Radioprotective and Radiomitigative Effects of Melatonin in Tissues with Different Proliferative Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Animals and Their Irradiation
2.3. Administration of Melatonin to Mice and Collection of Tissues for Analysis
2.4. DNA Isolation and Purification
2.5. Analysis of Damage and Repair of Mitochondrial DNA and Nuclear DNA
2.6. Quantitative Analysis of Mitochondrial DNA Copies Relative to the Nuclear DNA
2.7. Surveyor Nuclease Assay of mtDNA Mutant Copies
2.8. Determination of Hydrogen Peroxide Level
2.9. ATP Analysis
2.10. Determination of Lipid Peroxidation
2.11. Determination of Glutathione Level (GSH)
2.12. Statistical Analysis
3. Results
3.1. Damage and Repair of Nuclear DNA and Mitochondrial DNA following Irradiation
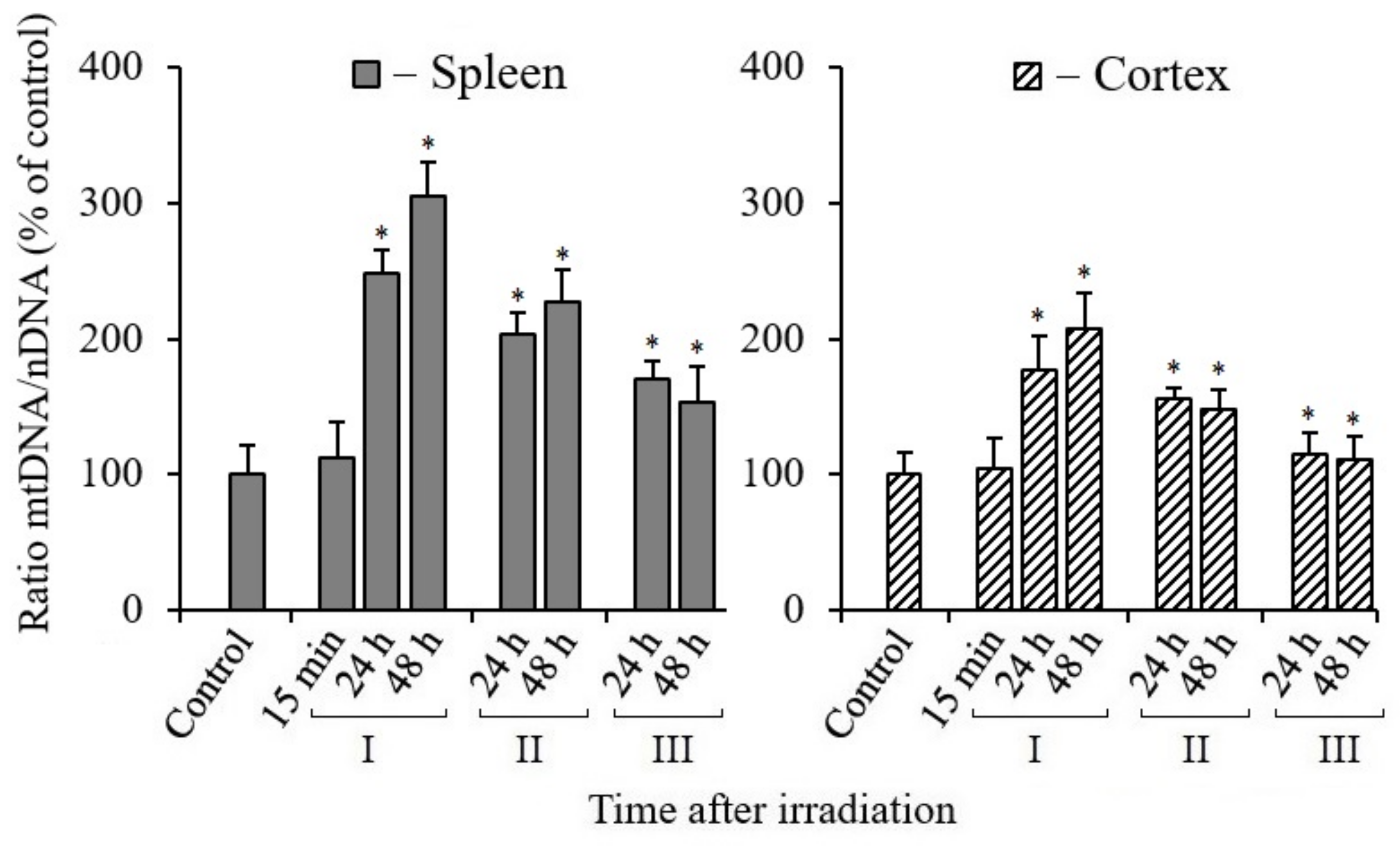
3.2. Effect of Melatonin on Mitochondrial Biogenesis in Tissues of X-Irradiated Mice
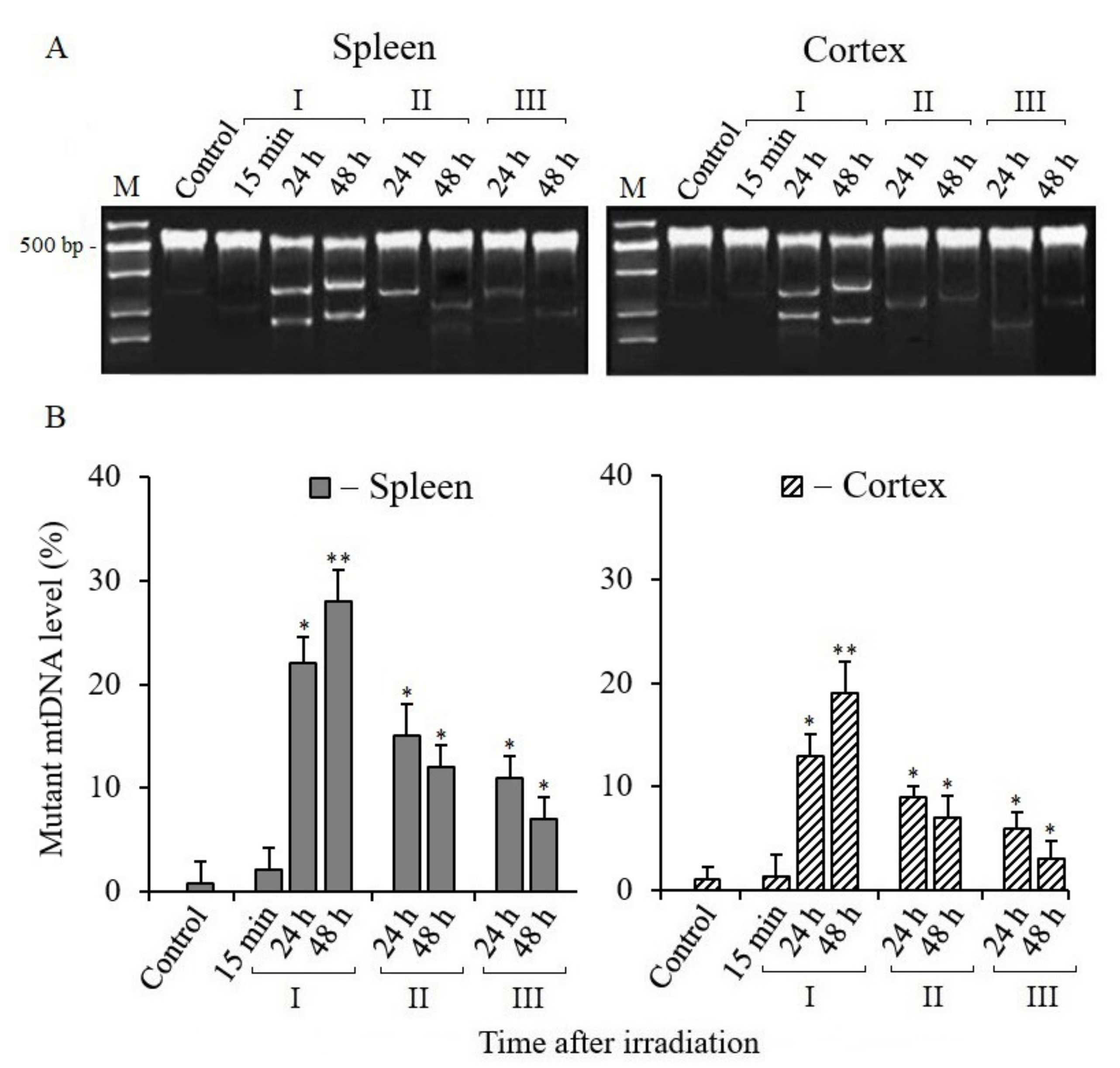
3.3. Analysis of Mitochondrial DNA Mutant Copies
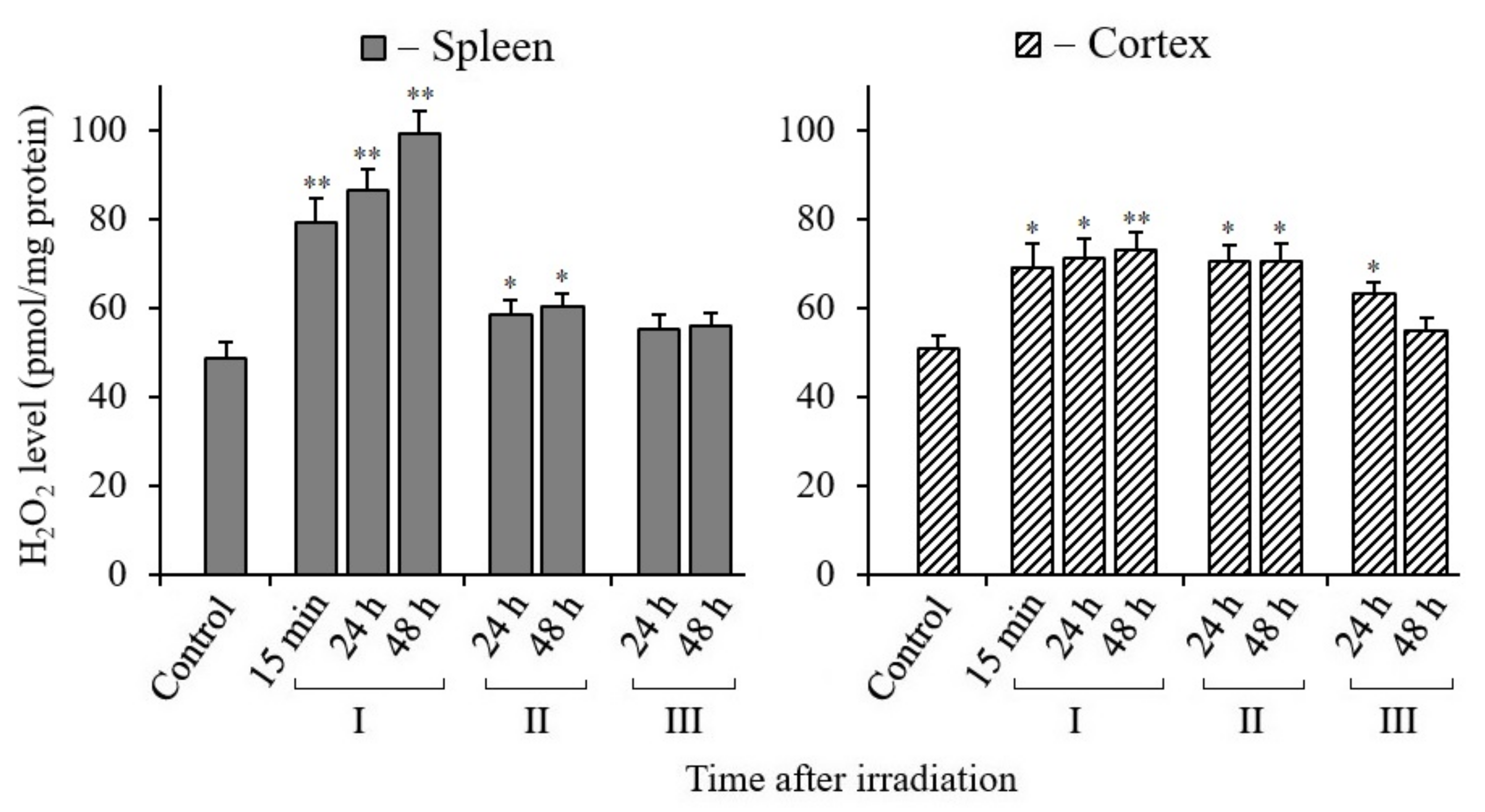
3.4. Changes in H2O2 Content in Tissues of X-Irradiated Mice
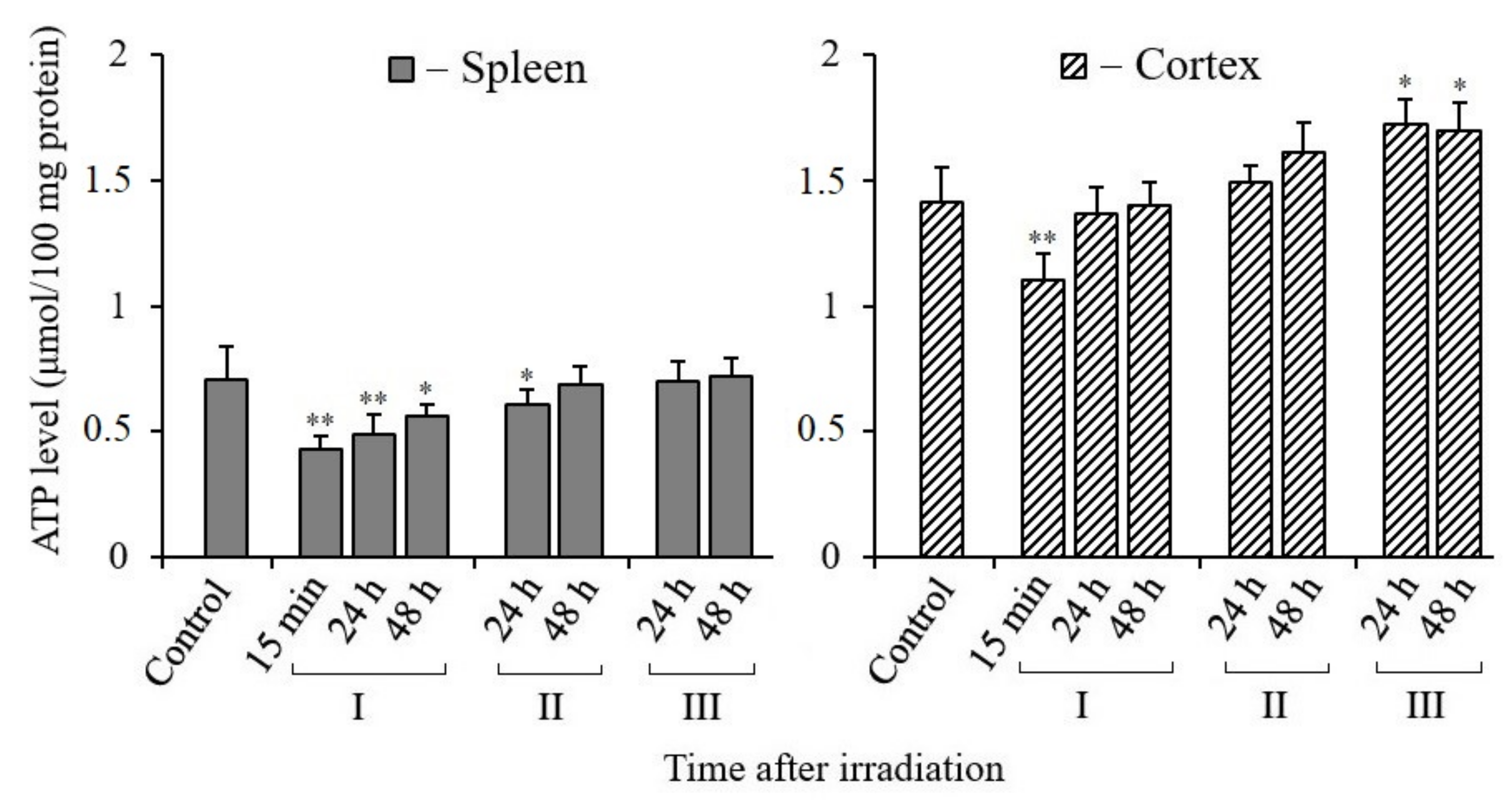
3.5. Changes in ATP Content in Tissues of X-Irradiated Mice
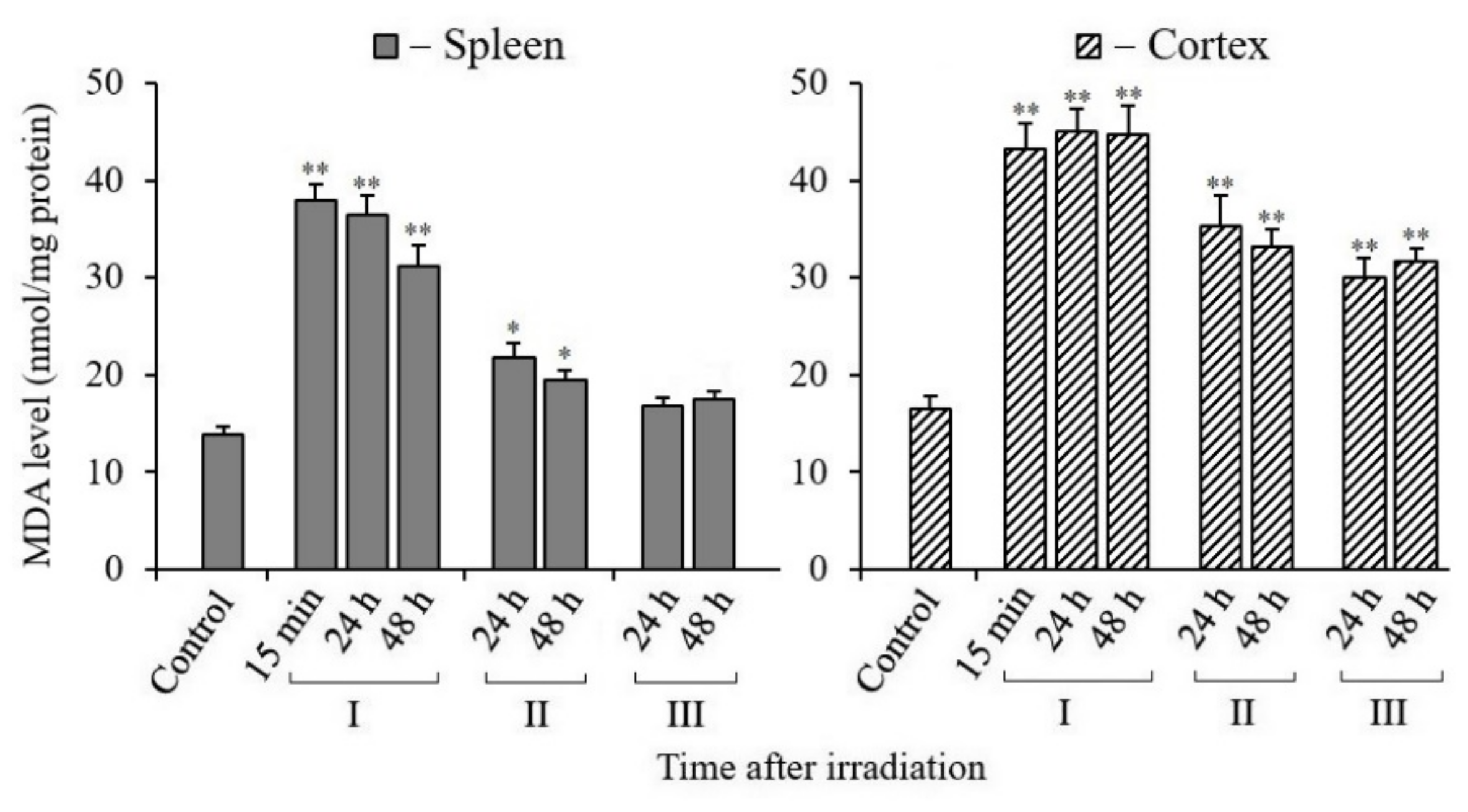
3.6. Changes in MDA Content in Tissues of X-Irradiated Mice
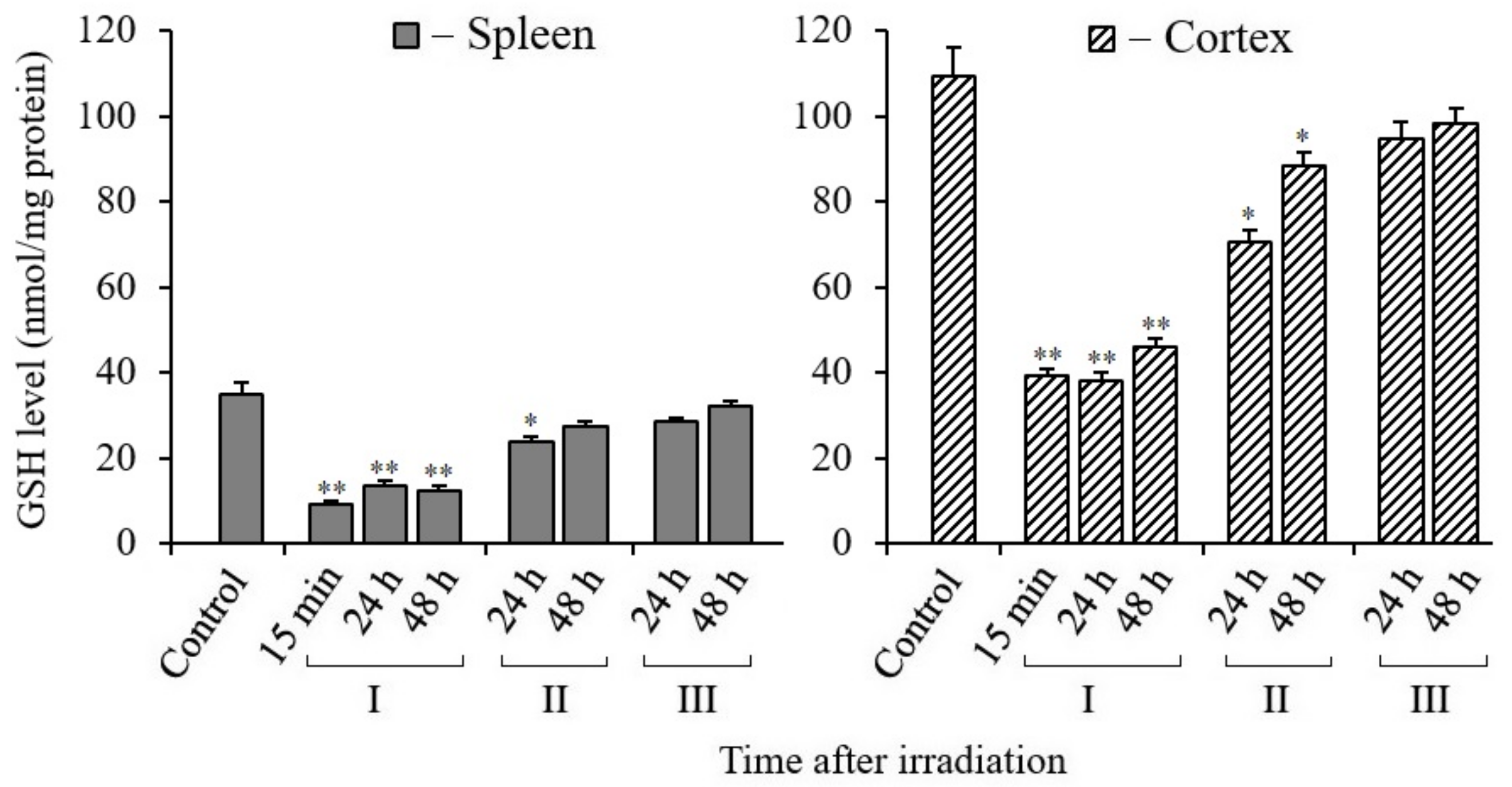
3.7. Changes in Glutathione Content in Tissues of X-Irradiated Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karri, J.; Lachman, L.; Hanania, A.; Marathe, A.; Singh, M.; Zacharias, N.; Orhurhu, V.; Gulati, A.; Abd-Elsayed, A. Radiotherapy-specific chronic pain syndromes in the cancer population: An evidence-based narrative review. Adv. Ther. 2021, 38, 1425–1446. [Google Scholar] [CrossRef] [PubMed]
- Chancellor, J.C.; Scott, G.B.; Sutton, J.P. Space radiation: The number one risk to astronaut health beyond low Earth orbit. Life 2014, 4, 491–510. [Google Scholar] [CrossRef] [Green Version]
- Hall, E.J.; Giaccia, A.J. Radiobiology for the Radiologist, 8th ed.; Wolters Kluwer: Philadelphia, PA, USA, 2019. [Google Scholar]
- Rosen, E.M.; Day, R.; Singh, V.K. New approaches to radiation protection. Front. Oncol. 2015, 4, 381. [Google Scholar] [CrossRef] [Green Version]
- Reiter, R.J.; Tan, D.X.; Herman, T.S.; Thomas, C.R., Jr. Melatonin as a radioprotective agent: A review. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, 639–653. [Google Scholar]
- Zetner, D.; Andersen, L.P.; Rosenberg, J. Melatonin as protection against radiation injury: A systematic review. Drug Res. 2016, 66, 281–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zharinov, G.M.; Bogomolov, O.A.; Chepurnaya, I.V.; Neklasova, N.Y.; Anisimov, V.N. Melatonin increases overall survival of prostate cancer patients with poor prognosis after combined hormone radiation treatment. Oncotarget 2020, 11, 3723–3729. [Google Scholar] [CrossRef]
- Gurunathan, S.; Qasim, M.; Kang, M.H.; Kim, J.H. Role and therapeutic potential of melatonin in various type of cancers. Onco Targets Ther. 2021, 14, 2019–2052. [Google Scholar] [CrossRef]
- Moloudizargari, M.; Moradkhani, F.; Hekmatirad, S.; Fallah, M.; Asghari, M.H.; Reiter, R.J. Therapeutic targets of cancer drugs: Modulation by melatonin. Life Sci. 2021, 267, 118934. [Google Scholar] [CrossRef]
- Liu, M.T.; Reiter, R.J. Melatonin protection against ionizing radiation in space. J. Cell. Sci. Apo. 2019, 2, 112. [Google Scholar]
- Simonsen, L.C.; Slaba, T.C.; Guida, P.; Rusek, A. NASA’s first ground-based galactic cosmic ray simulator: Enabling a new era in space radiobiology research. PLoS Biol. 2020, 18, e3000669. [Google Scholar] [CrossRef] [PubMed]
- Laiakis, E.C.; Shuryak, I.; Deziel, A.; Wang, Y.-W.; Barnette, B.L.; Yu, Y.; Ullrich, R.L.; Fornace, A.J., Jr.; Emmett, M.R. Effects of low dose space radiation exposures on the splenic metabolome. Int. J. Mol. Sci. 2021, 22, 3070. [Google Scholar] [CrossRef] [PubMed]
- Bergonié, J.; Tribondeau, L. De quelques résultats de la radiotherapie et essai de fixation d’une technique rationnelle. CR Acad. Sci. 1906, 143, 983–985. [Google Scholar]
- Betlazar, C.; Middleton, R.J.R.; Banati, B.; Liu, G.-J. The impact of high and low dose ionizing radiation on the central nervous system. Redox Biol. 2016, 9, 144–156. [Google Scholar] [CrossRef] [Green Version]
- Choudharya, S.; Kumara, A.; Sahac, N.; Chaudhury, N.K. PK-PD based optimal dose and time for orally administered suprapharmacological dose of melatonin to prevent radiation induced mortality in mice. Life Sci. 2019, 219, 31–39. [Google Scholar] [CrossRef]
- Gonzalez-Hunt, C.P.; Rooney, J.P.; Ryde, I.T.; Anbalagan, C.; Joglekar, R.; Meyer, J.N. PCR-based analysis of mitochondrial DNA copy number, mitochondrial DNA damage, and nuclear DNA damage. Curr. Protoc. Toxicol. 2016, 67, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Furda, A.; Santos, J.H.; Meyer, J.N.; Van Houten, B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 2014, 1105, 419–437. [Google Scholar] [PubMed] [Green Version]
- Abdullaev, S.; Gubina, N.; Bulanova, T.; Gaziev, A.I. Assessment of nuclear and mitochondrial DNA, expression of mitochondria-related genes in different brain regions in rats after whole-body X-ray irradiation. Int. J. Mol. Sci. 2020, 21, 1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, L.H.; Rouanet, J.P.; Howlett, E.H.; Leuthner, T.C.; Rooney, J.P.; Greenamyre, J.T.; Meyer, J.N. Newly revised protocol for quantitative PCR-based assay to measure mitochondrial and nuclear DNA damage. Curr. Protoc. Toxicol. 2018, 76, e50. [Google Scholar] [CrossRef]
- Rooney, J.P.; Ryde, I.T.; Sanders, L.H.; Howlett, E.H.; Colton, M.D.; Germ, K.E.; Mayer, G.D.; Greenamyre, J.T.; Meyer, J.N. PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol. Biol. 2015, 1241, 23–38. [Google Scholar]
- Bannwarth, S.; Procaccio, V.; Paquis-Flucklinger, V. Rapid identification of unknown heteroplasmic mitochondrial DNA mutations with mismatch-specific surveyor nuclease. Methods Mol. Biol. 2009, 554, 301–313. [Google Scholar]
- Abdullaev, S.; Bulanova, T.; Timoshenko, G.; Gaziev, A.I. Increase of mtDNA number and its mutant copies in rat brain after exposure to 150 MeV protons. Mol. Biol. Rep. 2020, 47, 4815–4820. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Chida, J.; Kido, H. Extraction and quantification of adenosine triphosphate in mammalian tissues and cells. Methods Mol. Biol. 2014, 1098, 21–32. [Google Scholar] [PubMed]
- Buege, J.A.; Aust, S.D. Microsomal lipid peroxidation. Methods Enzymol. 1978, 52, 302–310. [Google Scholar]
- Ellman, G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959, 8, 70–77. [Google Scholar] [CrossRef]
- Galano, A.; Reiter, R.J. Melatonin and its metabolites vs oxidative stress: From individual actions to collective protection. J. Pineal Res. 2018, 65, e12514. [Google Scholar] [CrossRef] [Green Version]
- Majidinia, M.; Reiter, R.J.; Shakouri, S.K.; Yousefi, B. The role of melatonin,a multitasking molecule, in retarding the processes of ageing. Ageing Res. Rev. 2018, 47, 198–213. [Google Scholar] [CrossRef]
- Van Houten, B.; Hunter, S.E.; Meyer, J.N. Mitochondrial DNA damage induced autophagy, cell death, and disease. Front. Biosci. 2016, 21, 42–54. [Google Scholar] [CrossRef]
- Moretton, A.; Morel, F.; Macao, B.; Lachaume, P.; Ishak, L.; Lefebvre, M.; Garreau-Balandier, I.; Vernet, P.; Falkenberg, M.; Farge, G. Selective mitochondrial DNA degradation following double strand breaks. PLoS ONE 2017, 12, e0176795. [Google Scholar] [CrossRef] [Green Version]
- Peeva, V.; Blei, D.; Trombly, G.; Corsi, S.; Szukszto, M.J.; Rebelo-Guiomar, P.; Gammage, P.A.; Kudin, A.P.; Becker, C.; Altmüller, J.; et al. Linear mitochondrial DNA is rapidly degraded by components of the replication machinery. Nat. Commun. 2018, 9, 1727. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L. Mitochondrial DNA degradation: A quality control measure for mitochondrial genome maintenance and stress response. Enzymes 2019, 45, 311–341. [Google Scholar] [PubMed]
- Lee, H.C.; Wei, Y.H. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int. J. Biochem. Cell Biol. 2005, 37, 822–834. [Google Scholar] [CrossRef]
- Andres, A.M.; Tucker, K.C.; Thomas, A.; Taylor, D.J.; Sengstock, D.; Jahania, S.M.; Dabir, R.; Pourpirali, S.; Brown, J.A.; Westbrook, D.G.; et al. Mitophagy and mitochondrial biogenesis in atrial tissue of patients undergoing heart surgery with cardiopulmonary bypass. JCI Insight 2017, 2, e89303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeBalsi, K.L.; Hoff, K.E.; Copeland, W.C. Role of the mitochondrial DNA replication machinery in mitochondrial DNA mutagenesis, aging and age-related diseases. Ageing Res. Rev. 2017, 33, 89–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisnovsky, S.; Sack, T.; Pagliarini, D.J.; Laposa, R.R.; Kelley, S.O. DNA polymerase θ increases mutational rates in mitochondrial DNA. ACS Chem. Biol. 2018, 13, 900–908. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.D.; Dizdaroglu, M.; Cooke, M.S. Oxidative DNA damage and disease: Induction, repair and significance. Mutat. Res. 2004, 567, 1–61. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [Green Version]
- Azzam, E.I.; Jay-Gerin, J.P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2011, 327, 48–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galano, A.; Tan, D.-X.; Reiter, R.J. Melatonin: A versatile protector against oxidative DNA damage. Molecules 2018, 23, 530. [Google Scholar] [CrossRef] [Green Version]
- Eskandari, A.; Mahmoudzadeh, A.; Shirazi, A.; Esmaely, F.; Carnovale, C.; Cheki, M. Melatonin a promising candidate for DNA double-stranded breaks reduction in patients undergoing abdomen-pelvis computed tomography examinations. Anticancer Agents Med. Chem. 2020, 2, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Esmaely, F.; Mahmoudzadeh, A.; Cheki, M.; Shirazi, A. The radioprotective effect of melatonin against radiation-induced DNA double-strand breaks in radiology. J. Cancer Res. Ther. 2020, 16, 59–63. [Google Scholar]
- Jafarpour, S.M.; Shekarchi, B.; Bagheri, H.; Farhood, B. The Radioprotective effects of melatonin and nanoselenium on DNA double-strand breaks in peripheral lymphocytes caused by I-131. Indian J. Nucl. Med. 2021, 36, 134–139. [Google Scholar]
- Iyama, T.; Wilson, D.M., III. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair 2013, 12, 620–636. [Google Scholar] [CrossRef] [Green Version]
- Mata-Garrido, J.; Tapia, O.; Casafont, I.; Berciano, M.T.; Cuadrado, A.; Lafarga, M. Persistent accumulation of unrepaired DNA damage in rat cortical neurons: Nuclear organization and ChIP-seq analysis of damaged DNA. Acta Neuropathol. Commun. 2018, 6, 68. [Google Scholar] [CrossRef]
- Mebius, R.E.; Kraal, G. Structure and function of the spleen. Nat. Rev. Immunol. 2005, 5, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Goto, S.; Kawakatsu, M.; Urata, Y.; Li, T.-S. Mitochondrial dysfunction, a probable cause of persistent oxidative stress after exposure to ionizing radiation. Free Radic. Res. 2012, 46, 147–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Inaba, Y.; Sogo, Y.; Ito, A.; Bekal, M.; Chida, K.; Moritake, T. Total body irradiation causes a chronic decrease in antioxidant levels. Sci. Rep. 2021, 11, 6716. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Inaba, Y.; Sato, K.; Hirayama, A.; Tsuboi, K.; Okazaki, R.; Chida, K.; Moritake, T. I Dose-dependent decrease in antioxidant capacity of whole blood after irradiation: A novel potential marker for biodosimetry. Sci. Rep. 2018, 8, 7425. [Google Scholar] [CrossRef] [Green Version]
- Malakhova, L.V.; Bezlepkin, V.G.; Antipova, V.N.; Gaziev, A.I. The increase in copy number of mitochondrial DNA in tissues of γirradiated mice. Cell. Mol. Biol. Lett. 2005, 10, 592–603. [Google Scholar]
- Nugent, S.M.; Mothersill, C.E.; Seymour, C.; McClean, B.; Lyng, F.M.; Murphy, J.E. Increased mitochondrial mass in cells with functionally compromised mitochondria after exposure to both direct gamma radiation and bystander factors. Radiat. Res. 2007, 168, 134–142. [Google Scholar] [CrossRef] [Green Version]
- Yamamori, T.; Sasagawa, T.; Ichii, O.; Hiyoshi, M.; Bo, T.; Yasui, H.; Kon, Y.; Inanami, O. Analysis of the mechanism of radiation-induced upregulation of mitochondrial abundance in mouse fibroblasts. J. Radiat. Res. 2017, 58, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Herbers, E.; Kekäläinen, N.J.; Hangas, A.; Pohjoismäki, J.L.; Goffart, S. Tissue specific differences in mitochondrial DNA maintenance and expression. Mitochondrion 2019, 44, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Anisimov, V.N.; Popovich, I.G.; Zabezhinski, M.A.; Anisimov, S.V.; Vesnushkin, G.M.; Vinogradova, I.A. Melatonin as antioxidant, geroprotector and anticarcinogen. Biochim. Biophys. Acta 2006, 1757, 573–589. [Google Scholar] [CrossRef] [Green Version]
- Tan, D.-X.; Manchester, L.C.; Qin, L.; Reiter, R.J. Melatonin: A mitochondrial targeting molecule involving mitochondrial protection and dynamics. Int. J. Mol. Sci. 2016, 17, 2124. [Google Scholar] [CrossRef]
- Slane, B.G.; Aykin-Burns, N.; Smith, B.J.; Kalen, A.L.; Goswami, P.C.; Domann, F.E.; Spitz, D.R. Mutation of succinate dehydrogenase subunit C results in increased, oxidative stress, and genomic instability. Cancer Res. 2006, 66, 7615–7620. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, R.; Marples, B.; Balasubramaniam, M.; Thomas, R.; Tucker, J. Mitochondrial gene expression changes in normal and mitochondrial mutant cells after exposure to ionizing radiation. Radiat. Res. 2010, 173, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Lawless, C.; Greaves, L.; Reeve, A.K.; Turnbull, D.M.; Vincent, A.E. The rise and rise of mitochondrial DNA mutations. Open Biol. 2020, 10, 200061. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signaling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Bedard, K.; Krause, K.H. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, H.; Xing, X.; Tian, M.; Hu, X.; Liu, F.; Feng, J.; Chang, S.; Liu, P.; Zhang, H. Ionizing radiation induces ferroptosis in splenic lymphocytes of mice. Int. J. Radiat. Res. 2021, 19, 99–111. [Google Scholar] [CrossRef]
- Masuyama, M.; Iida, R.; Takatsuka, H.; Yasuda, T.; Matsuki, T. Quantitative change in mitochondrial DNA content in various mouse tissues during aging. Biochim. Biophys. Acta 2005, 1723, 302–308. [Google Scholar] [CrossRef]
- Erol, F.S.; Topsakal, C.; Ozveren, M.F.; Kaplan, M.; Ilhan, N.; Ozercan, I.H.; Yildiz, O.G. Protective effects of melatonin and vitamin E in brain damage due to gamma radiation. Neurosurg. Rev. 2004, 27, 65–69. [Google Scholar] [CrossRef]
- Shirazi, A.R.; Fardid, R.; Mihandoost, E. Protective effect of low dose melatonin on radiation-induced damage to rat liver. J. Biomed. Phys. Eng. 2012, 2, 66–70. [Google Scholar]
- Tahamtan, R.; Shabestani, M.A.; Tahamtani, Y.; Tavassoli, A.R.; Akmali, M.; Mosleh-Shirazi, M.A.; Naghizadeh, M.M.; Ghasemi, D.; Tahamtan, R.; Keshavarz, M.; et al. Radioprotective effect of melatonin on radiation-induced lung injury and lipid peroxidation in rats. Cell J. 2015, 17, 111–120. [Google Scholar]
- Limoli, L.; Giedzinski, E.; Baure, J.; Rola, R.; Fike, J.R. Redox changes induced in hippocampal precursor cells by heavy ion irradiation. Radiat. Environ. Biophys. 2007, 46, 167–172. [Google Scholar] [CrossRef]

| Locus | Primer, Probes | Accession Number | 5′→3′ Sequence | Size |
|---|---|---|---|---|
| mtDNA | NC_005089.1 | Primers for LA-QPCR | ||
| for | GCCAGCCTGACCCATAGCCATAATAT | |||
| rev | GAGAGATTTTATGGGTGTAATGCGG | 10.9 kb | ||
| nDNA | for | NC_000073.7 X14061.1 | TTGAGACTGTGATTGGCAATGCCT | |
| rev | CCTTTAATGCCCATCCCGGACT | 8.7 kb | ||
| mtDNA | for | NC_005089.1 | CCCAGCTACTACCATCATTCAAGT | |
| rev | GATGGTTTGGGAGATTGGTTGATGT | 117 bp | ||
| nDNA | for | NC_000071.7 NM_007393.5 | CTGCCTGACGGCCAGG | |
| rev | GGAAAAGAGCCTCAGGGCAT | 110 bp | ||
| ND4 | NC_005089.1 | Primers for quantitative analysis of mtDNA/nDNA | ||
| for | ATTATTATTACCCGATGAGGGAACC | |||
| rev | ATTAAGATGAGGGCAATTAGCAGT | |||
| probe | FAM-ACGCCTAAACGCAGGGATTTATTTCCTA-BHQ1 | 115 bp | ||
| GAPDH | for | NC_000072.7 NM_001289726.1 | GTGAGGGAGATGCTCAGTGT | |
| rev | CTGGCATTGCTCTCAATGAC | |||
| probe | ROX-TAAGAAACCCTGGACCACCCACCCC-BHQ2 | 214 bp | ||
| ND3 | NC_005089.1 | Primers for mtDNA mutant copies | ||
| for | AGCTCTCCATTTATTGATGAGG | |||
| rev | GAGGTTGAAGAAGGTAGATGGC | 534 bp |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullaev, S.A.; Glukhov, S.I.; Gaziev, A.I. Radioprotective and Radiomitigative Effects of Melatonin in Tissues with Different Proliferative Activity. Antioxidants 2021, 10, 1885. https://doi.org/10.3390/antiox10121885
Abdullaev SA, Glukhov SI, Gaziev AI. Radioprotective and Radiomitigative Effects of Melatonin in Tissues with Different Proliferative Activity. Antioxidants. 2021; 10(12):1885. https://doi.org/10.3390/antiox10121885
Chicago/Turabian StyleAbdullaev, Serazhutdin A., Sergey I. Glukhov, and Azhub I. Gaziev. 2021. "Radioprotective and Radiomitigative Effects of Melatonin in Tissues with Different Proliferative Activity" Antioxidants 10, no. 12: 1885. https://doi.org/10.3390/antiox10121885
APA StyleAbdullaev, S. A., Glukhov, S. I., & Gaziev, A. I. (2021). Radioprotective and Radiomitigative Effects of Melatonin in Tissues with Different Proliferative Activity. Antioxidants, 10(12), 1885. https://doi.org/10.3390/antiox10121885






