Structural Investigation of Biologically Active Phenolic Compounds Isolated from European Tree Species
Abstract
:Introduction
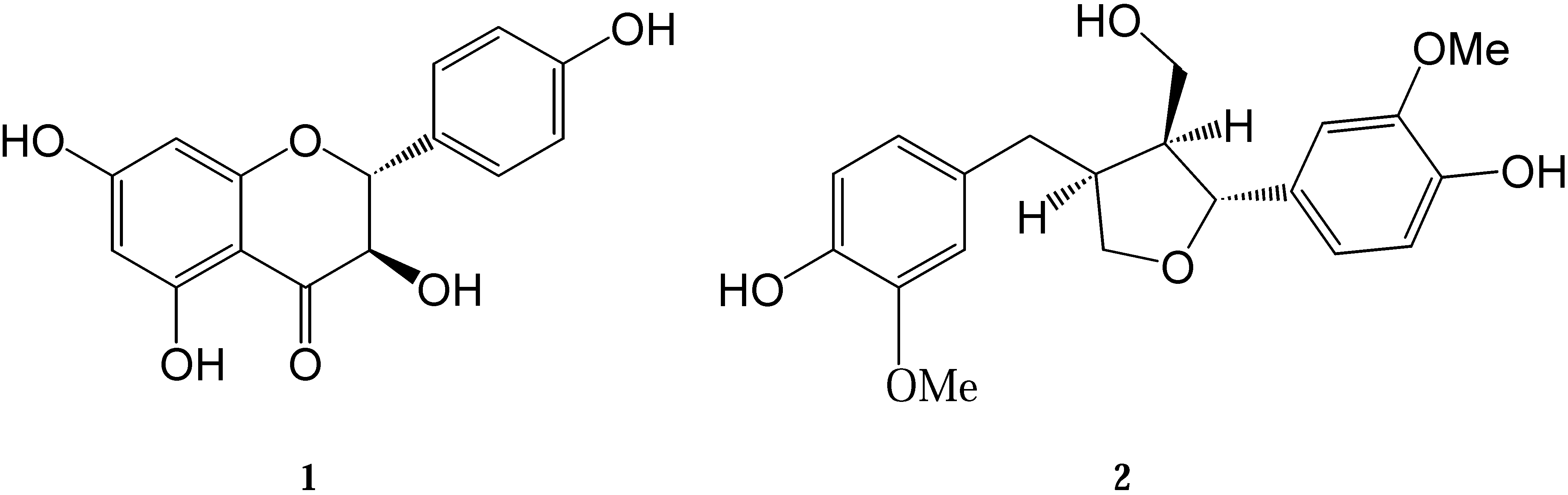
Results and Discussion
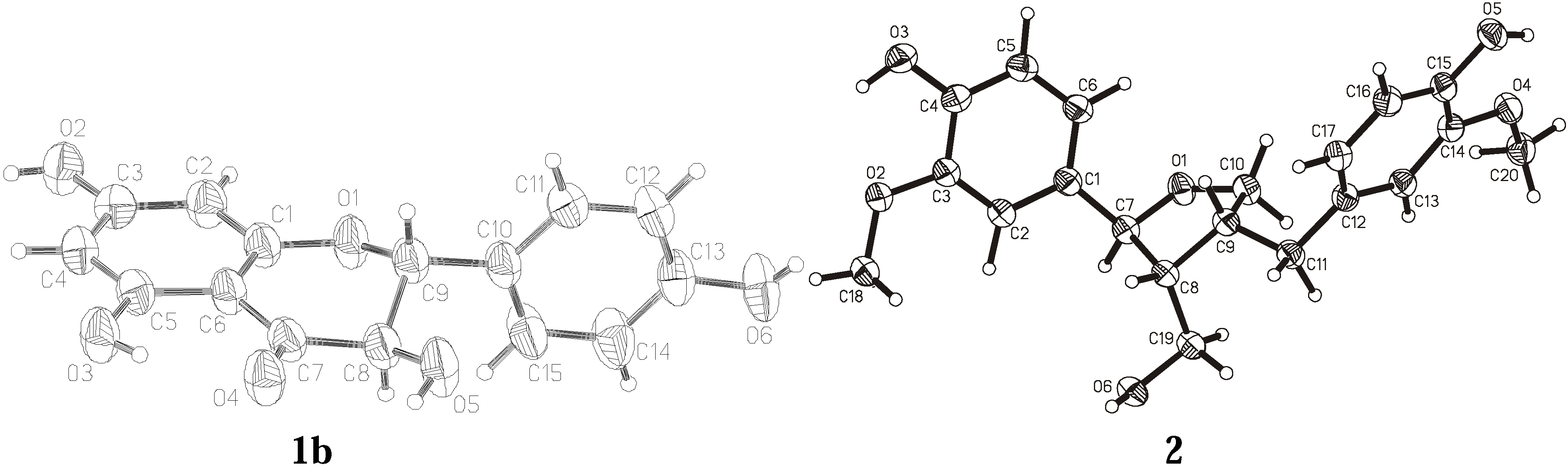
| Compound | 1a | 1b | 2 | ||||
|---|---|---|---|---|---|---|---|
| Molecular formula | C15H12O6*CH3CH2OH | C15H12O6*CH3OH | C20H24O6 | ||||
| Formula weight | 334.31 | 320.29 | 360.39 | ||||
| CCDC No. | 719360 | 719361 | 719362 | ||||
| Crystallographic system | triclinic | triclinic | monoclinic | ||||
| Space group | P1 | P1 | P21 | ||||
| a [Å] | 7.617(5) | 7.581(2) | 10.718(6) | ||||
| b [Å] | 10.349(3) | 10.275(2) | 5.656(3) | ||||
| c [Å] | 11.488(3) | 11.120(2) | 14.264(8) | ||||
| α [o] | 63.92(2) | 65.28(3) | |||||
| β [o] | 85.36(4) | 81.80(3) | 92.75(5) | ||||
| γ [o] | 79.18(3) | 76.61(3) | |||||
| V [Å3] | 798.9(6) | 764.4(3) | 863.7(8) | ||||
| Z | 2 | 2 | 2 | ||||
| Dc [g/cm3] | 1.390 | 1.392 | 1.386 | ||||
| μ [mm-1] | 0.918 | 0.936 | 0.102 | ||||
| Crystal dimensions [mm] | 0.60x0.40x0.02 | 0.56x0.12x0.1 | 1.00x0.06x0.02 | ||||
| Radiation, λ (Å) | CuKα, 1.54178 | CuKα, 1.54178 | synchrotron, 0.80420 | ||||
| hkl ranges: | h = | -9 | 0 | 0 | 9 | -14 | 14 |
| k = | -12 | 12 | -12 | 12 | -6 | 6 | |
| l = | -14 | 14 | -13 | 13 | -19 | 19 | |
| EAC correction: | min. | 0.8867 | 0.9392 | NA | |||
| max. | 0.9933 | 0.9980 | |||||
| ave. | 0.9294 | 0.9679 | |||||
| No. of reflections: | unique | 3545 | 3396 | 4342 | |||
| with I>0σ(I) | 3353 | 3210 | 3372 | ||||
| obs. with I>2σ(I) | 2982 | 2982 | 4007 | ||||
| No. of parameters refined | 472 | 454 | 332 | ||||
| Robs | 0.0691 | 0.0430 | 0.0431 | ||||
| wRobs | 0.1871 | 0.1376 | 0.1137 | ||||
| Rint | 0.0000 | 0.0000 | 0.0000 | ||||
| Sobs | 1.098 | 1.094 | 1.051 | ||||
| 1b | |||||
| molecule | 1 | 1’ | molecule | 1 | 1’ |
| ΔCsC6=ΔCsC9 | 12.2(8) | 12.4(8) | ΔC2C1-C6=ΔC2C8-C9 | 13.3(9) | 16.6(9) |
| 2 | |||||
| ΔCsC8 | 11.2(3) | ΔC2C8-C9 | 4.6(3) | ||
| ΔCsC9 | 17.8(3) | ΔC2C9-C10 | 40.2(3) | ||
| 1b | ||||||||||||
| molecule | 1 | 1’ | molecule | 1 | 1’ | |||||||
| C1 | C2 | C3 | O2 | -177.3(3) | -179.2(4) | C1 | O1 | C9 | C10 | 172.7(3) | -179.7(3) | |
| O2 | C3 | C4 | C5 | 176.5(3) | -179.7(4) | O5 | C8 | C9 | C10 | 60.4(4) | 54.0(4) | |
| C3 | C4 | C5 | O3 | -178.7(3) | 179.1(3) | C7 | C8 | C9 | C10 | -176.4(3) | 176.8(3) | |
| O3 | C5 | C6 | C1 | 180.0(3) | -178.8(3) | O1 | C9 | C10 | C15 | -65.8(4) | -66.5(4) | |
| O3 | C5 | C6 | C7 | 2.9(5) | 4.4(5) | C8 | C9 | C10 | C15 | 52.8(5) | 53.1(5) | |
| C6 | C7 | C8 | O5 | 160.4(3) | 161.4(3) | O1 | C9 | C10 | C11 | 118.9(4) | 114.5(4) | |
| C5 | C6 | C7 | O4 | -7.3(6) | -7.6(6) | C8 | C9 | C10 | C11 | -122.4(4) | -125.9(4) | |
| O4 | C7 | C8 | O5 | -22.5(5) | -20.0(5) | O6 | C13 | C14 | C15 | 179.4(5) | -179.5(4) | |
| O4 | C7 | C8 | C9 | -144.9(3) | -142.1(3) | |||||||
| 2 | ||||||||||||
| C18 | O2 | C3 | C2 | 0.4(2) | O1 | C7 | C8 | C19 | -88.0(1) | |||
| C18 | O2 | C3 | C4 | 179.8(1) | C1 | C7 | C8 | C19 | 149.7(1) | |||
| O2 | C3 | C4 | O3 | -0.1(2) | O1 | C7 | C8 | C9 | 33.6(1) | |||
| C2 | C3 | C4 | O3 | 179.3(1) | C19 | C8 | C9 | C11 | -43.2(2) | |||
| O2 | C3 | C4 | C5 | 180.0(1) | C9 | C11 | C12 | C17 | 84.7(2) | |||
| O3 | C4 | C5 | C6 | -178.6(1) | C9 | C11 | C12 | C13 | -92.5(2) | |||
| C10 | O1 | C7 | C1 | 107.9(1) | C20 | O4 | C14 | C13 | -0.2(2) | |||
| C10 | O1 | C7 | C8 | -15.4(1) | C20 | O4 | C14 | C15 | 177.7(1) | |||
| C6 | C1 | C7 | O1 | -19.2(2) | C12 | C13 | C14 | O4 | -179.6(1) | |||
| C2 | C1 | C7 | O1 | 162.0(1) | O4 | C14 | C15 | O5 | -1.1(2) | |||
| C6 | C1 | C7 | C8 | 99.3(2) | C7 | C8 | C19 | O6 | -67.7(2) | |||
| C2 | C1 | C7 | C8 | -79.4(2) | ||||||||
| 1b | 2 | |||
|---|---|---|---|---|
| Plane 1 C1, C2, C3, C4, C5, C6 | Plane 1 C1, C2, C3, C4, C5, C6 | |||
| Plane 2 C10, C11, C12, C13, C14, C15 | Plane 2 C12, C13, C14, C15, C16, C17 | |||
| Plane 3 C7, C8, C9 | Plane 3 O1, C7, C9, C10 | |||
| Plane 4 O1, C8, C9 | Plane 4 C7, C8, C9 | |||
| Plane 5 O1, C1, C6, C7 | Plane 5 O1, C7, C10 | |||
| molecule | 1 | 1’ | ||
| 1 / 2 | 85.76(14) | 87.22(16) | 1 / 2 | 38.61(4) |
| 1 / 3 | 38.63(38) | 40.55(22) | 1 / 3 | 86.53(5) |
| 2 / 3 | 56.54(36) | 52.61(25) | 2 / 3 | 55.71(5) |
| 1 / 4 | 44.27(39) | 50.10(21) | 1 / 4 | 68.13(7) |
| 2 / 4 | 88.23(19) | 89.68(23) | 2 / 4 | 75.63(8) |
| 3 / 4 | 58.58(42) | 63.41(28) | 3 / 4 | 36.70(10) |
| 1 / 5 | 3.06(20) | 3.02(18) | 1 / 5 | 80.48(8) |
| 2 / 5 | 88.57(20) | 89.78(17) | 2 / 5 | 61.55(8) |
| 3 / 5 | 35.64(42) | 37.53(28) | 3 / 5 | 6.16(9) |
| 4 / 5 | 45.21(38) | 50.32(20) | 4 / 5 | 32.72(12) |
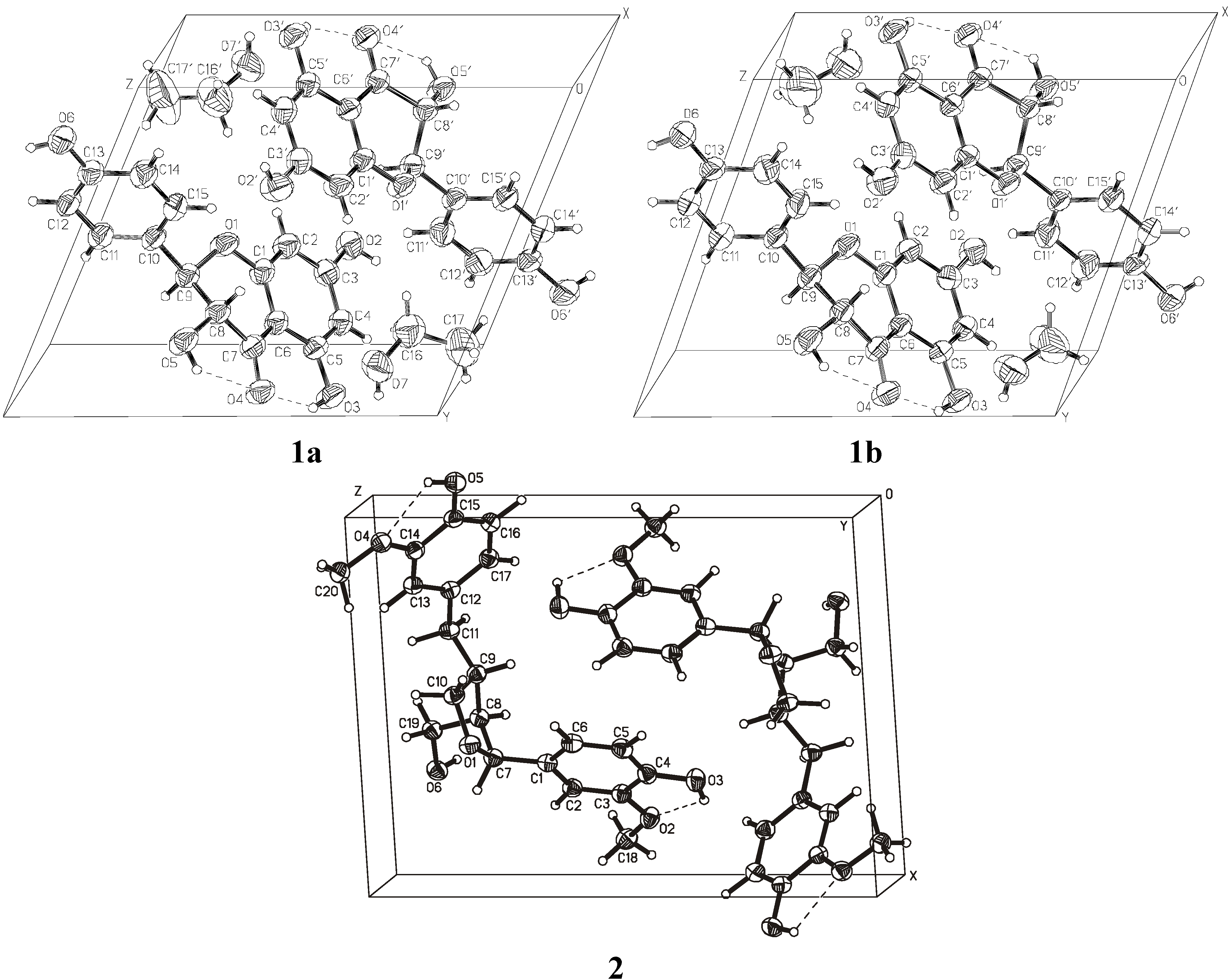

| D―H···A | D―H | H···A | D···A | D―H···A |
| 1a | ||||
| O3―H3O···O4 | 0.820(27) | 1.926(30) | 2.646(5) | 146.0(36) |
| O5―H5O···O4 | 0.820(26) | 2.265(28) | 2.698(5) | 113.4(28) |
| O3'―H3'O···O4' | 0.820(14) | 1.955(34) | 2.656(5) | 143.0(35) |
| O5'―H5'O···O4' | 0.820(23) | 2.231(20) | 2.701(4) | 116.7(23) |
| O5―H5O···O4' i | 0.820(26) | 2.068(33) | 2.767(5) | 142.9(31) |
| O6―H6O···O5' ii | 0.820(36) | 1.895(39) | 2.698(6) | 165.7(45) |
| O5'―H5'O···O4 iii | 0.820(23) | 2.088(20) | 2.751(4) | 137.7(25) |
| O6'―H6'O···O5 iv | 0.820(43) | 1.929(44) | 2.663(7) | 148.7(40) |
| O2―H2O···O7 v | 0.820(42) | 1.814(46) | 2.616(6) | 165.6(59) |
| O7―H7O···O6vi | 0.820(18) | 2.005(32) | 2.796(7) | 161.8(63) |
| O2'―H2'O···O7' vii | 0.820(19) | 1.934(56) | 2.630(6) | 142.1(42) |
| O7'―H7'O···O6' viii | 0.820(16) | 2.008(22) | 2.817(7) | 168.7(38) |
| C8―H8···O2 vii | 0.980(8) | 2.406(6) | 3.133(7) | 130.5(5) |
| 1b | ||||
| O3―H3O···O4 | 0.820(18) | 1.925(22) | 2.646(4) | 146.3(28) |
| O5―H5O···O4 | 0.820(23) | 2.303(27) | 2.689(4) | 109.4(28) |
| O3'―H3'O···O4' | 0.820(14) | 1.945(23) | 2.665(4) | 146.0(28) |
| O5'―H5'O···O4' | 0.820(21) | 2.257(23) | 2.698(3) | 114.1(23) |
| O5―H5O···O4' i | 0.820(23) | 2.104(35) | 2.773(4) | 138.5(32) |
| O6―H6O···O5' ii | 0.820(25) | 1.871(26) | 2.676(4) | 166.5(30) |
| O5'―H5'O···O4 iii | 0.820(21) | 2.066(13) | 2.762(3) | 142.5(26) |
| O6'―H6'O···O5 iv | 0.820(52) | 1.893(52) | 2.651(5) | 153.2(51) |
| O2―H2O···O7 v | 0.820(18) | 1.815(19) | 2.627(4) | 169.9(20) |
| O7―H7O···O6 vi | 0.820(38) | 1.979(33) | 2.769(5) | 161.4(43) |
| O2'―H2'O···O7' vii | 0.820(49) | 1.809(49) | 2.626(5) | 174.3(57) |
| O7'―H7'O···O6' viii | 0.820(18) | 1.995(19) | 2.789(6) | 162.6(33) |
| C8―H8···O2 vii | 0.980(6) | 2.441(5) | 3.124(5) | 126.5(4) |
| 2 | ||||
| O3―H3O···O2 | 0.889(30) | 2.128(30) | 2.652(2) | 117.0(24) |
| O5―H5O···O4 | 0.874(35) | 2.187(27) | 2.660(2) | 113.6(26) |
| O6―H6O···O1ix | 0.975(31) | 1.848(30) | 2.787(2) | 160.8(27) |
| O5―H5O···O6 i | 0.874(35) | 2.312(28) | 2.880(2) | 122.7(27) |
| C2―H2···O5 iii | 1.075(24) | 2.386(24) | 3.454(2) | 172.3(18) |
| C13―H13···O6 x | 1.018(23) | 2.546(24) | 3.452(2) | 148.1(19) |
| C20―H201···O4 xi | 0.965(30) | 2.544(28) | 3.464(3) | 159.5(22) |
Experimental
General
Conclusions
- Sample Availability: Samples of the compounds are available from the authors.
References
- Ekman, R.; Willför, S.; Sjöholm, R.; Reunanen, M.; Mäki, J.; Lehtilä, R.; Eckerman, C. Identification of the lignan Nortrachelogenin in knot and branch heartwood of Scots pine (Pinus sylvestris L.). Holzforschung 2002, 56, 253–256. [Google Scholar]
- Willför, S.; Hemming, J.; Reunanen, M.; Eckerman, C.; Holmbom, B. Lignans and lipophilic extractives in Norway spruce knots and stemwood. Holzforschung 2003, 57, 27–36. [Google Scholar]
- Willför, S.; Hemming, J.; Reunanen, M.; Holmbom, B. Phenolic and lipophilic extractives in scots pine knots and stemwood. Holzforschung 2003, 57, 359–372. [Google Scholar]
- Holmbom, B.; Eckerman, C.; Eklund, P.; Hemming, J.; Reunanen, M.; Sjöholm, R.; Sundberg, A.; Willför, S. Knots in trees – a new rich source of lignans. Phytochem. Rev. 2003, 2, 331–340. [Google Scholar]
- Pietarinen, S.P.; Willför, S.M.; Vikström, F.A.; Holmbom, B.R. Aspenknots, a rich source of flavonoids. J. Wood Chem. Technol. 2006, 26, 245–258. [Google Scholar] [CrossRef]
- Välimaa, A.L.; Honkalampi-Hämäläinen, U.; Pietarinen, S.; Willför, S.; Holmbom, B.; von Wright, A. Antimicrobial and cytotoxic knotwood extracts and related pure compounds and their effects on food-associated microorganisms. Int. J. Food Microbiol. 2007, 115, 235–243. [Google Scholar]
- Saarinen, N.M.; Power, K.A.; Chen, J.; Thompson, L.U. Lignans are accessible to human breast cancer xenografts in athymic mice. Nutr. Cancer 2008, 60, 245–250. [Google Scholar]
- Kangas, L.; Saarinen, N.; Mutanen, M.; Ahotupa, M.; Hirsinummi, R.; Unkila, M.; Perälä, M.; Soininen, P.; Laatikainen, R.; Korte, H.; Santti, R. Antioxidant and antitumor effects of hydroxymatairesinol (HM-3000, HMR), a lignan isolated from the knots of spruce. Eur. J. Cancer Prev. 2002, 11 (Suppl. 2), S48–S57. [Google Scholar]
- Saarinen, N.M.; Wärri, A.; Dings, R.P.M.; Airio, M.; Smeds, A.I.; Mäkelä, S. Dietary lariciresinol attenuates mammary tumor growth and reduces blood vessel density in human MCF-7 breast cancer xenografts and carcinogen-induced mammary tumors in rats. Int. J. Cancer 2008, 123, 1196–1204. [Google Scholar]
- Yang, X.W.; Li, S.M.; Shen, Y.H.; Zhang, W.D. Phytochemical and biological studies of Abies species. Chem. Biodivers. 2008, 5, 56–81. [Google Scholar] [CrossRef]
- Holmbom, B.; Eckerman, C.; Hemming, J.; Reunanen, M.; Sundberg, K.; Willför, S. Method for isolating chemical substances from wood. Pat. Appl. PCT/FI02/00418, 2002. [Google Scholar]
- Willför, S.M.; Ahotupa, M.O.; Hemming, J.E.; Reunanen, M.H.T.; Eklund, P.C.; Sjöholm, R.E.; Eckerman, C.S.E.; Pohjamo, S.P.; Holmbom, B.R. Antioxidant activity of knotwood extractives and phenolic compounds of selected tree species. J. Agric. Food Chem. 2003, 51, 7600–7606. [Google Scholar] [CrossRef]
- Arima, Y.; Hatanaka, A.; Fujimoto, K.; Fukuda, K.; Sakurai, H. Scavenging activities of α-, β- and γ-thujaplicins against active oxygen species. Chem. Pharm. Bull. 1997, 45, 1881–1886. [Google Scholar]
- Kähkönen, M.P.; Hopia, A.I.; Vuorela, H.J.; Rauha, J.P.; Pihlaja, K.; Kujala, T.S.; Heinonen, M. Antioxidant activity of plant extracts containing phenolic compounds. J. Agric. Food Chem. 1999, 47, 3954–3962. [Google Scholar] [CrossRef]
- Chang, S.T.; Wu, J.H.; Wang, S.Y.; Kang, P.L.; Yang, N.S.; Shyur, L.F. Antioxidant activity of extracts from Acacia confusa bark and heartwood. J. Agric. Food Chem. 2001, 49, 3420–3424. [Google Scholar] [CrossRef]
- Shimizu, K.; Kondo, R.; Sakai, K. Antioxidant activity of heartwood extracts of Papua New Guinean woods. J. Wood Sci. 2002, 48, 446–450. [Google Scholar] [CrossRef]
- Hopia, A.I.; Heinonen, M.J. Comparison of antioxidant activity of flavonoid aglycones and their glycosides in methyl linoleate. J. Am. Oil Chem. Soc. 1999, 76, 139–144. [Google Scholar]
- Willför, S.; Reunanen, M.; Eklund, P.; Sjöholm, R.; Kronberg, L.; Fardim, P.; Pietarinen, S.; Holmbom, B. Oligolignans in Norway spruce and Scots pine knots and Norway spruce stemwood. Holzforschung 2004, 58, 345–354. [Google Scholar]
- Pietarinen, S.P.; Willför, S.M.; Ahotupa, M.O.; Hemming, J.E.; Holmbom, B.R. Knotwood and bark extracts: strong antioxidants from waste materials. J. Wood Sci. 2006, 52, 436–444. [Google Scholar]
- Neacsu, M.; Eklund, P.C.; Sjöholm, R.E.; Pietarinen, S.P.; Ahotupa, M.O.; Holmbom, B.R.; Willför, S.M. Antioxidant flavonoids from knotwood of Jack pine and European aspen. Holz Roh. Werkst. 2007, 65, 1–6. [Google Scholar] [CrossRef]
- Siemens Analytical Xray Inst, XP - Interactive Molecular Grafics. 1992.
- Fábián, L.; Kálmán, A. Isostructurality in one and two dimensions: isostructurality of polymorphs. Acta Cryst. 2004, B60, 547–558. [Google Scholar]
- Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ. Available online: http://www.ccdc.cam.ac.uk/ accessed on 10 October 2009.
- Duax, W.L.; Weeks, C.M.; Rohrer, D.C. Topics in Stereochemistry; Eliel, E.L., Allinger, N.L., Eds.; Wiley-Interscience: New York, NY, USA, 1976; Volume 9, p. 271. [Google Scholar]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Frenz, B.A. SDP—Structure Determination Package; Enraf-Nonius: Delft, The Netherland, 1984. [Google Scholar]
- North, A.C.T.; Philips, D.C.; Mathews, F.S. A semi-empirical method of absorption correction. Acta Cryst. 1968, A24, 351–359. [Google Scholar]
- Sheldrick, G.M.; Kruger, G.M.; Goddard, R. SHELXS-86. In Crystallographic Computing 3; Oxford University Press: Oxford, UK, 1985; pp. 175–189. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. 2008, A64, 112–122. [Google Scholar]
- Sheldrick, G.M. SHELXL-93, Program for Crystal Structure Refinement; University of Göttingen: Göttingen, Germany, 1993. [Google Scholar]
- Vickovic, J. CSU, a highly automatic and selective program for the calculation and presentation of geometrical parameters and their e.s.d.`s. J. Appl. Cryst. 1988, 21, 987–990. [Google Scholar]
- Issa, A.Y.; Volate, S.R.; Wargovich, J.M. The role of phytochemicalsin inhibition of cancer and inflammation: New directions and perspectives. J. Food Compos. Anal. 2006, 19, 405–419. [Google Scholar] [CrossRef]
- Fresco, P.; Borges, F.; Diniz, C.; Marques, M.P.M. New insights on the anticancer properties of dietary polyphenols. Med. Res. Rev. 2006, 26, 747–766. [Google Scholar] [CrossRef]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Redzynia, I.; Ziółkowska, N.E.; Majzner, W.R.; Willför, S.; Sjöholm, R.; Eklund, P.; Bujacz, G.D. Structural Investigation of Biologically Active Phenolic Compounds Isolated from European Tree Species. Molecules 2009, 14, 4147-4158. https://doi.org/10.3390/molecules14104147
Redzynia I, Ziółkowska NE, Majzner WR, Willför S, Sjöholm R, Eklund P, Bujacz GD. Structural Investigation of Biologically Active Phenolic Compounds Isolated from European Tree Species. Molecules. 2009; 14(10):4147-4158. https://doi.org/10.3390/molecules14104147
Chicago/Turabian StyleRedzynia, Izabela, Natasza E. Ziółkowska, Wiesław R. Majzner, Stefan Willför, Rainer Sjöholm, Patrik Eklund, and Grzegorz D. Bujacz. 2009. "Structural Investigation of Biologically Active Phenolic Compounds Isolated from European Tree Species" Molecules 14, no. 10: 4147-4158. https://doi.org/10.3390/molecules14104147
APA StyleRedzynia, I., Ziółkowska, N. E., Majzner, W. R., Willför, S., Sjöholm, R., Eklund, P., & Bujacz, G. D. (2009). Structural Investigation of Biologically Active Phenolic Compounds Isolated from European Tree Species. Molecules, 14(10), 4147-4158. https://doi.org/10.3390/molecules14104147




