Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses
Abstract
:1. Introduction
2. Distribution of Group II LEA Proteins in Plants
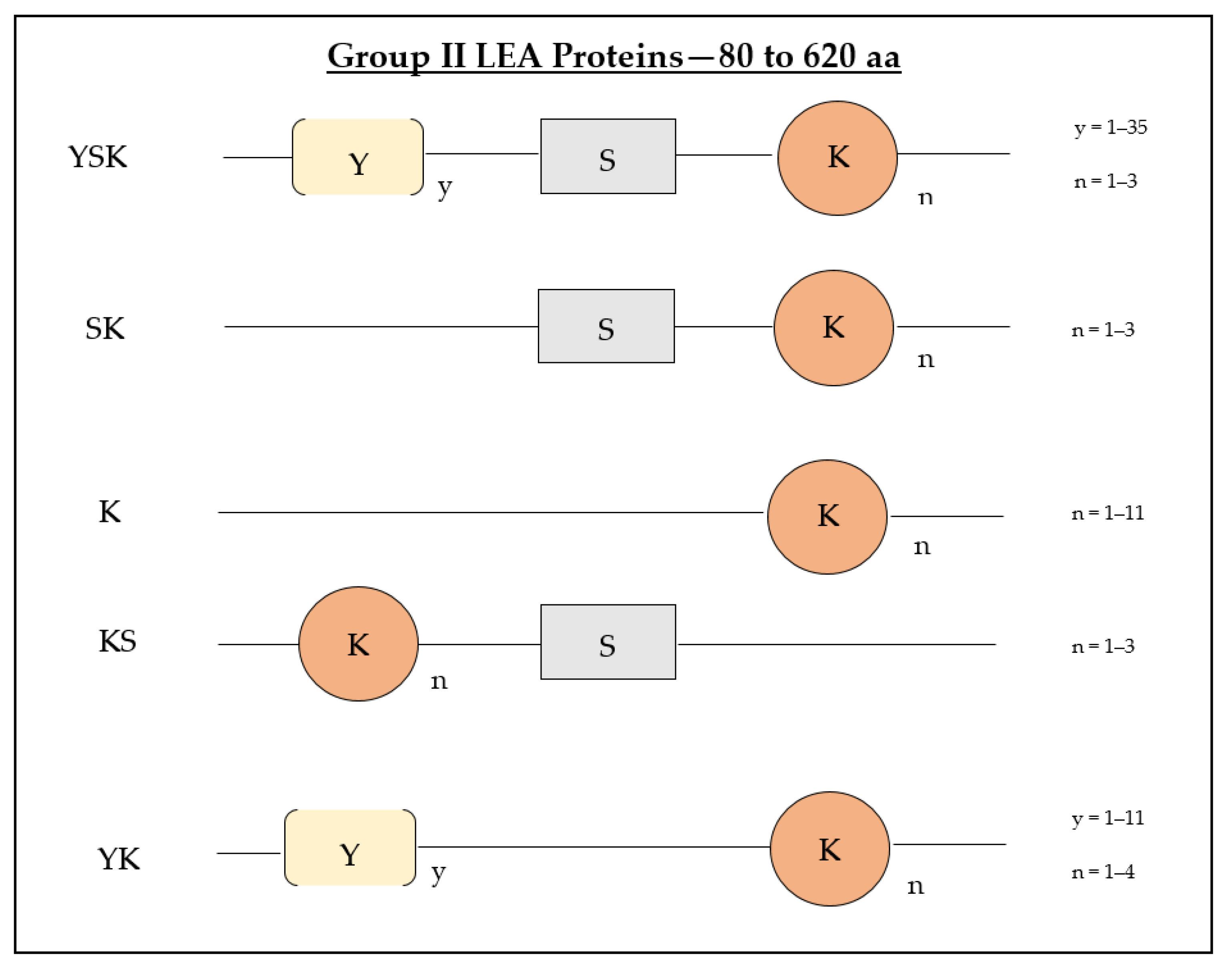
3. Sequence and Domain Architecture of Intrinsically Disordered Group II LEA Proteins
Evolution of the Structural Architecture of Group II LEA Proteins in Certain Plant Species
4. Genomic Diversification of Group II LEA Proteins
Genome-Wide Association Studies (GWAS) of Group II LEA Proteins
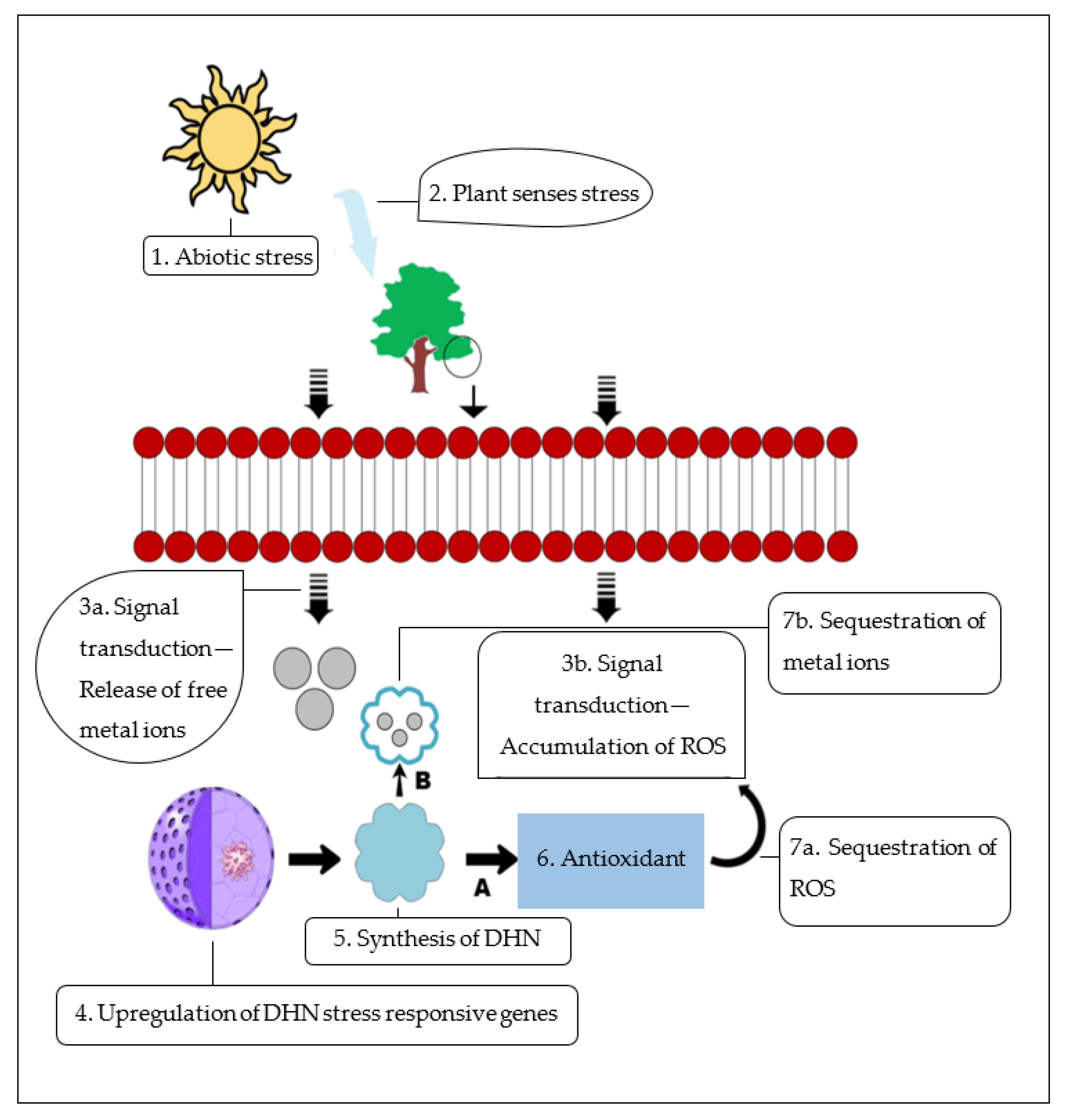
5. Group II LEA Gene Expression and Regulation Patterns under Abiotic Stresses
5.1. Expression of Group II LEA Genes under Salinity Stress
5.2. Expression of Group II LEA Genes under Drought Stress
5.3. Expression of Group II LEA Genes under Temperature Stress
5.4. Expression of Group II LEA Genes under Osmotic Stress
6. Group II LEA Protein or DHN Responses to Biotic Stresses
7. Accumulation of Group II LEA Proteins in Phoenix dactylifera
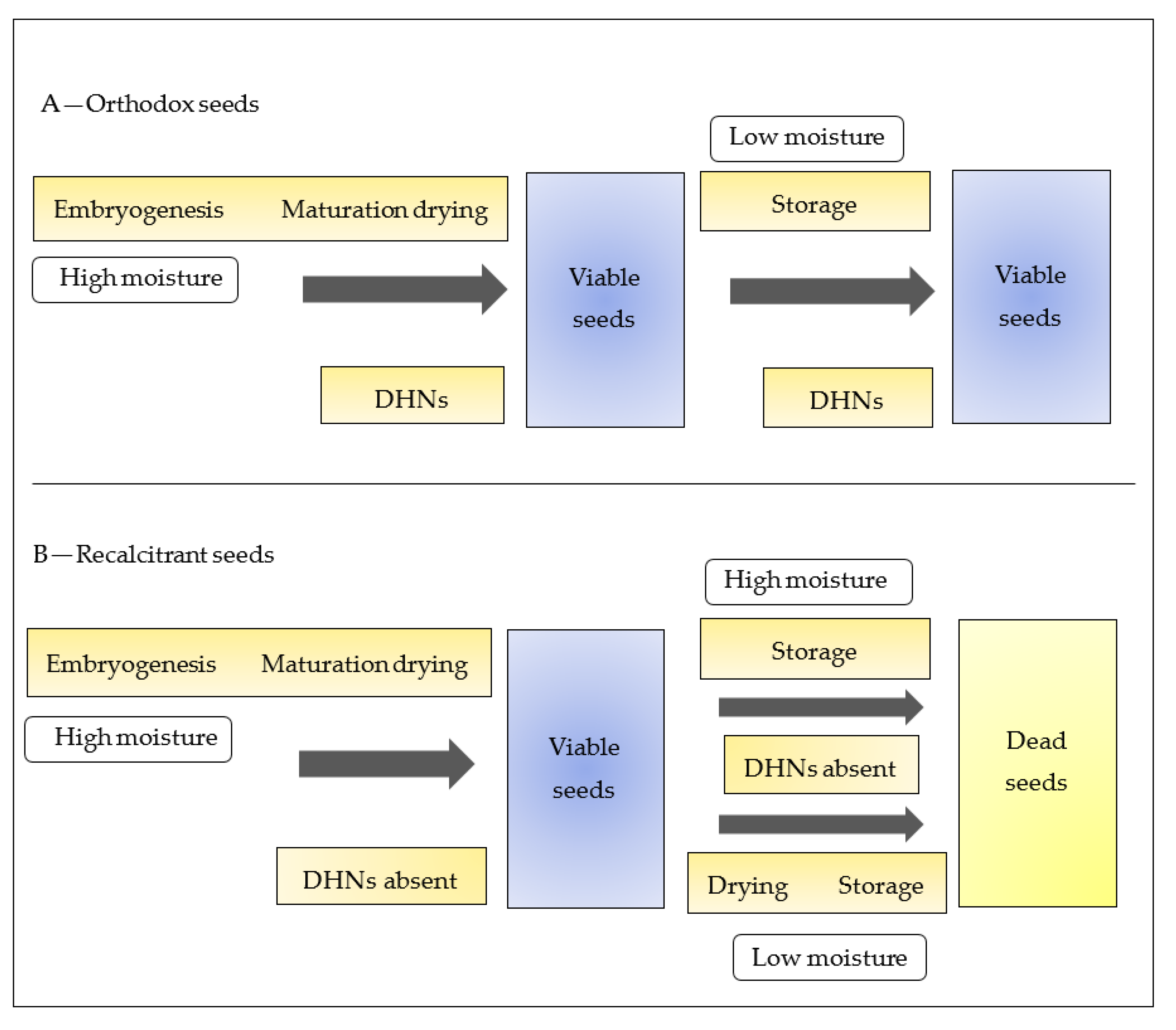
8. DHNs Relation in Storage and Conservation of Orthodox and Recalcitrant Seeds
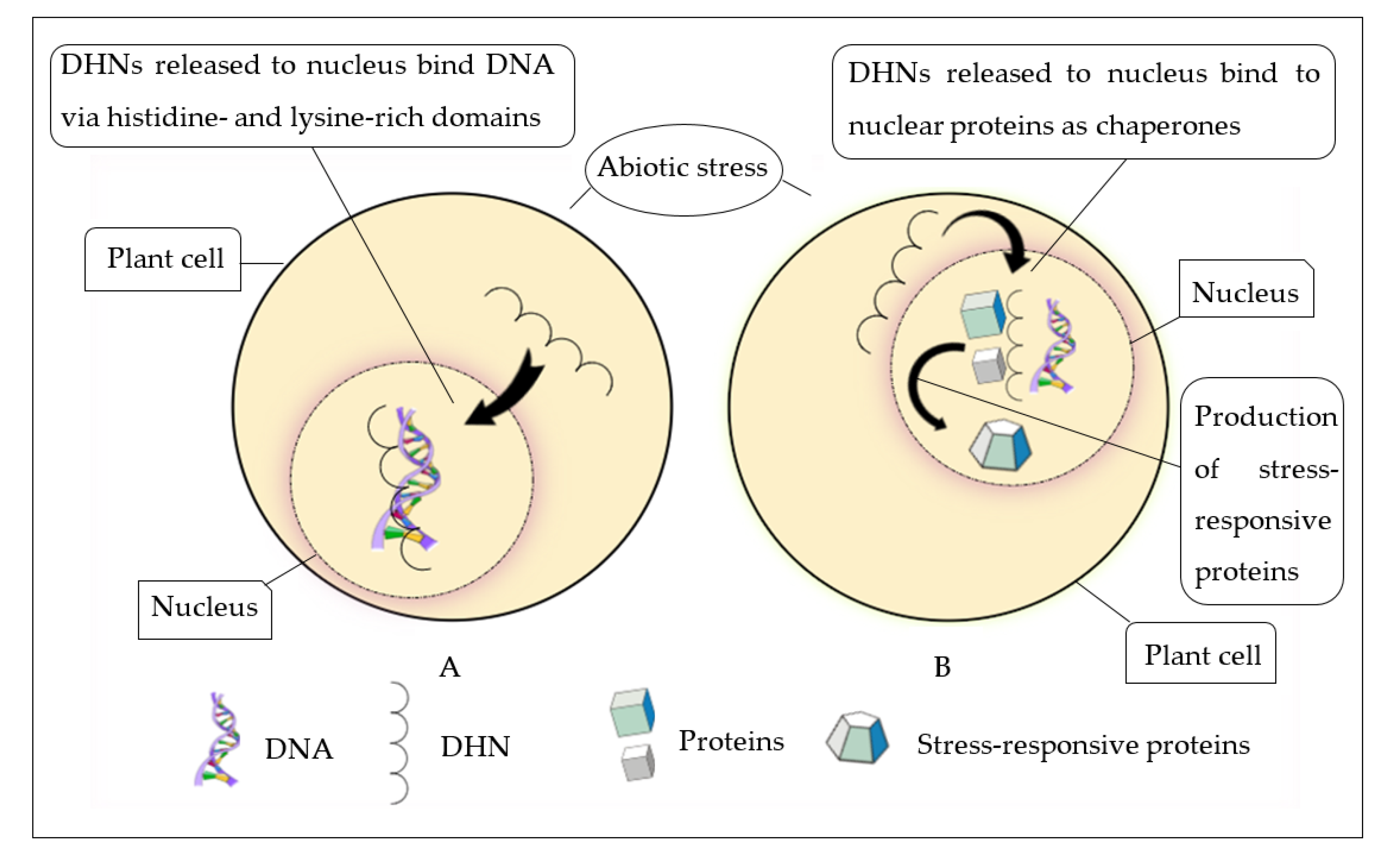
9. Group II LEA Proteins’ Functional Heterogeneity
9.1. Biomolecule Preservation
9.2. Scavenging Reactive Oxygen Species (ROS)
9.3. Metal-Ion-Binding Protein
9.4. Phospholipid-Binding Protein
10. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Blum, A. The Interdrought Conference in Perspective. J. Exp. Bot. 2011, 63, 5773–5774. [Google Scholar]
- Hong-Bo, S.; Zong-Suo, L.; Ming-An, S. LEA proteins in higher plants: Structure, function, gene expression and regulation. Colloids Surf. B. 2005, 45, 131–135. [Google Scholar] [CrossRef]
- Tunnacliffe, A.; Wise, M. The continuing conundrum of the LEA proteins. Sci. Nat. 2007, 94, 791–812. [Google Scholar] [CrossRef]
- Battaglia, M.; Covarrubias, A. Late embryogenesis abundant (LEA) proteins in legumes. Front. Plant Sci. 2013, 4, 190. [Google Scholar] [CrossRef] [Green Version]
- Hara, M. The multifunctionality of dehydrins: An overview. Plant Signal. Behav. 2010, 5, 503–508. [Google Scholar] [CrossRef]
- Abdul Aziz, M. Cloning and Sequencing of Five Lea2 Genes from Date Palm cv. Khalas and Functional Characterization to Heat and Salt Stress Tolerance Using Yeast Knockout Mutants. Master’s Thesis, United Arab Emirates University, Al Ain, United Arab Emirates, 22 December 2020. [Google Scholar]
- Jin, X.; Cao, D.; Wang, Z.; Ma, L.; Tian, K.; Liu, Y.; Gong, Z.; Zhu, X.; Jiang, C.; Li, Y. Genome-wide identification and expression analyses of the LEA protein gene family in tea plant reveal their involvement in seed development and abiotic stress responses. Sci. Rep. 2019, 9, 14123. [Google Scholar] [CrossRef]
- Zheng, J.; Su, H.; Lin, R.; Zhang, H.; Xia, K.; Jian, S.; Zhang, M. Isolation and characterization of an atypical LEA gene (IpLEA) from Ipomoea pes-caprae conferring salt/drought and oxidative stress tolerance. Sci. Rep. 2019, 9, 14838. [Google Scholar] [CrossRef] [PubMed]
- Allagulova, C.; Gimalov, F.; Shakirova, M.; Vakhitov, V. The plant dehydrins: Structure and putative functions. Biochemistry 2003, 68, 945–951. [Google Scholar] [PubMed]
- Zaman Khan, N.; Lal, S.; Ali, W.; Aasim, M.; Mumtaz, S.; Kamil, A.; Shad Bibi, N. Distribution and classification of dehydrins in selected plant species using bioinformatics approach. Iran. J. Biotechnol. 2020, 18, e2680. [Google Scholar]
- Rorat, T.; Grygorowicz, W.; Irzykowski, W.; Rey, P. Expression of KS-type dehydrins is primarily regulated by factors related to organ type and leaf developmental stage during vegetative growth. Planta 2004, 218, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Zhu, B.; Close, T. The barley (Hordeum vulgare L.) dehydrin multigene family: Sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor. Appl. Genet. 1999, 98, 1234–1247. [Google Scholar] [CrossRef]
- Robertson, M.; Chandler, P. A dehydrin cognate protein from pea (Pisum sativum L.) with an atypical pattern of expression. Plant Mol. Biol. 1994, 26, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Kiyosue, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kamada, H.; Harada, H. cDNA cloning of Ecp40, an embryogenic-cell protein in carrot, and its expression during somatic and zygotic embryogenesis. Plant Mol. Biol. 1993, 21, 1053–1068. [Google Scholar] [CrossRef]
- Momma, M.; Haraguchi, K.; Saito, M.; Chikuni, K.; Harada, K. Purification and characterization of the acid soluble 26-kDa polypeptide from soybean seeds. Biosci. Biotechnol. Biochem. 1997, 61, 1286–1291. [Google Scholar] [CrossRef] [PubMed]
- Nylander, M.; Svensson, J.; Palva, E.; Welin, B. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 2001, 45, 263–279. [Google Scholar] [CrossRef]
- Wisniewski, M.; Webb, R.; Balsamo, R.; Close, T.; Yu, X.; Griffith, M. Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: A dehydrin from peach (Prunus persica). Physiol. Plant. 1999, 105, 600–608. [Google Scholar] [CrossRef]
- Danyluk, J.; Perron, A.; Houde, M.; Limin, A.; Fowler, B.; Benhamou, N.; Sarhan, F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 1998, 10, 623–638. [Google Scholar] [CrossRef] [Green Version]
- Bravo, L.; Close, T.; Corcuera, L.; Guy, C. Characterization of an 80− kDa dehydrin-like protein in barley responsive to cold acclimation. Physiol. Plant 1999, 106, 177–183. [Google Scholar] [CrossRef]
- Houde, M.; Daniel, C.; Lachapelle, M.; Allard, F.; Laliberte, S.; Sarhan, F. Immunolocalization of freezing-tolerance-associated proteins in the cytoplasm and nucleoplasm of wheat crown tissues. Plant J. 1995, 8, 583–593. [Google Scholar] [CrossRef]
- Schneider, K.; Wells, B.; Schmelzer, E.; Salamini, F.; Bartels, D. Desiccation leads to the rapid accumulation of both cytosolic and chloroplastic proteins in the resurrection plant Craterostigma plantagineum Hochst. Planta 1993, 189, 120–131. [Google Scholar] [CrossRef]
- Godoy, J.; Lunar, R.; Torresschumann, S.; Moreno, J.; Rodrigo, R.; Pintortoro, J. Expression, tissue distribution and subcellular-localization of dehydrin Tas14 in salt-stressed tomato plants. Plant Mol. Biol. 1994, 26, 1921–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silveira Falavigna, V.; Malabarba, J.; Silveira, C.; Buffon, V.; Mariath, J.; Pasquali, G.; Margis-Pinheiro, M.; Revers, L. Characterization of the nucellus-specific dehydrin MdoDHN11 demonstrates its involvement in the tolerance to water deficit. Plant Cell Rep. 2019, 38, 1099–1107. [Google Scholar] [CrossRef]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.; Avelange-Macherel, M.; Macherel, D. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef] [Green Version]
- Dure, L., 3rd; Crouch, M.; Harada, J.; Ho, T.; Mundy, J.; Quatrano, R.; Thomas, T.; Sung, Z. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol. Biol. 1989, 12, 475–486. [Google Scholar] [CrossRef]
- Xie, H.; Vucetic, S.; Iakoucheva, L.; Oldfield, C.; Dunker, A.; Uversky, V.; Obradovic, Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res. 2007, 6, 1882–1898. [Google Scholar] [CrossRef] [Green Version]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.; Brown, C.; Dunker, A. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Tompa, P.; Szász, C.; Buday, L. Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 2005, 30, 484–489. [Google Scholar] [CrossRef]
- Garay-Arroyo, A.; Colmenero-Flores, J.; Garciarrubio, A.; Covarrubias, A. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 2000, 275, 5668–5674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strimbeck, G. Hiding in plain sight: The F segment and other conserved features of seed plant SKn dehydrins. Planta 2017, 245, 1061–1066. [Google Scholar] [CrossRef] [Green Version]
- Close, T. Dehydrins: Emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant 1996, 97, 795–803. [Google Scholar] [CrossRef]
- Malik, A.; Veltri, M.; Boddington, K.; Singh, K.; Graether, S. Genome analysis of conserved dehydrin motifs in vascular plants. Front. Plant Sci. 2017, 8, 709. [Google Scholar] [CrossRef] [Green Version]
- Drira, M.; Saibi, W.; Amara, I.; Masmoudi, K.; Hanin, M.; Brini, F. Wheat dehydrin K-segments ensure bacterial stress tolerance, antiaggregation and antimicrobial effects. Appl. Biochem. 2015, 175, 3310–3321. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Terashima, S.; Kuboi, T. Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J. Plant Physiol. 2001, 58, 1333–1339. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Q.; Li, D.; Yang, X.; Li, D. Multifunctional roles of plant dehydrins in response to environmental stresses. Front. Plant Sci. 2017, 8, 1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, S.; Graether, S. Cryoprotective mechanism of a small intrinsically disordered dehydrin protein. Protein Sci. 2011, 20, 42–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, S.; Close, T. Dehydrins: Genes, proteins, and associations with phenotypic traits. New Phytol. 1997, 137, 61–74. [Google Scholar] [CrossRef]
- Hincha, D.; Thalhammer, A. LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem. Soc. Trans. 2012, 40, 1000–1003. [Google Scholar] [CrossRef] [Green Version]
- Cuevas-Velazquez, C.; Rendón-Luna, D.; Covarrubias, A. Dissecting the cryoprotection mechanisms for dehydrins. Front. Plant Sci. 2014, 5, 583. [Google Scholar] [CrossRef] [Green Version]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef]
- Dalal, K.; Pio, F. Thermodynamics and stability of the PAAD/DAPIN/PYRIN domain of IFI-16. FEBS Lett. 2006, 580, 3083–3090. [Google Scholar] [CrossRef] [Green Version]
- Koag, M.; Wilkens, S.; Fenton, R.; Resnik, J.; Vo, E.; Close, T. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 2009, 150, 1503–1514. [Google Scholar] [CrossRef] [Green Version]
- Rosales, R.; Romero, I.; Escribano, M.I.; Merodio, C.; Sanchez-Ballesta, M. The crucial role of Φ-and K-segments in the in vitro functionality of Vitis vinifera dehydrin DHN1a. Phytochemistry 2014, 108, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Riley, A.; Ashlock, D.; Graether, S. Evolution of the modular, disordered stress proteins known as dehydrins. PLoS ONE 2019, 14, e0211813. [Google Scholar] [CrossRef] [Green Version]
- Hundertmark, M.; Hincha, D. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Abedini, R.; GhaneGolmohammadi, F.; PishkamRad, R.; Pourabed, E.; Jafarnezhad, A.; Shobbar, Z.; Shahbazi, M. Plant dehydrins: Shedding light on structure and expression patterns of dehydrin gene family in barley. J. Plant Res. 2017, 130, 747–763. [Google Scholar] [CrossRef]
- Liang, D.; Xia, H.; Wu, S.; Ma, F. Genome-wide identification and expression profiling of dehydrin gene family in Malus domestica. Mol. Biol. Rep. 2012, 39, 10759–10768. [Google Scholar] [CrossRef]
- Liang, Y.; Xiong, Z.; Zheng, J.; Xu, D.; Zhu, Z.; Xiang, J.; Gan, J.; Raboanatahiry, N.; Yin, Y.; Li, M. Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus. Sci. Rep. 2016, 6, 24265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Li, C.; Liu, B.; Ge, S.; Dong, X.; Li, W.; Zhu, H.; Wang, B.; Yang, C. Genome-wide identification and characterization of a dehydrin gene family in poplar (Populus trichocarpa). Plant Mol. Biol. Rep. 2012, 30, 848–859. [Google Scholar] [CrossRef]
- Charfeddine, S.; Saïdi, M.; Charfeddine, M.; Gargouri-Bouzid, R. Genome-wide identification and expression profiling of the late embryogenesis abundant genes in potato with emphasis on dehydrins. Mol. Biol. Rep. 2015, 42, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Verma, G.; Dhar, Y.; Srivastava, D.; Kidwai, M.; Chauhan, P.; Bag, S.; Asif, M.; Chakhrabarty, D. Genome-wide analysis of rice dehydrin gene family: Its evolutionary conservedness and expression pattern in response to PEG induced dehydration stress. PLoS ONE 2017, 12, e0176399. [Google Scholar]
- Gabaldón, T.; Koonin, E. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013, 14, 360–366. [Google Scholar] [CrossRef] [Green Version]
- Das, M.; Haberer, G.; Panda, A.; Laha, S.; Ghosh, T.; Schäffner, A. Expression pattern similarities support the prediction of orthologs retaining common functions after gene duplication events. Plant Physiol. 2016, 171, 2343–2357. [Google Scholar] [CrossRef] [Green Version]
- Kerstens, M.; Schranz, M.; Bouwmeester, K. Phylogenomic analysis of the APETALA2 transcription factor subfamily across angiosperms reveals both deep conservation and lineage-specific patterns. Plant J. 2020, 103, 1516–1524. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Schranz, M. Network approaches for plant phylogenomic synteny analysis. Curr. Opin. Plant Biol. 2017, 36, 129–134. [Google Scholar] [CrossRef]
- Pochon, S.; Simoneau, P.; Pigné, S.; Balidas, S.; Bataillé-Simoneau, N.; Campion, C.; Jaspard, E.; Calmes, B.; Hamon, B.; Berruyer, R.; et al. Dehydrin-like proteins in the necrotrophic fungus Alternaria brassicicola have a role in plant pathogenesis and stress response. PLoS ONE 2013, 8, e75143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artur, M.; Zhao, T.; Ligterink, W.; Schranz, E.; Hilhorst, H. Dissecting the genomic diversification of Late Embryogenesis Abundant (LEA) protein gene families in plants. Genome Biol. Evol. 2019, 11, 459–471. [Google Scholar] [CrossRef]
- Cao, J.; Li, X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 2015, 241, 757–772. [Google Scholar] [CrossRef]
- Ni, L.; Wang, Z.; Fu, Z.; Liu, D.; Yin, Y.; Li, H.; Gu, C. Genome-wide analysis of basic helix-loop-helix family genes and expression analysis in response to drought and salt stresses in Hibiscus hamabo Sieb. et Zucc. Int. J. Mol. Sci. 2021, 22, 8748. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, X.; Yao, W.; Zhao, K.; Liu, L.; Fan, G.; Zhou, B.; Jiang, T. Genome-wide search and structural and functional analyses for late embryogenesis-abundant (LEA) gene family in poplar. BMC Plant Biol. 2021, 21, 110. [Google Scholar] [CrossRef]
- Jing, H.; Li, C.; Ma, F.; Ma, J.; Khan, A.; Wang, X.; Zhao, L.; Gong, Z.; Chen, R. Genome-wide identification, expression diversication of dehydrin gene family and characterization of CaDHN3 in pepper (Capsicum annuum L.). PLoS ONE 2016, 11, e0161073. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, H.; Jin, G.; Liu, H.; Wu, W.; Zhu, J. Genome-scale identification and analysis of LEA genes in rice (Oryza Sativa L.). Plant Sci. 2007, 172, 414–420. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Qudeimat, E.; Thimma, M.; Chaiboonchoe, A.; Jijakli, K.; Alzahmi, A.; Arnoux, M.; Salehi-Ashtiani, K. Genome-wide expression analysis offers new insights into the origin and evolution of Physcomitrella patens stress response. Sci. Rep. 2015, 5, 17434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaraju, M.; Kumar, S.; Reddy, P.; Kumar, A.; Rao, D.; Kishor, P. Genome-scale identification, classification, and tissue specific expression analysis of late embryogenesis abundant (LEA) genes under abiotic stress conditions in Sorghum bicolor L. PLoS ONE 2019, 14, e0209980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Xia, H.; Liang, D.; Lin, L.; Deng, H.; Lv, X.; Wang, Z.; Zhang, X.; Wang, J.; Xiong, B. Genome-wide identification and expression profiling of the dehydrin gene family in Actinidia chinensis. Sci. Hortic. 2021, 280, 109930. [Google Scholar] [CrossRef]
- Yang, Y.; He, M.; Zhu, Z.; Li, S.; Xu, Y.; Zhang, C.; Singer, S.; Wang, Y. Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol. 2012, 12, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sena, J.; Giguère, I.; Rigault, P.; Bousquet, J.; Mackay, J. Expansion of the dehydrin gene family in the Pinaceae is associated with considerable structural diversity and drought-responsive expression. Tree Physiol. 2018, 38, 442–456. [Google Scholar] [CrossRef]
- Decena, M.; Galvez-Rojas, S.; Agostini, F.; Sancho, R.; Contreras-Moreira, B.; Des Marais, D.; Hernández, P.; Catalán, P. Evolution and functional dynamics of dehydrins in model Brachypodium grasses. bioRxiv 2021, 458816. [Google Scholar] [CrossRef]
- Graether, S.; Boddington, K. Disorder and function: A review of the dehydrin protein family. Front. Plant Sci. 2014, 5, 576. [Google Scholar] [CrossRef] [Green Version]
- Hara, M.; Shinoda, Y.; Kubo, M.; Kashima, D.; Takahashi, I.; Kato, T.; Horiike, T.; Kuboi, T. Biochemical characterization of the Arabidopsis KS-type dehydrin protein, whose gene expression is constitutively abundant rather than stress dependent. Acta Physiol. Plant 2011, 33, 2103–2116. [Google Scholar] [CrossRef]
- Huang, G.; Ma, S.; Bai, L.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Brini, F.; Hanin, M.; Lumbreras, V.; Amara, I.; Khoudi, H.; Hassairi, A.; Pagès, M.; Masmoudi, K. Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep. 2007, 26, 2017–2026. [Google Scholar] [CrossRef]
- Shekhawat, U.; Srinivas, L.; Ganapathi, T. MusaDHN1, a novel multiple stress-inducible SK3-type dehydrin gene, contributes affirmatively to drought- and salt-stress tolerance in banana. Planta 2011, 234, 915. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, X.; Lv, Q.; Zhu, D.; Qiu, T.; Xu, Y.; Bao, F.; He, Y.; Hu, Y. Physcomitrella patens dehydrins (PpDHNA and PpDHNC) confer salinity and drought tolerance to transgenic Arabidopsis plants. Front. Plant Sci. 2017, 8, 1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Liu, S.; Ma, J.; Wang, K.; Meng, Y.; Zhang, Y.; Chen, R. CaDHN4, a salt and cold stress-responsive dehydrin gene from pepper decreases abscisic acid sensitivity in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 26. [Google Scholar] [CrossRef] [Green Version]
- Avalbaev, A.; Allagulova, C.; Maslennikova, D.; Fedorova, K.; Shakirova, F. Methyl jasmonate and cytokinin mitigate the salinity-induced oxidative injury in wheat seedlings. J. Plant Growth Regul. 2020, 40, 1741–1752. [Google Scholar] [CrossRef]
- Cao, Y.; Xiang, X.; Geng, M.; You, Q.; Huang, X. Effect of HbDHN1 and HbDHN2 genes on abiotic stress responses in Arabidopsis. Front. Plant Sci. 2017, 8, 470. [Google Scholar] [CrossRef] [Green Version]
- Jyothi-Prakash, P.; Mohanty, B.; Wijaya, E.; Lim, T.; Lin, Q.; Loh, C.; Kumar, P. Identification of salt gland-associated genes and characterization of a dehydrin from the salt secretor mangrove Avicennia officinalis. BMC Plant Biol. 2014, 14, 6. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zheng, J.; Su, H.; Xia, K.; Jian, S.; Zhang, M. Molecular cloning and functional characterization of the dehydrin (IpDHN) gene from Ipomoea pes-caprae. Front. Plant Sci. 2018, 9, 1454. [Google Scholar] [CrossRef]
- Kosová, K.; Prášil, I.; Vítámvás, P. Role of dehydrins in plant stress response. In Handbook of Plant and Crop Stress, 4th ed.; Pessarakli, M., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 239–285. ISBN 978-0-81-539082-4. [Google Scholar]
- Kosová, K.; Vítámvás, P.; Prášil, I. Wheat and barley dehydrins under cold, drought, and salinity—What can LEA-II proteins tell us about plant stress response? Front. Plant Sci. 2014, 9, 1454. [Google Scholar] [CrossRef] [Green Version]
- Reyes, J.; Rodrigo, M.; Colmenero-Flores, J.; Gil, J.; Garay-Arroyo, A.; Campos, F.; Salamini, F.; Bartels, D.; Covarrubias, A.A. Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ. 2005, 28, 709–718. [Google Scholar] [CrossRef] [Green Version]
- Shakirova, F.; Allagulova, C.; Maslennikova, D.; Fedorova, K.; Yuldashev, R.; Lubyanova, A.; Bezrukova, M.; Avalbaev, A. Involvement of dehydrins in 24-epibrassinolide-induced protection of wheat plants against drought stress. Plant Physiol. Biochem. 2016, 108, 539–548. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, H.; Zhao, Z.; An, L. Abscisic acid is involved in brassinosteroids-induced chilling tolerance in the suspension cultured cells from Chorispora bungeana. J. Plant Physiol. 2011, 168, 853–862. [Google Scholar] [CrossRef]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Wani, S.; Singh, B.; Bohra, A.; Dar, Z.; Lone, A.; Pareek, A.; Singla-Pareek, S. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Liu, Q.; Feng, H.; Deng, J.; Zhang, R.; Wen, J.; Dong, J.; Wang, T. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 2020, 16, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Mehrabad Pour-Benab, S.; Fabriki-Ourang, S.; Mehrabi, A. Expression of dehydrin and antioxidant genes and enzymatic antioxidant defense under drought stress in wild relatives of wheat. Biotechnol. Biotechnol. Equip. 2019, 33, 1063–1073. [Google Scholar] [CrossRef] [Green Version]
- Sharma, E.; Jain, M.; Khurana, J. Differential quantitative regulation of specific gene groups and pathways under drought stress in rice. Genomics 2019, 111, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Yang, J.; Fu, J.; Xiong, L. Synergistic regulation of drought-responsive genes by transcription factor OsbZIP23 and histone modification in rice. J. Integr. Plant Biol. 2020, 62, 723–729. [Google Scholar] [CrossRef]
- Liu, H.; Yu, C.; Li, H.; Ouyang, B.; Wang, T.; Zhang, J.; Wang, X.; Ye, Z. Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato. Plant Sci. 2015, 231, 198–211. [Google Scholar] [CrossRef]
- Qiu, H.; Zhang, L.; Liu, C.; He, L.; Wang, A.; Liu, H.; Zhu, J. Cloning and characterization of a novel dehydrin gene, SiDhn2, from Saussurea involucrata Kar. et Kir. Plant Mol. Biol. 2014, 84, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhang, L.; Lv, H.; Zhang, H.; Zhang, D.; Wang, X.; Chen, J. The dehydrin wzy2 promoter from wheat defines its contribution to stress tolerance. Funct. Integr. 2014, 14, 111–125. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, X.; Zhang, L. Structural and Functional Dynamics of Dehydrins: A Plant Protector Protein under Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 3420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, V.; Soengas, P.; Alonso-Villaverde, V.; Sotelo, T.; Cartea, M.; Velasco, P. Effect of temperature stress on the early vegetative development of Brassica oleracea L. BMC Plant Biol. 2015, 15, 145. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, T.; Hong, T.; Ellis, R.; Batts, G.; Morison, J.; Hadley, P. The duration and rate of grain growth, and harvest index, of wheat (Triticum aestivum L.) in response to temperature and CO2. J. Exp. Bot. 1996, 47, 623–630. [Google Scholar] [CrossRef] [Green Version]
- Liliane, T.; Charles, M. Factors Affecting Yield of Crops; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Sakai, A.; Larcher, W. Frost Survival of Plants Responses and Adaptation to Freezing Stress; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1–321. ISBN 978-3-642-71747-5. [Google Scholar]
- Yang, W.; Zhang, L.; Lv, H.; Li, H.; Zhang, Y.; Xu, Y.; Yu, J. The K-segments of wheat dehydrin WZY2 are essential for its protective functions under temperature stress. Front. Plant Sci. 2015, 6, 406. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; He, P.; Xu, Y.; Liu, Q.; Yang, Y.; Liu, S. Overexpression of CsLEA11, a Y3SK 2-type dehydrin gene from cucumber (Cucumis sativus), enhances tolerance to heat and cold in Escherichia coli. Amb. Express. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Qin, F. Dehydrins from wheat x Thinopyrum ponticum amphiploid increase salinity and drought tolerance under their own inducible promoters without growth retardation. Plant Physiol. Biochem. 2016, 99, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Maruyama, K.; Todaka, D.; Mizoi, J.; Yoshida, T.; Kidokoro, S.; Matsukura, S.; Takasaki, H.; Sakurai, T.; Yamamoto, Y.Y.; Yoshiwara, K.; et al. Identification of cis-acting promoter elements in cold-and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 2012, 19, 37–49. [Google Scholar] [CrossRef]
- Bao, F.; Du, D.; An, Y.; Yang, W.; Wang, J.; Cheng, T.; Zhang, Q. Overexpression of Prunus mume dehydrin genes in tobacco enhances tolerance to cold and drought. Front. Plant Sci. 2017, 8, 151. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Ohta, A.; Takagi, M.; Imai, R. Expression of plant group 2 and group 3 lea genes in Saccharomyces cerevisiae revealed functional divergence among LEA proteins. J. Biochem. 2000, 127, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Mundy, J.; Chua, N. Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 1988, 7, 2279–2286. [Google Scholar] [CrossRef]
- Ismail, A.; Hall, A.; Close, T. Allelic variation of a dehydrin gene cosegregates with chilling tolerance during seedling emergence. Proc. Natl. Acad. Sci. USA 1999, 96, 13566–13570. [Google Scholar] [CrossRef] [Green Version]
- Cellier, F.; Conéjéro, G.; Breitler, J.; Casse, F. Molecular and physiological responses to water deficit in drought-tolerant and drought-sensitive lines of sunflower. Plant Physiol. 1998, 116, 319–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuriko, O.; Keishi, O.; Kazuo, S.; Tran, L. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar]
- Saavedra, L.; Svensson, J.; Carballo, V.; Izmendi, D.; Welin, B.; Vidal, S. A dehydrin gene in Physcomitrella patens is required for salt and osmotic stress tolerance. Plant J. 2006, 45, 237–249. [Google Scholar] [CrossRef]
- Ruibal, C.; Salamó, I.; Carballo, V.; Castro, A.; Bentancor, M.; Borsani, O.; Szabados, L.; Vidal, S. Differential contribution of individual dehydrin genes from Physcomitrella patens to salt and osmotic stress tolerance. Plant Sci. 2012, 190, 89–102. [Google Scholar] [CrossRef]
- Dean, R.; Fu, S.; Stocker, R.; Davies, M. Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J. 1997, 324, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeWald, D.; Torabinejad, J.; Jones, C.; Shope, J.; Cangelosi, A.; Thompson, J.E.; Prestwich, G.; Hama, H. Rapid accumulation of phosphatidylinositol 4, 5-bisphosphate and inositol 1, 4, 5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol. 2001, 126, 759–769. [Google Scholar] [CrossRef] [Green Version]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A new insight of salt stress signaling in plant. Mol. Cells 2016, 39, 447. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Reddy, P.; Reddy, C.; Reddy, M. Molecular cloning and characterization of salt inducible dehydrin gene from the C4 plant Pennisetum glaucum. Plant Gene 2015, 4, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Drira, M.; Hanin, M.; Masmoudi, K.; Brini, F. Comparison of full-length and conserved segments of wheat dehydrin DHN-5 overexpressed in Arabidopsis thaliana showed different responses to abiotic and biotic stress. Funct. Plant Biol. 2016, 43, 1048–1060. [Google Scholar] [CrossRef]
- Yu, X.; Yue, W.; Yang, Q.; Zhang, Y.; Han, X.; Yang, F.; Wang, R.; Li, G. Identification of the LEA family members from Caragana korshinskii (Fabaceae) and functional characterization of CkLEA2-3 in response to abiotic stress in Arabidopsis. Rev. Bras. Bot. 2019, 42, 227–238. [Google Scholar] [CrossRef]
- Yang, Z.; Sheng, J.; Lv, K.; Ren, L.; Zhang, D. Y2SK2 and SK3 type dehydrins from Agapanthus praecox can improve plant stress tolerance and act as multifunctional protectants. Plant Sci. 2019, 284, 143–160. [Google Scholar] [CrossRef]
- Halder, T.; Upadhyaya, G.; Ray, S. YSK2 type dehydrin (SbDhn1) from Sorghum bicolor showed improved protection under high temperature and osmotic stress condition. Front. Plant Sci. 2017, 8, 918. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Wang, Y.; Yu, T.; Chen, S.; Chen, Y.; Lu, C. Heterologous expression of three Ammopiptanthus mongolicus dehydrin genes confers abiotic stress tolerance in Arabidopsis thaliana. Plants 2020, 9, 193. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, P.; Tossounian, M.; Kovacs, D.; Thu, T.; Stijlemans, B.; Vertommen, D.; Pauwels, J.; Geveart, K.; Angenon, G.; Messens, J.; et al. Dehydrin ERD14 activates glutathione transferase Phi9 in Arabidopsis thaliana under osmotic stress. Biochim. Biophys. Acta 2020, 1864, 129506. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Hou, X.; Zhang, Y.; Meng, Y.; Zhang, H.; Liu, S.; Wang, X.; Chen, R. CaDHN5, a dehydrin gene from pepper, plays an important role in salt and osmotic stress responses. Int. J. Mol. Sci. 2019, 20, 1989. [Google Scholar] [CrossRef] [Green Version]
- Ganguly, M.; Datta, K.; Roychoudhury, A.; Gayen, D.; Sengupta, D.; Datta, S. Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance. Plant Signal. Behav. 2012, 7, 502–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Zhang, R.; Qu, Y.; Miao, Z.; Zhang, Y.; Shen, X.; Wang, T.; Dong, J. Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol. 2012, 195, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Mayor, A.; Pineda, B.; Garcia-Abellán, J.O.; Antón, T.; Garcia-Sogo, B.; Sanchez- Bel, P.; Flores, F.B.; Atarés, A.; Angosto, T.; Pintor-Toro, J.A.; et al. Overexpression of dehydrin tas14 gene improves the osmotic stress imposed by drought and salinity in tomato. J. Plant Physiol. 2012, 169, 459–468. [Google Scholar] [CrossRef]
- Duan, J.; Cai, W.; Park, S. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE 2012, 7, e45117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochoa-Alfaro, A.E.; Rodríguez-Kessler, M.; Pérez-Morales, M.B.; Delgado-Sánchez, P.; Cuevas-Velazquez, C.L.; Gómez-Anduro, G.; Jiménez-Bremont, J.F. Functional characterization of an acidic SK3 dehydrin isolated from an Opuntia streptacantha cDNA library. Planta 2012, 235, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Kim, H.Y.; Kim, Y.S.; Choi, H.G.; Kang, S.H.; Yoon, H.S. Expression of dehydrin gene from Arctic Cerastium arcticum increases abiotic stress tolerance and enhances the fermentation capacity of a genetically engineered Saccharomyces cerevisiae laboratory strain. Appl. Microbiol. Biotechnol. 2013, 97, 8997–9009. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Bai, S.; Li, Q.; Gao, C.; Liu, G.; Li, G.; Tan, F.; Alvarez, M.L. Overexpression of TaLEA gene from Tamarix androssowii improves salt and drought tolerance in transgenic poplar (Populus simonii × P. nigra). PLoS ONE 2013, 8, e67462. [Google Scholar] [CrossRef]
- Imamura, T.; Higuchi, A.; Takahashi, H. Dehydrins are highly expressed in overwintering buds and enhance drought and freezing tolerance in Gentiana triflora. Plant Sci. 2013, 213, 55–66. [Google Scholar] [CrossRef]
- Sun, J.; Nie, L.; Sun, G.; Guo, J.; Liu, Y. Cloning and characterization of dehydrin gene from Ammopiptanthus mongolicus. Mol. Biol. Rep. 2013, 40, 2281–2291. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, X.; Yang, S.; Li, X.; Yang, Y. Molecular cloning and characterization of a novel SK3-type dehydrin gene from Stipa purpurea. Biochem. Biophys. Res. Commun. 2014, 448, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Lee, S.C.; Kim, J.Y.; Kim, S.J.; Aye, S.S.; Kim, S.R. Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 2014, 57, 383–393. [Google Scholar] [CrossRef]
- Jia, F.; Qi, S.; Li, H.; Liu, P.; Li, P.; Wu, C.; Zheng, C.; Huang, J. Overexpression of Late Embryogenesis Abundant 14 enhances Arabidopsis salt stress tolerance. Biochem. Biophys. Res. Commun. 2014, 454, 505–511. [Google Scholar] [CrossRef]
- Saibi, W.; Feki, K.; Mahmoud, R.B.; Brini, F. Durum wheat dehydrin (DHN-5) confers salinity tolerance to transgenic Arabidopsis plants through the regulation of proline metabolism and ROS scavenging system. Planta 2015, 242, 1187–1194. [Google Scholar] [CrossRef]
- Chiappetta, A.; Muto, A.; Bruno, L.; Woloszynska, M.; Van Lijsebettens, M.; Bitonti, M.B. A dehydrin gene isolated from feral olive enhances drought tolerance in Arabidopsis transgenic plants. Front. Plant Sci. 2015, 6, 392. [Google Scholar] [CrossRef] [Green Version]
- Jardak-Jamoussi, R.; Zarrouk, O.; Ben Salem, A.; Zoghlami, N.; Mejri, S.; Gandoura, S.; Khiari, B.; Mliki, A.; Chaves, M.; Ghorbel, A.; et al. Overexpressing Vitis vinifera YSK2 dehydrin in tobacco improves plant performance. Agric. Water Manag. 2016, 164, 176–189. [Google Scholar] [CrossRef]
- Aguayo, P.; Sanhueza, J.; Noriega, F.; Ochoa, M.; Lefeuvre, R.; Navarrete, D.; Fernández, M.; Valenzuela, S. Overexpression of an SK n-dehydrin gene from Eucalyptus globulus and Eucalyptus nitens enhances tolerance to freezing stress in Arabidopsis. Trees 2016, 30, 1785–1797. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.i.; Zhu, J.; Liu, H.; Wang, A. Cloning and characterization of SiDHN, a novel dehydrin gene from Saussurea involucrata Kar. et Kir. that enhances cold and drought tolerance in tobacco. Plant Sci. 2017, 256, 160–169. [Google Scholar] [CrossRef]
- Lv, A.; Su, L.; Liu, X.; Xing, Q.; Huang, B.; An, Y.; Zhou, P. Characterization of Dehydrin protein, CdDHN4-L and CdDHN4-S, and their differential protective roles against abiotic stress in vitro. BMC Plant Biol. 2018, 18, 299. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.X.; Li, X.Y.; Xu, C.J.; Chen, J.W. Overexpression of Loquat dehydrin gene EjDHN1 promotes Cold Tolerance in Transgenic Tobacco. Russ. J. Plant Physiol. 2018, 65, 69–77. [Google Scholar] [CrossRef]
- Zeng, X.; Ling, H.; Yang, J.; Li, Y.; Guo, S. LEA proteins from Gastrodia elata enhance tolerance to low temperature stress in Escherichia coli. Gene 2018, 646, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Indoliya, Y.; Singh, P.K.; Singh, P.C.; Chauhan, P.S.; Pande, V.; Chakrabarty, D. Role of dehydrin-FK506-binding protein complex in enhancing drought tolerance through the ABA-mediated signaling pathway. Environ. Exp. Bot. 2019, 158, 136–149. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Song, Q.; Zhang, T.; Li, D.; Yang, X. The maize late embryogenesis abundant protein ZmDHN13 positively regulates copper tolerance in transgenic yeast and tobacco. Crop. J. 2019, 7, 403–410. [Google Scholar] [CrossRef]
- Shin, S.Y.; Kim, H.Y.; Kim, I.S.; Kim, J.J.; Kim, Y.S.; Yoon, H.S. The dehydrin gene of the Arctic plant Cerastium arcticum, CaDHN, increases tolerance to multiple stresses in Arabidopsis thaliana. Plant Biotechnol. Rep. 2020, 14, 387–395. [Google Scholar] [CrossRef]
- Shi, H.; He, X.; Zhao, Y.; Lu, S.; Guo, Z. Constitutive expression of a group 3 LEA protein from Medicago falcata (MfLEA3) increases cold and drought tolerance in transgenic tobacco. Plant Cell Rep. 2020, 39, 851–860. [Google Scholar] [CrossRef]
- Poku, S.; Chukwurah, P.; Aung, H.H.; Nakamura, I. Over-expression of a melon Y3SK2-type LEA gene confers drought and salt tolerance in transgenic tobacco plants. Plants 2020, 9, 1749. [Google Scholar] [CrossRef]
- Vazquez-Hernandez, M.; Romero, I.; Sanchez-Ballesta, M.T.; Merodio, C.; Escribano, M.I. Functional characterization of VviDHN2 and VviDHN4 dehydrin isoforms from Vitis vinifera (L.): An in silico and in vitro approach. Plant Physiol. Biochem. 2021, 158, 146–157. [Google Scholar] [CrossRef]
- Meng, Y.C.; Zhang, H.-F.; Pan, X.X.; Chen, N.; Hu, H.F.; Haq, S.; Khan, A.; Chen, R.G. CaDHN3, a pepper (Capsicum annuum L.) dehydrin gene enhances the tolerance against salt and drought stresses by reducing ROS accumulation. Int. J. Mol. Sci. 2021, 22, 3205. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Tang, M.; Hu, Y.; Lin, Z. Isolation and characterization of a dehydrin-like gene from drought-tolerant Boea crassifolia. Plant Sci. 2004, 166, 1167–1175. [Google Scholar] [CrossRef]
- Mota, A.; Oliveira, T.; Vinson, C.; Williams, T.; Costa, M.; Araujo, A.; Danchin, E.; Grossi-de-Sá, M.; Guimaraes, P.; Brasileiro, A. Contrasting effects of wild Arachis dehydrin under abiotic and biotic stresses. Front. Plant Sci. 2019, 10, 497. [Google Scholar] [CrossRef] [Green Version]
- Salleh, F.; Evans, K.; Goodall, B.; Machin, H.; Mowla, S.; Mur, L.; Runions, J.; Theodoulou, F.; Foyer, C.; Rogers, H. A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ. 2012, 35, 418–429. [Google Scholar] [CrossRef] [Green Version]
- Ait-El-Mokhtar, M.; Baslam, M.; Ben-Laouane, R.; Anli, M.; Boutasknit, A.; Mitsui, T.; Wahbi, S.; Meddich, A. Alleviation of detrimental effects of salt stress on date palm (Phoenix dactylifera L.) by the application of arbuscular mycorrhizal fungi and/or compost. Front. Sustain. Food Syst. 2020, 4, 131. [Google Scholar] [CrossRef]
- Al Kharusi, L.; Al Yahyai, R.; Yaish, M.W. Antioxidant response to salinity in salt-tolerant and salt-susceptible cultivars of date palm. Agriculture 2019, 9, 8. [Google Scholar] [CrossRef] [Green Version]
- Yaish, M.; Kumar, P. Salt tolerance research in date palm tree (Phoenix dactylifera L.), past, present, and future perspectives. Front. Plant Sci. 2015, 6, 348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anli, M.; Baslam, M.; Tahiri, A.; Raklami, A.; Symanczik, S.; Boutasknit, A.; Ait-El-Mokhtar, M.; Ben-Laouane, R.; Toubali, S.; Ait Rahou, Y.; et al. Biofertilizers as strategies to improve photosynthetic apparatus, growth, and drought stress tolerance in the date palm. Front. Plant Sci. 2020, 11, 516818. [Google Scholar] [CrossRef] [PubMed]
- Ghirardo, A.; Nosenko, T.; Kreuzwieser, J.; Winkler, J.B.; Kruse, J.; Albert, A.; Merl-Pham, J.; Lux, T.; Ache, P.; Zimmer, I.; et al. Protein expression plasticity contributes to heat and drought tolerance of date palm. Oecologia 2021, 1–17. [Google Scholar] [CrossRef]
- Al-Khateeb, S.; Al-Khateeb, A.; Sattar, M.; Mohmand, A. Induced in vitro adaptation for salt tolerance in date palm (Phoenix dactylifera L.) cultivar Khalas. Biol. Res. 2020, 53, 37. [Google Scholar] [CrossRef]
- Yaish, M.; Antony, I.; Glick, B. Isolation and characterization of endophytic plant growth-promoting bacteria from date palm tree (Phoenix dactylifera L.) and their potential role in salinity tolerance. Antonie Leeuwenhoek 2015, 107, 1519–1532. [Google Scholar] [CrossRef]
- Müller, H.; Schäfer, N.; Bauer, H.; Geiger, D.; Lautner, S.; Fromm, J.; Riederer, M.; Bueno, A.; Nussbaumer, T.; Mayer, K.; et al. The desert plant Phoenix dactylifera closes stomata via nitrate-regulated SLAC1 anion channel. New Phytol. 2017, 216, 150–162. [Google Scholar] [CrossRef] [Green Version]
- Al-Mssallem, I.S.; Hu, S.; Zhang, X.; Lin, Q.; Liu, W.; Tan, J.; Yu, X.; Liu, J.; Pan, L.; Zhang, T.; et al. Genome sequence of the date palm Phoenix dactylifera L. Nat. Commun. 2013, 4, 2274. [Google Scholar] [CrossRef] [Green Version]
- Mathew, L.; Spannagl, M.; Al-Malki, A.; George, B.; Torres, M.; Al-Dous, E.; Al-Azwani, E.; Hussein, E.; Mathew, S.; Mayer, K.; et al. A first genetic map of date palm (Phoenix dactylifera) reveals long-range genome structure conservation in the palms. BMC Genom. 2014, 15, 285. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Xia, W.; Yang, Y.; Mason, A.S.; Lei, X.; Ma, Z. Characterization and evolution of conserved microRNA through duplication events in date palm (Phoenix dactylifera). PLoS ONE 2013, 8, e71435. [Google Scholar] [CrossRef] [PubMed]
- Delahaie, J.; Hundertmark, M.; Bove, J.; Leprince, O.; Rogniaux, H.; Buitink, J. LEA polypeptide profiling of recalcitrant and orthodox legume seeds reveals ABI3-regulated LEA protein abundance linked to desiccation tolerance. J. Exp. Bot. 2013, 64, 4559–4573. [Google Scholar] [CrossRef]
- Walters, C. Orthodoxy, recalcitrance and in-between: Describing variation in seed storage characteristics using threshold responses to water loss. Planta 2015, 242, 397–406. [Google Scholar] [CrossRef]
- Obroucheva, N.; Sinkevich, I.; Lityagina, S. Physiological aspects of seed recalcitrance: A case study on the tree Aesculus hippocastanum. Tree Physiol. 2016, 36, 1127–1150. [Google Scholar] [CrossRef] [Green Version]
- Gayatri, G.; Kumar, K.; Nair, P.; Deth, G.; Baiju, K. Dynamics of water and abscisic acid during embryogeny and embryo drying in the recalcitrant seeds of Vateria indica L. J. Plant Growth Regul. 2021, 1–8. [Google Scholar] [CrossRef]
- Finch-Savage, W.; Pramanik, S.; Bewly, J. The expression of dehydrin proteins in desiccation-sensitive (recalcitrant) seeds of temperate trees. Planta 1994, 193, 478–485. [Google Scholar] [CrossRef]
- Solberg, Ø.; Yndgaard, F.; Andreasen, C.; von Bothmer, R.; Loskutov, G.; Asdal, Å. Long-term storage and longevity of orthodox seeds: A systematic review. Front. Plant Sci. 2020, 11, 1007. [Google Scholar] [CrossRef] [PubMed]
- Azarkovich, M.I. Dehydrins in orthodox and recalcitrant seeds. Russ. J. Plant Physiol. 2020, 67, 221–230. [Google Scholar] [CrossRef]
- Kleinwächter, M.; Radwan, A.; Hara, M.; Selmar, D. Dehydrin expression in seeds: An issue of maturation drying. Front. Plant Sci. 2014, 5, 402. [Google Scholar] [CrossRef] [Green Version]
- Galau, G.; Hughes, D.; Dure, L. Abscisic acid induction of cloned cotton late embryogenesis-abundant (Lea) mRNAs. Plant Mol. Biol. 1986, 7, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.; Buijs, G.; Ligterink, W.; Hilhorst, H. Evolutionary ecophysiology of seed desiccation sensitivity. Funct. Plant Biol. 2018, 45, 1083–1095. [Google Scholar] [CrossRef]
- Li, N.; Zhang, S.; Liang, Y.; Qi, Y.; Chen, J.; Zhu, W.; Zhang, L. Label-free quantitative proteomic analysis of drought stress-responsive late embryogenesis abundant proteins in the seedling leaves of two wheat (Triticum aestivum L.) genotypes. J. Proteomics 2018, 172, 122–142. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Boschetti, C.; Walton, L.; Sarkar, S.; Rubinsztein, D.; Tunnacliffe, A. Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc. Natl. Acad. Sci. USA 2007, 104, 18073–18078. [Google Scholar] [CrossRef] [Green Version]
- Boddington, K.; Graether, S. Binding of a Vitis riparia dehydrin to DNA. Plant Sci. 2019, 287, 110172. [Google Scholar] [CrossRef] [PubMed]
- Drira, M.; Saibi, W.; Brini, F.; Gargouri, A.; Masmoudi, K.; Hanin, M. The K-segments of the wheat dehydrin DHN-5 are essential for the protection of lactate dehydrogenase and β-glucosidase activities in vitro. Mol. Biotechnol. 2013, 54, 643–650. [Google Scholar] [CrossRef]
- Clarke, M.; Boddington, K.; Warnica, J.; Atkinson, J.; McKenna, S.; Madge, J.; Barker, C.; Graether, S. Structural and functional insights into the cryoprotection of membranes by the intrinsically disordered dehydrins. J. Biol. Chem. 2015, 290, 26900–26913. [Google Scholar] [CrossRef] [Green Version]
- Reczek, C.; Chandel, N. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015, 33, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Dietz, K.; Mittler, R.; Noctor, G. Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol. 2016, 171, 1535–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Monna, S.; Murata, T.; Nakano, T.; Amano, S.; Nachbar, M.; Wätzigb, H. The Arabidopsis KS-type dehydrin recovers lactate dehydrogenase activity inhibited by copper with the contribution of his residues. Plant Sci. 2016, 245, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Krüger, C.; Berkowitz, O.; Stephan, U.; Hell, R. A metal binding member of the late embryogenesis abundant protein family transports iron in the phloem of Ricinus communis L. J. Biol. Chem. 2002, 277, 25062–25069. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Marzinek, J.K.; Jefferies, D.; Bond, P.J.; Harryson, P.; Wohland, T. The disordered plant dehydrin Lti30 protects the membrane during water-related stress by cross-linking lipids. J. Biol. Chem. 2019, 294, 6468–6482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, S.; Kutzer, M.; Procek, J.; Gröbner, G.; Harryson, P. Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 2011, 23, 2391–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puhakainen, T.; Hess, M.; Makela, P.; Svensson, J.; Heino, P.; Palva, E. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol. Biol. 2004, 54, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Egerton-Warburton, L.; Balsamo, R.; Close, T. Temporal accumulation and ultrastructural localization of dehydrins in Zea mays. Physiol. Plant. 1997, 101, 545–555. [Google Scholar] [CrossRef]

| S No. | Source | Gene | Year | Target | Tolerance | Reference |
|---|---|---|---|---|---|---|
| 1. | Rice | Rab16A | 2012 | Rice | Salinity | [126] |
| Medicago Truncatula | MtCAS31 | Arabidopsis | Drought | [127] | ||
| Physcomitrella patens | PpDHNA and PpDHNB | Arabidopsis | Salt and osmotic | [114] | ||
| Tomato | tas14 | Tomato | Drought and salinity | [128] | ||
| Rice | OsLEA3-2 | Arabidopsis and rice | Drought and salinity | [129] | ||
| Opuntia streptacantha | OpsDHN1 | Arabidopsis | Freezing | [130] | ||
| 2. | Cerastium arcticum | CaDHN | 2013 | Saccharomyces | Salinity and freezing | [131] |
| Tamarix androssowii | TaLEA | Poplar | Drought and salinity | [132] | ||
| Ammopiptanthus mongolicus | AmDHN | Arabidopsis | Drought and salinity | [133] | ||
| Gentiana triflora | GtDHN1 and GtDHN2 | Gentiana trifloral | Drought and freezing | [134] | ||
| 3. | Stipa purpurea | SpDHN1 | 2014 | Arabidopsis | Drought | [135] |
| Rice | OsDhn1 | Rice | Drought and salinity | [136] | ||
| Arabidopsis | AtLEA14 | Arabidopsis and yeast | Salinity | [137] | ||
| Saussurea involucrata | SiDhn2 | Tobacco | Freezing and drought | [94] | ||
| 4. | Wheat | DHN-5 | 2015 | Arabidopsis | Salinity | [138] |
| Pennisetum glaucum | PgDHN | E. coli and yeast | Salt, osmotic, and heat | [118] | ||
| Solanum habrochaites | ShDHN | Tomato | Cold, drought, salt, and osmotic | [93] | ||
| Olea europaea | OesDHN | Arabidopsis | Drought | [139] | ||
| 5. | Vitis vinifera | VvDhn | 2016 | Tobacco | Drought and salinity | [140] |
| Wheat | DHN-5 | Arabidopsis | Salt and osmotic | [119] | ||
| Eucalyptus nitens | EniDHN2 | Arabidopsis | Cold | [141] | ||
| Wheat | TaDHN1 and TaDHN3 | Arabidopsis | Drought and salinity | [104] | ||
| 6. | Prunus mume | PmLEAs | 2017 | Tobacco | Drought and cold | [107] |
| Hevea brasiliensis | HbDHN1 and HbDHN2 | Arabidopsis | Salt, drought, and osmotic | [79] | ||
| Saussurea involucrata | SiDHN | Tobacco | Drought and cold | [142] | ||
| 7. | Bermudagrass | CdDHN4 | 2018 | Arabidopsis and E. coli | Salinity, cold, and heat | [143] |
| Ipomoea pes-caprae | IpDHN | Arabidopsis | Salt and drought | [81] | ||
| Eriobotrya japonica | EjDHN | Tobacco | Cold | [144] | ||
| Gastrodia elata | GeLEA | E. coli | Cold | [145] | ||
| 8. | Malus domestica | MdoDHN11 | 2019 | Arabidopsis | Drought | [24] |
| Oryza sativa | OsDhnRab16 | Rice | Drought | [146] | ||
| Capsicum annuum | CaDHN5 | Arabidopsis | Salt and osmotic | [125] | ||
| Korshinsk pea shrub | CkLEA2-3 | Arabidopsis | Salt and osmotic | [120] | ||
| African lily | ApY2SK2 and ApSK3 | Arabidopsis | Salt, osmotic, cold, and drought | [121] | ||
| Zea mays | ZmDHN13 | Yeast and tobacco | Oxidative stress | [147] | ||
| 9. | Ammopiptanthus mongolicus | AmDHN132, AmDHN154 and AmDHN200 | 2020 | Arabidopsis | Salt, osmotic, and cold | [123] |
| Cerastium arcticum | CaDHN | Arabidopsis and E. coli | Salt, cold, and drought | [148] | ||
| Medicago falcate | MfLEA3 | Tobacco | Cold and drought | [149] | ||
| Capsicum annuum | CaDHN4 | Capsicum annuum | Salt | [77] | ||
| Cucumis melo | CmLEA-S | Tobacco | Salinity and drought | [150] | ||
| 10. | Vitis vinifera | VviDHN2 and VviDHN4 | 2021 | E. coli | Freezing and drought | [151] |
| Capsicum annuum | CaDHN3 and CaHIRD11 | Arabidopsis | Salt and drought | [152] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdul Aziz, M.; Sabeem, M.; Mullath, S.K.; Brini, F.; Masmoudi, K. Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses. Biomolecules 2021, 11, 1662. https://doi.org/10.3390/biom11111662
Abdul Aziz M, Sabeem M, Mullath SK, Brini F, Masmoudi K. Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses. Biomolecules. 2021; 11(11):1662. https://doi.org/10.3390/biom11111662
Chicago/Turabian StyleAbdul Aziz, Mughair, Miloofer Sabeem, Sangeeta Kutty Mullath, Faical Brini, and Khaled Masmoudi. 2021. "Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses" Biomolecules 11, no. 11: 1662. https://doi.org/10.3390/biom11111662
APA StyleAbdul Aziz, M., Sabeem, M., Mullath, S. K., Brini, F., & Masmoudi, K. (2021). Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses. Biomolecules, 11(11), 1662. https://doi.org/10.3390/biom11111662








