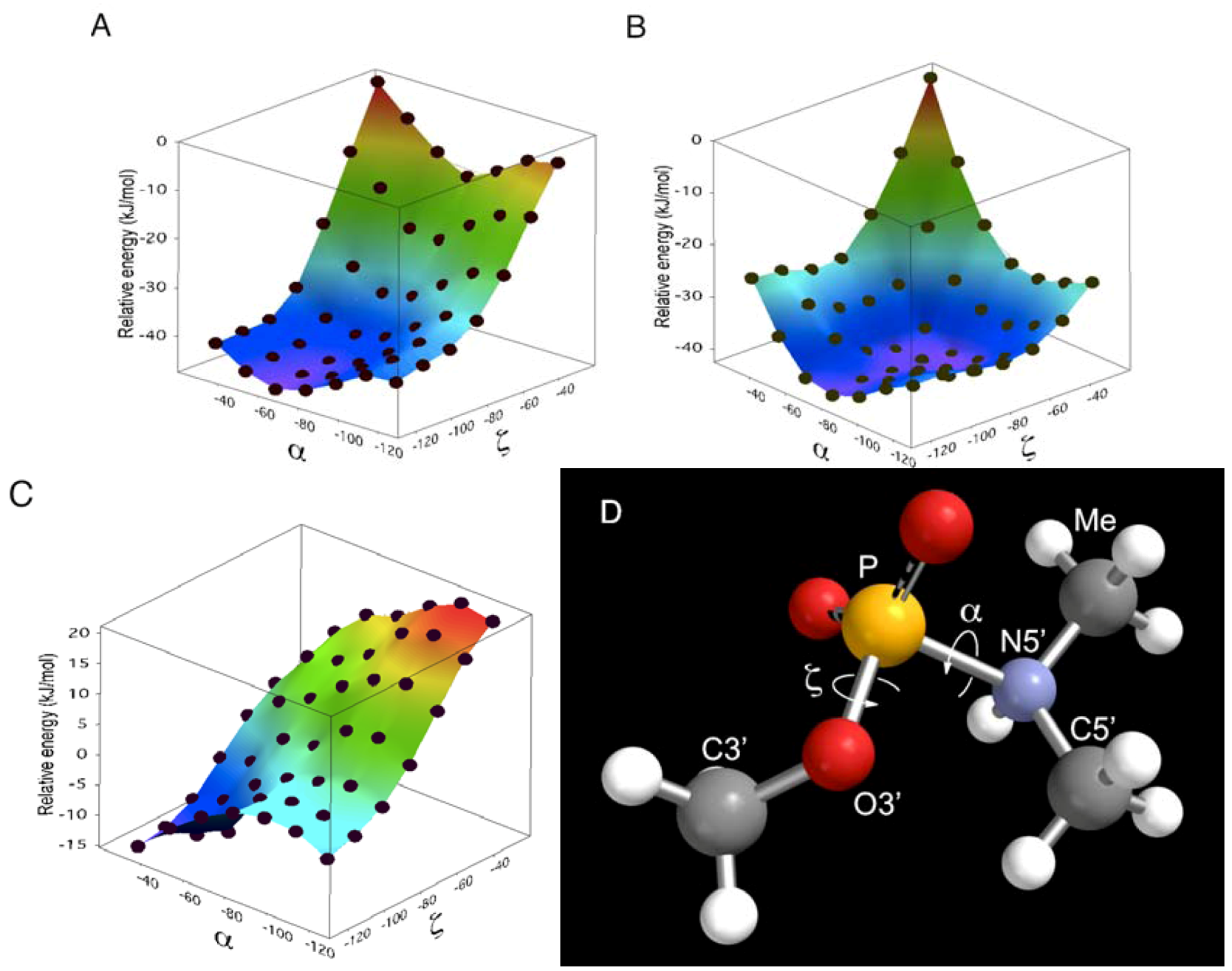
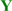 is known to affect the sugar conformation of 3’-adjacent nucleotide inducing C3’-endo conformation [50,51]. Although such an effect is not the case in ON-2 and ON-4 because
is known to affect the sugar conformation of 3’-adjacent nucleotide inducing C3’-endo conformation [50,51]. Although such an effect is not the case in ON-2 and ON-4 because  is pre-locked to C3’-endo conformation due to the 2’,4’-bridge moiety [26],
is pre-locked to C3’-endo conformation due to the 2’,4’-bridge moiety [26], 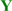 at 5’-adjacent of the phosphoramidate will have structurally affected the preferred conformation of the phosphoramidate, which resulted in inactivation of ON-2 and ON-4 in the presence of templates.
at 5’-adjacent of the phosphoramidate will have structurally affected the preferred conformation of the phosphoramidate, which resulted in inactivation of ON-2 and ON-4 in the presence of templates.4. Experimental Section
4.1. Preparation of Oligonucleotides
2.2. UV Melting Experiments
4.3. Hydrolysis Experiments
4.4. Molecular Modeling and Computation
 . The structures generated were exported to MacroModel 9.1TM (Schrödinger, LLC, New York, NY, USA). An energy minimization calculation was performed for each structure using 1) AMBER* as a force field, 2) the GB/SA solvation model of water, and 3) the PRCG method to obtain structures optimized to within a gradient of 0.05 kJ/molÅ. Finally, the 5’-oxygen of 2’,4’-BNA/LNA in ON-1 of the optimized structures was replaced by nitrogen attached to a methyl group to obtain the molecular models (Figure 5, Figure S5).
. The structures generated were exported to MacroModel 9.1TM (Schrödinger, LLC, New York, NY, USA). An energy minimization calculation was performed for each structure using 1) AMBER* as a force field, 2) the GB/SA solvation model of water, and 3) the PRCG method to obtain structures optimized to within a gradient of 0.05 kJ/molÅ. Finally, the 5’-oxygen of 2’,4’-BNA/LNA in ON-1 of the optimized structures was replaced by nitrogen attached to a methyl group to obtain the molecular models (Figure 5, Figure S5).5. Conclusions
Supplementary Materials
Acknowledgments
Conflict of Interest
References
- Xu, Y.; Karalkar, N.B.; Kool, E.T. Nonenzymatic autoligation in direct three-color detection of RNA and DNA point mutations. Nat. Biotechnol. 2001, 19, 148–152. [Google Scholar] [CrossRef]
- Sando, S.; Kool, E.T. Quencher as leaving group: Efficient detection of DNA-joining reactions. J. Am. Chem. Soc. 2002, 124, 2096–2097. [Google Scholar] [CrossRef]
- Abe, H.; Kool, E.T. Destabilizing universal linkers for signal amplification in self-ligating probes for RNA. J. Am. Chem. Soc. 2004, 126, 13980–13986. [Google Scholar] [CrossRef]
- Ficht, S.; Dose, C.; Seitz, O. As fast and selective as enzymatic ligations: Unpaired nucleobases increase the selectivity of DNA-controlled native chemical PNA ligation. ChemBioChem 2005, 6, 2098–2103. [Google Scholar] [CrossRef]
- Dose, C.; Ficht, S.; Seitz, O. Reducing product inhibition in DNA-template-controlled ligation reactions. Angew. Chem. Int. Ed. 2006, 45, 5369–5373. [Google Scholar] [CrossRef]
- Abe, H.; Kool, E.T. Flow cytometric detection of specific RNAs in native human cells with quenched autoligating FRET probes. Proc. Natl. Acad. Sci. USA 2006, 103, 263–268. [Google Scholar] [CrossRef]
- Ogasawara, S.; Fujimoto, K. SNP genotyping by using photochemical ligation. Angew. Chem. Int. Ed. 2006, 45, 4512–4515. [Google Scholar] [CrossRef]
- Yoshimura, Y.; Noguchi, Y.; Sato, H.; Fujimoto, K. Template-directed DNA photoligation in rapid and selective detection of RNA point mutations. ChemBioChem 2006, 7, 598–601. [Google Scholar] [CrossRef]
- Peng, X.; Greenberg, M.M. Facile SNP detection using bifunctional, cross-linking oligonucleotide probes. Nucleic Acids Res. 2008, 26, e31. [Google Scholar]
- Abe, H.; Kondo, Y.; Jinmei, H.; Abe, N.; Furukawa, K.; Uchiyama, A.; Tsuneda, S.; Aikawa, K.; Matsumoto, I.; Ito, Y. Rapid DNA chemical ligation for amplification of RNA and DNA signal. Bioconjug. Chem. 2008, 19, 327–333. [Google Scholar] [CrossRef]
- Dose, C.; Seitz, O. Single nucleotide specific detection of DNA by native chemical ligation of fluorescence labeled PNA-probes. Bioorg. Med. Chem. 2008, 16, 65–77. [Google Scholar] [CrossRef]
- Grossmann, T.N.; Seitz, O. DNA-catalyzed transfer of a reporter group. J. Am. Chem. Soc. 2006, 128, 15596–15597. [Google Scholar] [CrossRef]
- Grossmann, T.N.; Röglin, L.; Seitz, O. Angew. Chem. Int. Ed. 2008, 47, 7119–7123. [CrossRef]
- Grossmann, T.N.; Seitz, O. Nucleic acid templated reactions: Consequences of probe reactivity and readout strategy for amplified signaling and sequence selectivity. Chem. Eur. J. 2009, 15, 6723–6730. [Google Scholar] [CrossRef]
- Brunner, J.; Mokhir, A.; Kraemer, R. DNA-templated metal catalysis. J. Am. Chem. Soc. 2003, 125, 12410–12411. [Google Scholar] [CrossRef]
- Boll, I.; Krämer, R.; Brunner, J.; Mokhir, A. Templated metal catalysis for single nucleotide specific DNA sequence detection. J. Am. Chem. Soc. 2005, 127, 7849–7856. [Google Scholar] [CrossRef]
- Franzini, R.M.; Kool, E.T. Efficient nucleic acid detection by templated reductive quencher release. J. Am. Chem. Soc. 2009, 131, 16021–16023. [Google Scholar] [CrossRef]
- Cai, J.; Li, X.; Yue, X.; Taylor, J.S. Nucleic acid-triggered fluorescent probe activation by the Staudinger reaction. J. Am. Chem. Soc. 2004, 126, 16324–16325. [Google Scholar]
- Cai, J.; Li, X.; Taylor, J.S. Improved nucleic acid triggered probe activation through the use of a 5-thiomethyluracil peptide nucleic acid building block. Org. Lett. 2005, 7, 751–754. [Google Scholar] [CrossRef]
- Pianowski, Z.L.; Winssinger, N. Fluorescence-based detection of single nucleotide permutation in DNA via catalytically templated reaction. Chem. Commun. 2007, 3820–3822. [Google Scholar] [CrossRef]
- Franzini, R.M.; Kool, E.T. 7-Azidomethoxy-coumarins as profluorophores for templated nucleic acid detection. ChemBioChem 2008, 9, 2981–2988. [Google Scholar] [CrossRef]
- Franzini, R.M.; Kool, E.T. Organometallic activation of a fluorogen for templated nucleic acid detection. Org. Lett. 2008, 10, 2935–2938. [Google Scholar] [CrossRef]
- Furukawa, K.; Abe, H.; Wang, J.; Uda, M.; Koshino, H.; Tsuneda, S.; Ito, Y. Reduction-triggered red fluorescent probes for dual-color detection of oligonucleotide sequences. Org. Biomol. Chem. 2009, 7, 671–677. [Google Scholar] [CrossRef]
- Prusty, D.K.; Herrmann, A. A fluorogenic reaction based on heavy-atom removal for ultrasensitive DNA detection. J. Am. Chem. Soc. 2010, 132, 12197–12199. [Google Scholar] [CrossRef]
- Li, X.; Liu, D.R. DNA-templated organic synthesis: Nature’s strategy for controlling chemical reactivity applied to synthetic molecules. Angew. Chem. Int. Ed. 2004, 43, 4848–4870. [Google Scholar] [CrossRef]
- Obika, S.; Nakagawa, O.; Hiroto, A.; Hari, Y.; Imanishi, T. Synthesis and properties of a novel bridged nucleic acid with a P3’→N5’ phosphoramidate linkage, 5’-amino-2’,4’-BNA. Chem. Commun. 2003, 2202–2203. [Google Scholar]
- Obika, S.; Tomizu, M.; Negoro, Y.; Osakai, T.; Orita, A.; Ueyama, Y.; Nakagawa, O.; Imanishi, T. Acid-mediated cleavage of oligonucleotide P3’→N5’ phosphoramidates triggered by sequence-specific triplex formation. Nucleos. Nucleot. Nucleic Acids 2007, 26, 893–896. [Google Scholar] [CrossRef]
- Obika, S.; Tomizu, M.; Negoro, Y.; Orita, A.; Nakagawa, O.; Imanishi, T. Double-stranded DNA-templated digestion triggered by triplex formation. ChemBioChem 2007, 8, 1924–1928. [Google Scholar] [CrossRef]
- Ito, K.R.; Kodama, T.; Tomizu, M.; Negoro, Y.; Orita, A.; Osaki, T.; Hosoki, N.; Tanaka, T.; Imanishi, T.; Obika, S. Double-stranded DNA-templated cleavage of oligonucleotides containing a P3’→N5’ linkage triggered by triplex formation: The effects of chemical modifications and remarkable enhancement in reactivity. Nucleic Acids Res. 2010, 38, 7332–7342. [Google Scholar]
- Obika, S.; Nanbu, D.; Hari, Y.; Andoh, J.; Morio, K.; Doi, T.; Imanishi, T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2’-O,4’-C-methyleneribonucleosides. Tetrahedron Lett. 1998, 39, 5401–5404. [Google Scholar] [CrossRef]
- Singh, S.K.; Nielsen, P.; Koshkin, A.A.; Wengel, J. LNA (locked nucleic acids): Synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998, 455–456. [Google Scholar]
- Bhattacharyya, J.; Maiti, S.; Muhuri, S.; Nakano, S.; Miyoshi, D.; Sugimoto, N. Effect of locked nucleic acid modifications on the thermal stability of noncanonical DNA structure. Biochemistry 2011, 50, 7414–7425. [Google Scholar] [CrossRef]
- Rizzo, C.J.; Dougherty, J.P.; Breslow, R. 3’-Deoxy-2’-phosphoramidites of adenosine and 5-methyluridine used for the solid phase synthesis of unnatural 3’-deoxy-2’-5’’-oligonucleotides. Tetrahedron Lett. 1992, 33, 4129–4132. [Google Scholar]
- Dougherty, J.P.; Rizzo, C.J.; Breslow, R. Oligodeoxynucleotides that contain 2’,5’’ linkages: Synthesis and hybridization properties. J. Am. Chem. Soc. 1992, 114, 6254–6255. [Google Scholar] [CrossRef]
- Giannaris, P.A.; Damha, M.J. Oligoribonucleotides containing 2’,5’-phosphodiester linkages exhibit binding selectivity for 3’,5’-RNA over 3’,5’-ssDNA. Nucleic Acids Res. 1993, 21, 4742–4749. [Google Scholar] [CrossRef]
- Prakash, T.P.; Jung, K.; Switzer, C. RNA recognition by the 2’-structural isomer of DNA. Chem. Commun. 1996, 1793–1794. [Google Scholar]
- Sheppard, T.L.; Breslow, R.C. Selective binding of RNA, not DNA, by complementary 2’,5’-linked DNA. J. Am. Chem. Soc. 1996, 118, 9810–9811. [Google Scholar] [CrossRef]
- Raghunathan, G.; Miles, H.T.; Sasisekharan, V. Parallel nucleic acid helices with Hoogsteen base pairing: Symmetry and structure. Biopolymers 1994, 34, 1573–1581. [Google Scholar] [CrossRef]
- Singleton, S.F.; Dervan, P.B. Influence of pH on the equilibrium association constants for oligodeoxyribonucleotide-directed triple helix formation at single DNAsites. Biochemistry 1992, 31, 10995–11003. [Google Scholar]
- Hashem, G.M.; Wen, J.; Do, Q.; Gray, D.M. Evidence from CD spectra and melting temperatures for stable Hoogsteen-paired oligomer duplexes derived from DNA and hybrid triplexes. Nucleic Acids Res. 1999, 3371–3379. [Google Scholar]
- Sugimoto, N.; Wu, P.; Hara, H.; Kawamoto, Y. pH and cation effects on the properties of parallel pyrimidine motif DNA triplexes. Biochemistry 2001, 40, 9396–9405. [Google Scholar] [CrossRef]
- Roberts, R.W.; Crothers, D.M. Stability and properties of double and triple helices: Dramatic effects of RNA or DNA backbone composition. Science 1992, 258, 1463–1466. [Google Scholar]
- Han, H.; Dervan, P.B. Sequence-specific recognition of double helical RNA and RNA•DNA by triple helix formation. Proc. Natl. Acad. Sci. USA 1993, 90, 3806–3810. [Google Scholar] [CrossRef]
- Escudé, C.; François, J.; Sun, J.; Ott, G.; Sprinzl, M.; Garestier, T.; Hélène, C. Stability of triple helices containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res. 1993, 21, 5547–5553. [Google Scholar] [CrossRef]
- Han, H.; Dervan, P.B. Different conformational families of pyrimidine•purine•pyrimidine triple helices depending on backbone composition. Nucleic Acids Res. 1994, 22, 2837–2844. [Google Scholar] [CrossRef]
- Xiong, Y.; Sundaralingam, M. Crystal structure and conformation of a DNA-RNA hybrid duplex with a polypurine RNA strand: d(TTCTTBr5CTTC)-r(GAAGAAGAA). Structure 1998, 6, 1493–1501. [Google Scholar] [CrossRef]
- MacKerell, A.D., Jr. Contribution of the intrinsic mechanical energy of the phosphodiester linkage to the relative stability of the A, BI, and BII forms of duplex DNA. J. Phys. Chem. B 2009, 113, 3235–3244. [Google Scholar] [CrossRef]
- Bhaumik, S.R.; Chary, K.V.R.; Govil, G.; Liu, K.; Miles, H.T. A novel palindromic triple-stranded structure fromed by homopyrimidine dodecamer d-CTTCTCCTCTTC and homopurine hexamer d-GAAGAG. Nucleic Acids Res. 1998, 26, 2981–2988. [Google Scholar] [CrossRef]
- Rhee, S.; Han, Z.; Liu, K.; Miles, H.T.; Davies, D.R. Structure of a triple helical DNA with a triplex-duplex junction. Biochemistry 1999, 38, 16810–16815. [Google Scholar] [CrossRef]
- Petersen, M.; Nielsen, C.B.; Nielsen, K.E.; Jensen, G.A.; Bondensgaard, K.; Singh, S.K.; Rajwanshi, V.K.; Koshkin, A.A.; Dahl, B.M.; Wengel, J.; et al. The conformations of locked nucleic acids (LNA). J. Mol. Recognit. 2000, 13, 44–53. [Google Scholar] [CrossRef]
- Jensen, G.A.; Singh, S.K.; Kumar, R.; Wengel, J.; Jacobsen, J.P. A comparison of the solution structures of an LNA:DNA duplex and the unmodified DNA:DNA duplex. J. Chem. Soc. Perkin Trans. 2 2001, 1224–1232. [Google Scholar]
- Sample Availability: Not available.
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ito, K.R.; Kodama, T.; Makimura, F.; Hosoki, N.; Osaki, T.; Orita, A.; Imanishi, T.; Obika, S. Cleavage of Oligonucleotides Containing a P3’→N5’ Phosphoramidate Linkage Mediated by Single-Stranded Oligonucleotide Templates. Molecules 2011, 16, 10695-10708. https://doi.org/10.3390/molecules161210695
Ito KR, Kodama T, Makimura F, Hosoki N, Osaki T, Orita A, Imanishi T, Obika S. Cleavage of Oligonucleotides Containing a P3’→N5’ Phosphoramidate Linkage Mediated by Single-Stranded Oligonucleotide Templates. Molecules. 2011; 16(12):10695-10708. https://doi.org/10.3390/molecules161210695
Chicago/Turabian StyleIto, Kosuke Ramon, Tetsuya Kodama, Futaba Makimura, Noritsugu Hosoki, Tomohisa Osaki, Ayako Orita, Takeshi Imanishi, and Satoshi Obika. 2011. "Cleavage of Oligonucleotides Containing a P3’→N5’ Phosphoramidate Linkage Mediated by Single-Stranded Oligonucleotide Templates" Molecules 16, no. 12: 10695-10708. https://doi.org/10.3390/molecules161210695
APA StyleIto, K. R., Kodama, T., Makimura, F., Hosoki, N., Osaki, T., Orita, A., Imanishi, T., & Obika, S. (2011). Cleavage of Oligonucleotides Containing a P3’→N5’ Phosphoramidate Linkage Mediated by Single-Stranded Oligonucleotide Templates. Molecules, 16(12), 10695-10708. https://doi.org/10.3390/molecules161210695




