Subcellular Localization of Seed-Expressed LEA_4 Proteins Reveals Liquid-Liquid Phase Separation for LEA9 and for LEA48 Homo- and LEA42-LEA48 Heterodimers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis of Seed-Expressed LEA_4 Proteins
2.2. Construction of Transgenic Lines Containing Seed-Expressed proLEA_4::LEA_4:Venus Constructs
2.3. Subcellular Localization of Seed-Expressed LEA_4 Proteins in Embryos
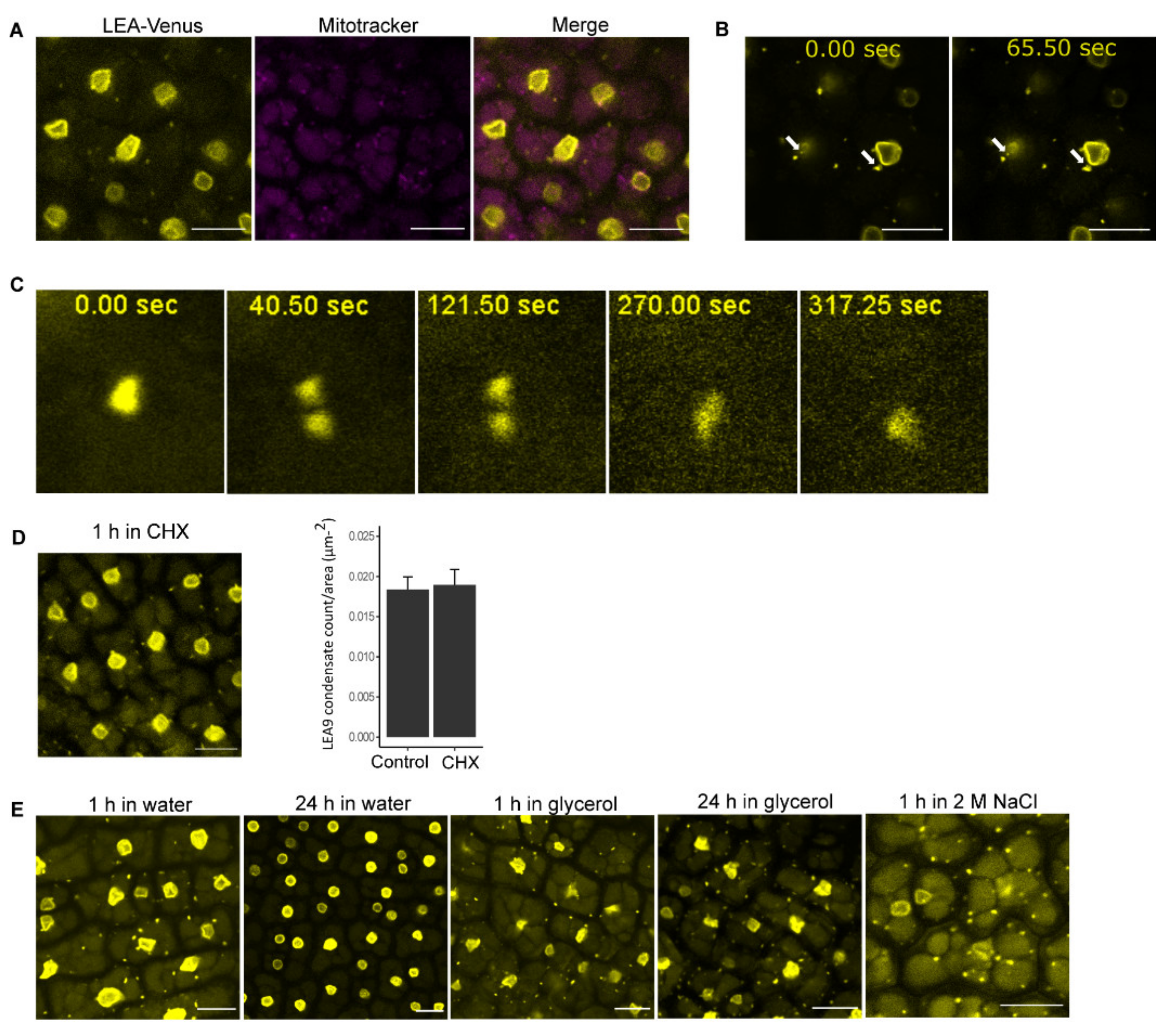
2.4. Investigation of Cytoplasmic Condensates of LEA9
2.5. Expression of Seed-Expressed LEA_4 Proteins in Seedlings
2.6. Construction of Expression Vectors for Seed-Expressed LEA_4
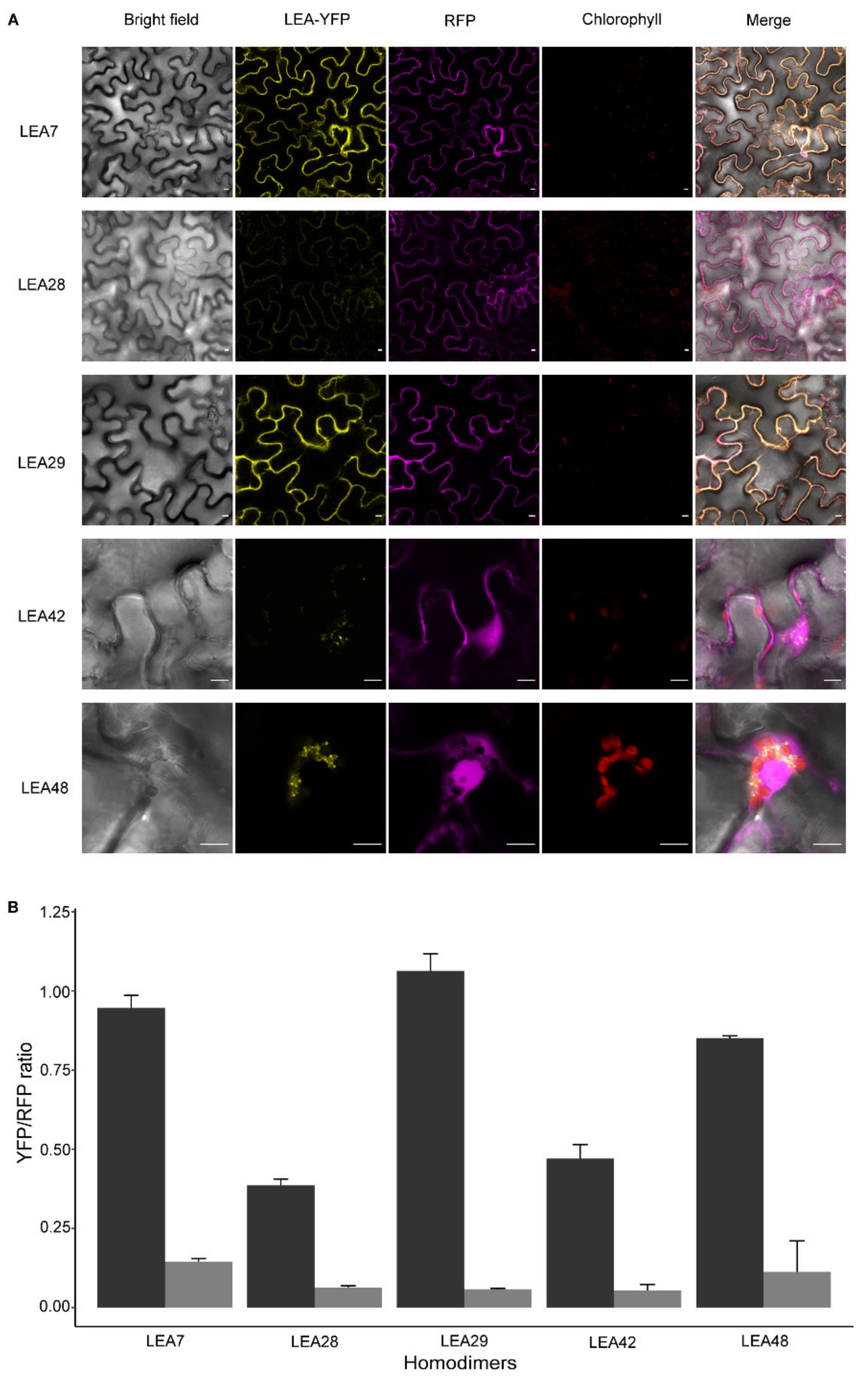
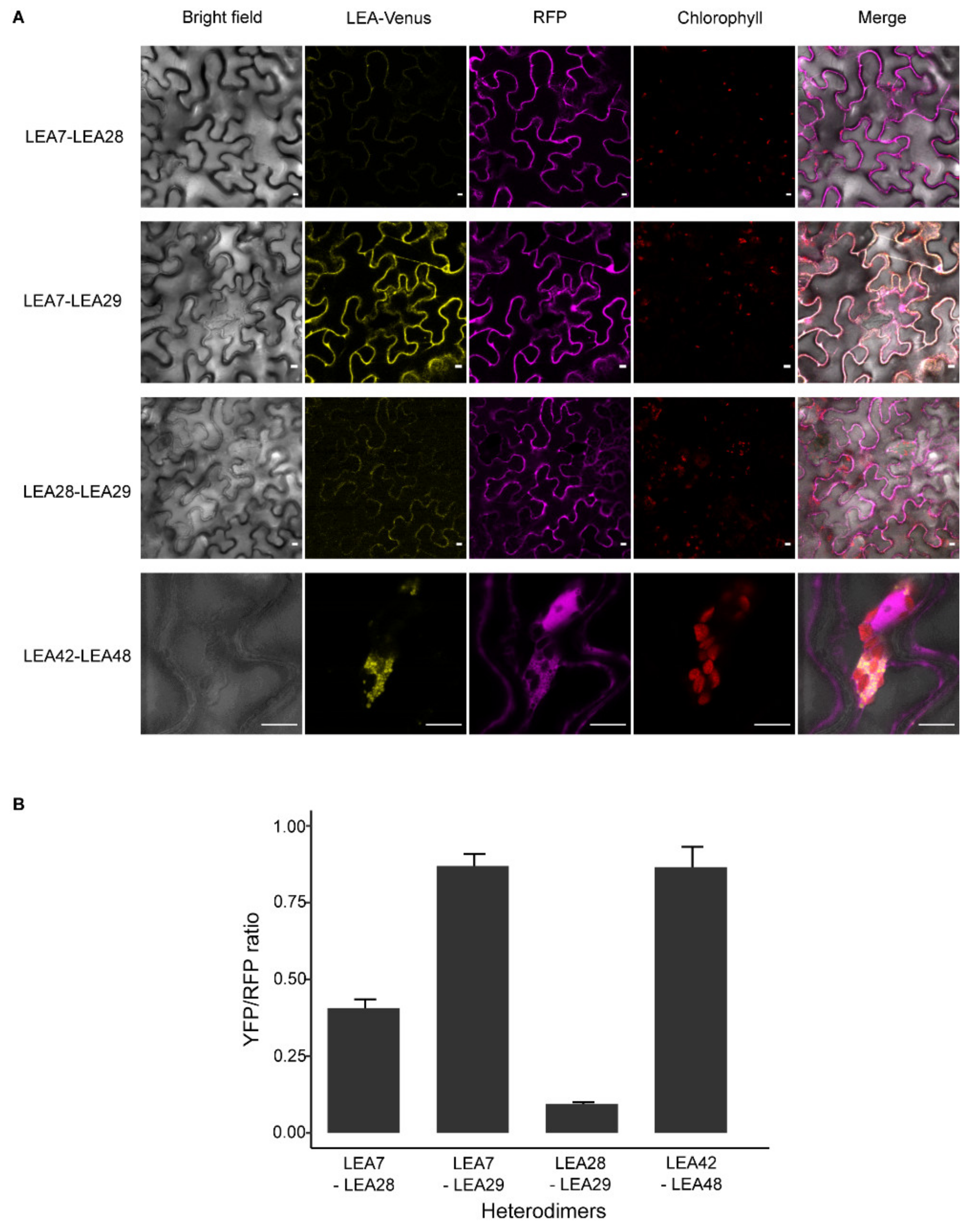
2.7. Homo- and Heterodimer Complex Formation of Seed-Expressed LEA_4 Proteins in Tobacco Leaves
3. Results
3.1. Localization Prediction of Seed-Expressed LEA_4 Proteins Is Varied
3.2. Differential Subcellular Localization of LEA_4 Proteins Expressed in Embryos from Dry Seeds
3.3. LEA9 Forms Hydration Dependent Cytoplasmic Condensates in Embryos
3.4. Cis-Acting Elements in the Promoter Regions of Seed-Expressed LEA_4 Genes
3.5. Analysis of Seed-Expressed LEA_4 Proteins in Young Seedlings
3.6. LEA Proteins with the Same Subcellular Localization Form Homo- and Heterodimers
4. Discussion
4.1. Subcellular Localization of Seed Expressed LEA_4 Proteins Differed Partly from Previous Studies
4.2. Homo- and Heterodimerization Was Found for LEA_4 Proteins with Same Subcellular Localization in Tobacco Leaves
4.3. LEA9, the Homodimers of LEA48 and the Heterodimers of LEA42-LEA48 Undergo LLPS In Vivo
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dure, L.; Chlan, C. Developmental biochemistry of cottonseed embryogenesis and germination: XII. Purification and properties of principal storage proteins. Plant Physiol. 1981, 68, 180–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dure, L.; Galau, G.A. Developmental biochemistry of cottonseed embryogenesis and germination: XIII. Regulation of biosynthesis of principal storage proteins. Plant Physiol. 1981, 68, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Dure, L., 3rd; Greenway, S.C.; Galau, G.A. Developmental biochemistry of cottonseed embryogenesis and germination: Changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 1981, 20, 4162–4168. [Google Scholar] [CrossRef] [PubMed]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar] [CrossRef]
- Tunnacliffe, A.; Hincha, D.; Leprince, O.; Macherel, D. LEA proteins: Versatility of forms and function. In Dormancy and Resistance in Harsh Environments; Lubzens, E., Cerda, J., Clark, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 91–108. [Google Scholar]
- Hand, S.C.; Menze, M.A.; Toner, M.; Boswell, L.; Moore, D. LEA proteins during water stress: Not just for plants anymore. Annu. Rev. Physiol. 2011, 73, 115–134. [Google Scholar] [CrossRef] [Green Version]
- Campos, F.; Cuevas-Velazquez, C.; Fares, M.A.; Reyes, J.L.; Covarrubias, A.A. Group 1 LEA proteins, an ancestral plant protein group, are also present in other eukaryotes, and in the archeae and bacteria domains. Mol. Genet. Genom. MGG 2013, 288, 503–517. [Google Scholar] [CrossRef]
- Janis, B.; Belott, C.; Menze, M.A. Role of intrinsic disorder in animal desiccation tolerance. Proteomics 2018, 18, e1800067. [Google Scholar] [CrossRef] [Green Version]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [Green Version]
- Boudet, J.; Buitink, J.; Hoekstra, F.A.; Rogniaux, H.; Larre, C.; Satour, P.; Leprince, O. Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiol. 2006, 140, 1418–1436. [Google Scholar] [CrossRef] [Green Version]
- Chatelain, E.; Hundertmark, M.; Leprince, O.; Le Gall, S.; Satour, P.; Deligny-Penninck, S.; Rogniaux, H.; Buitink, J. Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 2012, 35, 1440–1455. [Google Scholar] [CrossRef]
- Covarrubias, A.A.; Romero-Perez, P.S.; Cuevas-Velazquez, C.L.; Rendon-Luna, D.F. The functional diversity of structural disorder in plant proteins. Arch. Biochem. Biophys. 2020, 680, 108229. [Google Scholar] [CrossRef]
- Hincha, D.K.; Thalhammer, A. LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem. Soc. Trans. 2012, 40, 1000–1003. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Rikkerink, E.H.; Jones, W.T.; Uversky, V.N. Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 2013, 25, 38–55. [Google Scholar] [CrossRef] [Green Version]
- Mouillon, J.M.; Eriksson, S.K.; Harryson, P. Mimicking the plant cell interior under water stress by macromolecular crowding: Disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol. 2008, 148, 1925–1937. [Google Scholar] [CrossRef] [Green Version]
- Hundertmark, M.; Hincha, D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Lu, M.; Wang, Y.; Wang, Y.; Liu, Z.; Chen, S. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Dirk, L.M.A.; Abdel, C.G.; Ahmad, I.; Neta, I.C.S.; Pereira, C.C.; Pereira, F.E.C.B.; Unêda-Trevisoli, S.H.; Pinheiro, D.G.; Downie, A.B. Late embryogenesis abundant protein–client protein interactions. Plants 2020, 9, 814. [Google Scholar] [CrossRef]
- Hernández-Sánchez, I.E.; Maruri-López, I.; Molphe-Balch, E.P.; Becerra-Flora, A.; Jaimes-Miranda, F.; Jiménez-Bremont, J.F. Evidence for in vivo interactions between dehydrins and the aquaporin AtPIP2b. Biochem. Biophys. Res. Commun. 2019, 510, 545–550. [Google Scholar] [CrossRef]
- Bray, E.A. Molecular responses to water deficit. Plant Physiol. 1993, 103, 1035–1040. [Google Scholar] [CrossRef] [Green Version]
- Dure, L., 3rd; Crouch, M.; Harada, J.; Ho, T.H.; Mundy, J.; Quatrano, R.; Thomas, T.; Sung, Z.R. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol. Biol. 1989, 12, 475–486. [Google Scholar] [CrossRef]
- Roberts, J.K.; DeSimone, N.A.; Lingle, W.L.; Dure, L., 3rd. Cellular concentrations and uniformity of cell-type accumulation of two LEA proteins in cotton embryos. Plant Cell 1993, 5, 769–780. [Google Scholar] [CrossRef]
- Boswell, L.C.; Hand, S.C. Intracellular localization of group 3 LEA proteins in embryos of Artemia franciscana. Tissue Cell 2014, 46, 514–519. [Google Scholar] [CrossRef]
- Grelet, J.; Benamar, A.; Teyssier, E.; Avelange-Macherel, M.H.; Grunwald, D.; Macherel, D. Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol. 2005, 137, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Marie-Hélène, A.-M.; Candat, A.; Neveu, M.; Tolleter, D.; Macherel, D. Decoding the divergent subcellular location of two highly similar paralogous LEA proteins. Int. J. Mol. Sci. 2018, 19, 1620. [Google Scholar]
- Ukaji, N.; Kuwabara, C.; Takezawa, D.; Arakawa, K.; Fujikawa, S. Cold acclimation-induced WAP27 localized in endoplasmic reticulum in cortical parenchyma cells of mulberry tree was homologous to group 3 late-embryogenesis abundant proteins. Plant Physiol. 2001, 126, 1588–1597. [Google Scholar] [CrossRef] [Green Version]
- Houde, M.; Daniel, C.; Lachapelle, M.; Allard, F.; Laliberté, S.; Sarhan, F. Immunolocalization of freezing-tolerance-associated proteins in the cytoplasm and nucleoplasm of wheat crown tissues. Plant J. 1995, 8, 583–593. [Google Scholar] [CrossRef]
- Salleh, F.M.; Evans, K.; Goodall, B.; Machin, H.; Mowla, S.B.; Mur, L.A.; Runions, J.; Theodoulou, F.L.; Foyer, C.H.; Rogers, H.J. A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ. 2012, 35, 418–429. [Google Scholar] [CrossRef] [Green Version]
- Borrell, A.; Cutanda, M.C.; Lumbreras, V.; Pujal, J.; Goday, A.; Culiáñez-Macià, F.A.; Pagès, M. Arabidopsis thaliana AtRAB28: A nuclear targeted protein related to germination and toxic cation tolerance. Plant Mol. Biol. 2002, 50, 249–259. [Google Scholar] [CrossRef]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.H.; Macherel, D. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, L.; Zhang, Y.; Bai, Z.; Liu, H.; Zhang, D. Triticum aestivum WRAB18 functions in plastids and confers abiotic stress tolerance when overexpressed in Escherichia coli and Nicotiania benthamiana. PLoS ONE 2017, 12, e0171340. [Google Scholar] [CrossRef] [Green Version]
- Menze, M.A.; Boswell, L.; Toner, M.; Hand, S.C. Occurrence of mitochondria-targeted late embryogenesis abundant (LEA) gene in animals increases organelle resistance to water stress. J. Biol. Chem. 2009, 284, 10714–10719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. Busca: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Sonderby, C.K.; Sonderby, S.K.; Nielsen, H.; Winther, O. Deeploc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 4049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sperschneider, J.; Catanzariti, A.-M.; DeBoer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, 44598. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, T.; Hecht, M.; Hamp, T.; Karl, T.; Yachdav, G.; Ahmed, N.; Altermann, U.; Angerer, P.; Ansorge, S.; Balasz, K.; et al. Loctree3 prediction of localization. Nucleic Acids Res. 2014, 42, W350–W355. [Google Scholar] [CrossRef] [Green Version]
- Sahu, S.S.; Loaiza, C.D.; Kaundal, R. Plant-msubp: A computational framework for the prediction of single- and multi-target protein subcellular localization using integrated machine-learning approaches. AoB Plants 2020, 12, plz068. [Google Scholar] [CrossRef]
- Cheng, X.; Xiao, X.; Chou, K.C. Ploc-mplant: Predict subcellular localization of multi-location plant proteins by incorporating the optimal go information into general pseaac. Mol. Biosyst. 2017, 13, 1722–1727. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [Green Version]
- Mizianty, M.J.; Peng, Z.; Kurgan, L. Mfdp2: Accurate predictor of disorder in proteins by fusion of disorder probabilities, content and profiles. Intrinsically Disord Proteins 2013, 1, e24428. [Google Scholar] [CrossRef]
- Kumar, M.; Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Pancsa, R.; Glavina, J.; Diakogianni, A.; Valverde, J.A.; Bukirova, D.; Čalyševa, J.; et al. ELM—The eukaryotic linear motif resource in 2020. Nucleic Acids Res. 2019, 48, D296–D306. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, A.; Mejia-Guerra, M.K.; Kurz, K.; Liang, X.; Welch, L.; Grotewold, E. Agris: The Arabidopsis gene regulatory information server, an update. Nucleic Acids Res. 2011, 39, D1118–D1122. [Google Scholar] [CrossRef] [Green Version]
- RStudio Team. Rstudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2019. [Google Scholar]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Coutu, C.; Brandle, J.; Brown, D.; Brown, K.; Miki, B.; Simmonds, J.; Hegedus, D.D. Pore: A modular binary vector series suited for both monocot and dicot plant transformation. Transgenic. Res. 2007, 16, 771–781. [Google Scholar] [CrossRef]
- Birnboim, H.C.; Doly, J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979, 7, 1513–1523. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Nelson, B.K.; Cai, X.; Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Voinnet, O.; Rivas, S.; Mestre, P.; Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003, 33, 949–956. [Google Scholar] [CrossRef] [Green Version]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Harmon, T.S.; Holehouse, A.S.; Rosen, M.K.; Pappu, R.V. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife 2017, 6, e30294. [Google Scholar] [CrossRef]
- Majumdar, A.; Dogra, P.; Maity, S.; Mukhopadhyay, S. Liquid–liquid phase separation is driven by large-scale conformational unwinding and fluctuations of intrinsically disordered protein molecules. J. Phys. Chem. Lett. 2019, 10, 3929–3936. [Google Scholar] [CrossRef] [PubMed]
- Mahboubi, H.; Stochaj, U. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Abiotic stresses cause differential regulation of alternative splice forms of GATA transcription factor in rice. Front. Plant Sci. 2017, 8, 1944. [Google Scholar] [CrossRef] [Green Version]
- Ginsawaeng, O.; Gorka, M.; Erban, A.; Heise, C.; Brueckner, F.; Hoefgen, R.; Kopka, J.; Skirycz, A.; Hincha, D.K.; Zuther, E. Characterization of the heat-stable proteome during seed germination in Arabidopsis with special focus on LEA proteins. Int. J. Mol. Sci. 2021, 22, 8172. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Velichko, A.K.; Magnitov, M.D.; Luzhin, A.V.; Golov, A.K.; Ovsyannikova, N.; Kireev, I.I.; Gavrikov, A.S.; Mishin, A.S.; Garaev, A.K.; et al. Suppression of liquid-liquid phase separation by 1,6-hexanediol partially compromises the 3d genome organization in living cells. Nucleic Acids Res. 2021, 49, 10524–10541. [Google Scholar] [CrossRef] [PubMed]
- Amara, I.; Zaidi, I.; Masmoudi, K.; Ludevid, M.; Pagés, M.; Goday, A.; Brini, F. Insights into late embryogenesis abundant (LEA) proteins in plants: From structure to the functions. Am. J. Plant Sci. 2014, 5, 3440–3455. [Google Scholar] [CrossRef] [Green Version]
- Dingwall, C.; Laskey, R.A. Protein import into the cell nucleus. Annu. Rev. Cell Biol. 1986, 2, 367–390. [Google Scholar] [CrossRef]
- Knox-Brown, P.; Rindfleisch, T.; Gunther, A.; Balow, K.; Bremer, A.; Walther, D.; Miettinen, M.S.; Hincha, D.K.; Thalhammer, A. Similar yet different-structural and functional diversity among Arabidopsis thaliana LEA_4 proteins. Int. J. Mol. Sci. 2020, 21, 2794. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Sánchez, I.E.; Martynowicz, D.M.; Rodríguez-Hernández, A.A.; Pérez-Morales, M.B.; Graether, S.P.; Jiménez-Bremont, J.F. A dehydrin-dehydrin interaction: The case of SK3 from Opuntia streptacantha. Front. Plant Sci. 2014, 5, 520. [Google Scholar] [CrossRef]
- Offringa, R.; Huang, F. Phosphorylation-dependent trafficking of plasma membrane proteins in animal and plant cells. J. Integr. Plant Biol. 2013, 55, 789–808. [Google Scholar] [CrossRef]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, J.L.; Garciarrubio, A.; Covarrubias, A.A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010, 154, 373–390. [Google Scholar] [CrossRef] [Green Version]
- Kruger, C.; Berkowitz, O.; Stephan, U.W.; Hell, R. A metal-binding member of the late embryogenesis abundant protein family transports iron in the phloem of Ricinus communis L. J. Biol. Chem. 2002, 277, 25062–25069. [Google Scholar] [CrossRef] [Green Version]
- Krüger, C.; Hell, R.; Stephan, U.W. A metal-binding LEA protein trafficks micronutrients in the phloem of Ricinus communis L. In Plant Nutrition: Food Security and Sustainability of Agro-Ecosystems through Basic and Applied Research; Horst, W.J., Schenk, M.K., Bürkert, A., Claassen, N., Flessa, H., Frommer, W.B., Goldbach, H., Olfs, H.W., Römheld, V., Sattelmacher, B., et al., Eds.; Springer: Dordrecht, The Netherlands, 2001; pp. 194–195. [Google Scholar]
- Hernández-Sánchez, I.E.; Maruri-López, I.; Graether, S.P.; Jiménez-Bremont, J.F. In vivo evidence for homo- and heterodimeric interactions of Arabidopsis thaliana dehydrins AtCOR47, AtERD10, and AtRAB18. Sci. Rep. 2017, 7, 17036. [Google Scholar] [CrossRef] [Green Version]
- Rahman, L.N.; McKay, F.; Giuliani, M.; Quirk, A.; Moffatt, B.A.; Harauz, G.; Dutcher, J.R. Interactions of Thellungiella salsuginea dehydrins TsDHN-1 and TsDHN-2 with membranes at cold and ambient temperatures—Surface morphology and single-molecule force measurements show phase separation, and reveal tertiary and quaternary associations. Biochim. Biophys. Acta Biomembr. 2013, 1828, 967–980. [Google Scholar] [CrossRef] [Green Version]
- Kazuoka, T.; Oeda, K. Purification and characterization of cor85-oligomeric complex from cold-acclimated spinach. Plant Cell Physiol. 1994, 35, 601–611. [Google Scholar] [CrossRef]
- Wang, X.; Yu, Z.; Liu, H.; Zhang, Y.; Bai, Z.; Zhang, L. Effect of k-/s- segments on subcellular localization and dimerization of wheat dehydrin WZY1-2. Plant Signal. Behav. 2020, 15, 1827583. [Google Scholar] [CrossRef]
- Nakayama, K.; Okawa, K.; Kakizaki, T.; Honma, T.; Itoh, H.; Inaba, T. Arabidopsis COR15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol. 2007, 144, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Rivera-Najera, L.Y.; Saab-Rincón, G.; Battaglia, M.; Amero, C.; Pulido, N.O.; García-Hernández, E.; Solórzano, R.M.; Reyes, J.L.; Covarrubias, A.A. A group 6 late embryogenesis abundant protein from common bean is a disordered protein with extended helical structure and oligomer-forming properties. J. Biol. Chem. 2014, 289, 31995–32009. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lu, S.; Jiang, C.; Wang, Y.; Lv, B.; Shen, J.; Ming, F. RcLEA, a late embryogenesis abundant protein gene isolated from Rosa chinensis, confers tolerance to Escherichia coli and Arabidopsis thaliana and stabilizes enzyme activity under diverse stresses. Plant Mol. Biol. 2014, 85, 333–347. [Google Scholar] [CrossRef]
- Hughes, S.L.; Schart, V.; Malcolmson, J.; Hogarth, K.A.; Martynowicz, D.M.; Tralman-Baker, E.; Patel, S.N.; Graether, S.P. The importance of size and disorder in the cryoprotective effects of dehydrins. Plant Physiol. 2013, 163, 1376–1386. [Google Scholar] [CrossRef] [Green Version]
- Puhakainen, T.; Hess, M.W.; Mäkelä, P.; Svensson, J.; Heino, P.; Palva, E.T. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol. Biol. 2004, 54, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Receveur-Brechot, V.; Bourhis, J.M.; Uversky, V.N.; Canard, B.; Longhi, S. Assessing protein disorder and induced folding. Proteins 2006, 62, 24–45. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belott, C.; Janis, B.; Menze, M.A. Liquid-liquid phase separation promotes animal desiccation tolerance. Proc. Natl. Acad. Sci. USA 2020, 117, 27676–27684. [Google Scholar] [CrossRef] [PubMed]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegmann, S.; Eftekharzadeh, B.; Tepper, K.; Zoltowska, K.M.; Bennett, R.E.; Dujardin, S.; Laskowski, P.R.; MacKenzie, D.; Kamath, T.; Commins, C.; et al. TAU protein liquid-liquid phase separation can initiate TAU aggregation. EMBO J. 2018, 37, e98049. [Google Scholar] [CrossRef]
- Murakami, T.; Qamar, S.; Lin, J.Q.; Schierle, G.S.; Rees, E.; Miyashita, A.; Costa, A.R.; Dodd, R.B.; Chan, F.T.; Michel, C.H.; et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 2015, 88, 678–690. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.Y.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.M.; et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 2015, 162, 1066–1077. [Google Scholar] [CrossRef] [Green Version]
- Ray, S.; Singh, N.; Kumar, R.; Patel, K.; Pandey, S.; Datta, D.; Mahato, J.; Panigrahi, R.; Navalkar, A.; Mehra, S.; et al. A-synuclein aggregation nucleates through liquid–liquid phase separation. Nat. Chem. 2020, 12, 705–716. [Google Scholar] [CrossRef]
- Ambadipudi, S.; Biernat, J.; Riedel, D.; Mandelkow, E.; Zweckstetter, M. Liquid–liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein TAU. Nat. Commun. 2017, 8, 275. [Google Scholar] [CrossRef]
- Conicella, A.E.; Zerze, G.H.; Mittal, J.; Fawzi, N.L. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity c-terminal domain. Structure 2016, 24, 1537–1549. [Google Scholar] [CrossRef] [Green Version]
- Cuevas-Velazquez, C.L.; Dinneny, J.R. Organization out of disorder: Liquid-liquid phase separation in plants. Curr. Opin. Plant Biol. 2018, 45, 68–74. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Holehouse, A.S.; Strader, L.C. Emerging roles for phase separation in plants. Dev. Cell 2020, 55, 69–83. [Google Scholar] [CrossRef]
- Wallmann, A.; Kesten, C. Common functions of disordered proteins across evolutionary distant organisms. Int. J. Mol. Sci. 2020, 21, 2105. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.L.; Liu, Y.; Oldfield, C.J.; Uversky, V.N.; Darling, A.L.; Liu, Y.; Oldfield, C.J.; Uversky, V.N. Intrinsically disordered proteome of human membrane-less organelles. Proteomics 2018, 18, 1700193. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Holehouse, A.S.; Pappu, R.V. Functional implications of intracellular phase transitions. Biochemistry 2018, 57, 2415–2423. [Google Scholar] [CrossRef]
- Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Intrinsic disorder-based emergence in cellular biology: Physiological and pathological liquid-liquid phase transitions in cells. Polymers 2019, 11, 990. [Google Scholar] [CrossRef] [Green Version]
- McSwiggen, D.; Mir, M.; Darzacq, X.; Tjian, R.J.G. Evaluating phase separation in live cells: Diagnosis, caveats, and functional consequences. Genes Dev. 2019, 33, 1619–1634. [Google Scholar] [CrossRef]
- Moore, H.M.; Bai, B.; Boisvert, F.-M.; Latonen, L.; Rantanen, V.; Simpson, J.C.; Pepperkok, R.; Lamond, A.I.; Laiho, M. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol. Cell. Proteom. MCP 2011, 10, M111.009241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, H.; Triandafillou, C.; Drummond, D.A. Cellular sensing by phase separation: Using the process, not just the products. J. Biol. Chem. 2019, 294, 7151–7159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.-H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Silva, C.S.; Lai, X.; Pierre, E.; Geng, F.; et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 2020, 585, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Dorone, Y.; Boeynaems, S.; Flores, E.; Jin, B.; Hateley, S.; Bossi, F.; Lazarus, E.; Pennington, J.G.; Michiels, E.; De Decker, M.; et al. A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation. Cell 2021, 184, 4284–4298. [Google Scholar] [CrossRef]
- Kroschwald, S.; Munder, M.C.; Maharana, S.; Franzmann, T.M.; Richter, D.; Ruer, M.; Hyman, A.A.; Alberti, S. Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Rep. 2018, 23, 3327–3339. [Google Scholar] [CrossRef]


| LEA Protein | Subcellular Localization |
|---|---|
| LEA7 | Nucleus, ER |
| LEA9 | Nucleus, Cytoplasmic condensates |
| LEA19 | ER |
| LEA25 | ER |
| LEA28 | Nucleus, ER |
| LEA29 | Nucleus, ER |
| LEA30 | ER |
| LEA36 | ER |
| LEA42 | Mitochondria, Chloroplasts |
| LEA43 | ER |
| LEA48 | Mitochondria, Chloroplasts |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ginsawaeng, O.; Heise, C.; Sangwan, R.; Karcher, D.; Hernández-Sánchez, I.E.; Sampathkumar, A.; Zuther, E. Subcellular Localization of Seed-Expressed LEA_4 Proteins Reveals Liquid-Liquid Phase Separation for LEA9 and for LEA48 Homo- and LEA42-LEA48 Heterodimers. Biomolecules 2021, 11, 1770. https://doi.org/10.3390/biom11121770
Ginsawaeng O, Heise C, Sangwan R, Karcher D, Hernández-Sánchez IE, Sampathkumar A, Zuther E. Subcellular Localization of Seed-Expressed LEA_4 Proteins Reveals Liquid-Liquid Phase Separation for LEA9 and for LEA48 Homo- and LEA42-LEA48 Heterodimers. Biomolecules. 2021; 11(12):1770. https://doi.org/10.3390/biom11121770
Chicago/Turabian StyleGinsawaeng, Orarat, Carolin Heise, Rohit Sangwan, Daniel Karcher, Itzell Euridice Hernández-Sánchez, Arun Sampathkumar, and Ellen Zuther. 2021. "Subcellular Localization of Seed-Expressed LEA_4 Proteins Reveals Liquid-Liquid Phase Separation for LEA9 and for LEA48 Homo- and LEA42-LEA48 Heterodimers" Biomolecules 11, no. 12: 1770. https://doi.org/10.3390/biom11121770
APA StyleGinsawaeng, O., Heise, C., Sangwan, R., Karcher, D., Hernández-Sánchez, I. E., Sampathkumar, A., & Zuther, E. (2021). Subcellular Localization of Seed-Expressed LEA_4 Proteins Reveals Liquid-Liquid Phase Separation for LEA9 and for LEA48 Homo- and LEA42-LEA48 Heterodimers. Biomolecules, 11(12), 1770. https://doi.org/10.3390/biom11121770







