1. Introduction
Soil salinity is a major abiotic factor, influencing over 1000 million ha of land in several parts of the world, and is one of the main current constraints in developing irrigated agriculture [
1,
2]. An excess of salt causes several physiological and biochemical problems such as ion imbalance, osmotic stress, ion toxicity and oxidative stress [
3], thus reducing the growth and productivity of numerous crops [
4,
5]. In vitro culture, as an induced somaclonal variation, proved to be a promising and effective method to obtain salt-tolerant genotypes. The selection of salt-tolerant cell lines has been reported for numerous crops [
6,
7]. However, the results are not always predictable. The tolerance of cell lines is often transient, rather than a stable and heritable adaptation [
8,
9]. Therefore, the offspring of somaclonal variants should be further screened and selected under field conditions to confirm the stability of the salt tolerance obtained [
10,
11]. Somaclonal variation can be evaluated not only phenotypically, but also through the number of chromosomes, chromosome structure, proteins or direct DNA analysis [
12,
13,
14].
The investigation of salt tolerance during different growth stages is important for elucidating saline limits at each developmental phase [
15]. Salt stress reduced and delayed the germination of solanaceous vegetables such as tomatoes [
16] and eggplant [
17,
18]. The reduced seed germination induced by salinity could be caused by the induction of dormancy, by osmotic stress or by specific ion toxicity [
19].
Salts influence plant growth by increasing the osmotic pressure in the soil and interfering with plant nutrition [
20]. Carbohydrates, which are required for cell growth, are supplied mainly through the photosynthesis process, and the photosynthesis rates are usually lower in plants exposed to salt and especially to NaCl. The accumulation of compatible solutes, such as proline and polyols, in the cytoplasm is required to compensate for the decrease in water potential occurring in the vacuole due to ion accumulation in that compartment [
21]. Commonly used biochemical markers of abiotic stress, including salinity, are ROS and lipid peroxidation products [
22].
Abiotic stress contributes to the production of reactive oxygen species (ROS) in plants. ROS are continuously produced in chloroplasts, mitochondria and peroxisomes in higher plants [
23]. However, H
2O
2 produced in peroxisomes and chloroplasts diffuses to the cytosol and is converted to hydroxyl radicals by the Fenton reaction [
24]. The overproduction of ROS (such as superoxide (O
2−), singlet oxygen (O
2), hydrogen peroxide (H
2O
2) and hydroxyl radicals (OH
−)) in plant cells under stress contributes to the damage of cellular components, including DNA, proteins and membrane lipids, and leads to the destruction of photosynthetic pigments [
25,
26]. Abiotic stress may induce oxidative stress in plants and generates cellular adaptive responses such as the induction of stress proteins and acceleration of ROS-scavenging systems [
3]. According to [
27], to scavenge reactive oxygen species, plants have evolved both enzymatic and non-enzymatic defense systems. The enzymatic antioxidants including superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), guaiacol peroxidase (POD) and glutathione reductase (GR) [
28]. The authors in [
29] showed that ROS is converted by SOD into H
2O
2, which is further scavenged by CAT and various peroxidases (APX, POD) to H
2O.
On the other hand, salinity tolerance usually correlates with high amounts of compatible solutes and efficient compartmentation, as shown in a large set of studies [
30]. Both salt tolerance and the sensitivity of a specific crop depend on its ability to extract water and nutrients from saline soils and to avoid excessive tissue accumulation of salt ions [
31,
32,
33]. The ion toxicities have diverse consequences resulting in ionic imbalance, i.e., in terms of the uptake competition of Na
+ with K
+, Ca
2+ and Mg
2+, and may reduce the availability of the beneficial nutrients [
34].
Salinity also affects photosynthesis mainly through a reduction in leaf area, chlorophyll content and stomatal conductance, and to a lesser extent through a decrease in photosystem II efficiency [
35].
Additionally, in susceptible tomato cultivars [
36] chlorophyll fluorescence measurements showed a considerable decrease in the efficiency of PSII and ETR under increasing salt stress. The absorbed energy non-utilized in the photochemical pathway was dissipated as heat and this was confirmed by the concomitant increase in NPQ. Such an increase was suggested to cause a downregulation of PSII in order to avoid an over-reduction in Q
A.Plants develop several strategies to overcome NaCl stress, including the plant growth-promoting bacteria (PGPB) living in the rhizosphere and rhizoplane of the plants [
37,
38,
39]. The PGPB involve several beneficial bacteria which contribute efficiently to increasing plant salt tolerance by decreasing the ethylene level through hydrolysis of 1-aminocyclopropane-1-carboxylic acid (ACC) by the ACC deaminase enzyme and convert it into ammonia and α-ketobutyrate. ACC is the direct precursor of the hormone ethylene in plants [
40,
41]. Consequently, the PGPR with their ACC deaminase activity lower the abiotic stress engendered by ethylene synthesis and its impact on plants [
42,
43,
44,
45].
Eggplant (
Solanum melongena L.) is one of the most popular and economically important vegetable crops worldwide [
46]. It is considered to be moderately sensitive to salinity [
47]. Therefore, improving the salt tolerance of the current existing eggplant germplasm seems to be of immense importance in order to increase crop productivity. In several crops, spontaneous or induced somaclonal variation induced during the in vitro culture of plant cells occurs as a promising strategy for obtaining salt-tolerant mutants [
48,
49,
50]. However, the assessment of the genetic stability of salt-tolerant selected somaclones is highly required [
51,
52,
53]. Although successful eggplant somatic embryogenesis [
53,
54] and shoot organogenesis [
55,
56] was reported, the first attempt to produce and select a salt-tolerant eggplant cell line was only achieved recently.
In the present work, we analyzed the progeny of eggplant somaclonal variants, which had been obtained previously by a stepwise increasing salinity (40, 80, 120 or 160 mM NaCl) in vitro selection and screened for salt tolerance [
57]. These somaclones were regenerated on the calli lines that could tolerate up to 120 mM NaCl. From 32 plants tested in vitro, four plants with a superior number of leaves and root length were selected for further in vitro and in vivo evaluation.
The present study aimed to identify possible stable salt-tolerant genotypes among the progeny of somaclonal variants and characterize them through agronomic, physiological and biochemical parameters.
3. Discussion
Salinity is still considered the most important abiotic stress threatening agriculture [
58]. Somaclonal variation, a chromosomal/epigenetic rearrangement, can be used in breeding for salt tolerance. A phenotypic evaluation based on morphological, physiological and metabolic traits is indispensable to confirm stable mutations. Therefore, we evaluated the stability of salt tolerance in the progeny of four selected eggplant somaclonal variants (‘R18’, ‘R19’, ‘R23’ and ‘R30’) by assessing a number of agronomic, physiological and biochemical parameters at different developmental stages.
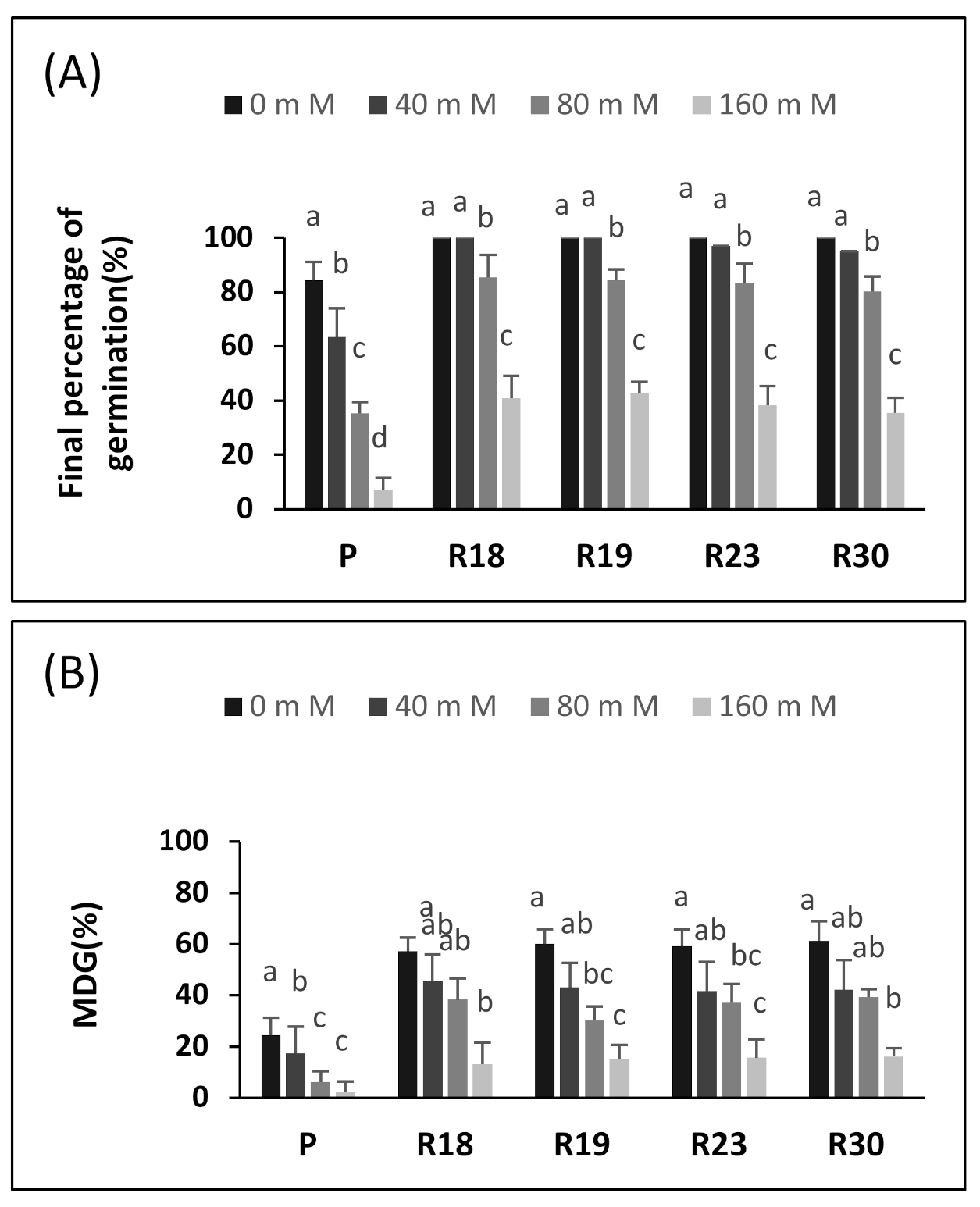
The germination of seeds is the most vulnerable stage of development; therefore, the germinating seeds of most plant species do not tolerate salt. This has already been demonstrated by numerous vegetables such as lettuce [
59] and beans [
60]. The germination parameters (GP, MGT and MDG) were clearly affected by salinity. The seeds derived from the regenerants germinated 1–2 days slower than the control seeds, depending on the line (
Figure 1). The salt limited water uptake by the seeds due to the lower osmotic potential of the germination medium, which contributed to an osmotically enforced “dormancy”. We propose the following explanation. Under salt stress, seeds develop an effective defense mechanism that prevents germination until more optimal conditions arise, such as rain in field conditions [
61].
It should be noted that in our experiment, the germination of the control seeds decreased significantly at 80 mM NaCl. The same finding was reported for eggplant, in which a strong inhibition of germination at 100 mM NaCl was observed [
62]. Moreover, the germination of susceptible tomato cultivars decreased strongly at 80 mM and drastically at 190 mM NaCl [
16]. In addition, previous work demonstrated that the germination of sensitive lettuce varieties was substantially decreased at 100 and 150 mM of NaCl.
Similar results, describing a drastic decrease in seed germination, were found in tomato at 100 mM NaCl and in red beet, bell pepper, melon and broccoli at 150 mM NaCl [
15]. The reduced MDT and MDG of our control seeds at 40 mM NaCl confirmed the salt sensitivity of eggplant. For comparison, decreases in MGT and MDG were described in Atriplex patula at 34 mM NaCl [
63] and in Phaseolus at 120 mM NaCl [
61]. According to [
16], tomato seeds showed a crucial requirement of 50% more days to ensure germination at 80 mM NaCl than in a free salt medium and of almost 100% more days at a higher salt concentration (190 mM).
The salt sensitivity of eggplant during germination was confirmed at further growth stages. The decrease in growth parameters (shoot height, number of leaves, FW and DW) was more pronounced for ‘P’ compared to ‘R18’, ‘R19’, ‘R23’ and ‘R30’ (
Table 1 and
Table 2). Indeed, refs. [
17,
62] highlighted a growth reduction in eggplant under saline conditions at 100 mM NaCl. Exposing plants to saline conditions significantly reduced water and nutrient uptake during the initial phase. This was followed by excessive Na+ accumulation, leading to nutritional imbalances and water deficit during a second phase [
30,
64,
65]. Ionic toxicity and osmotic adverse effects were associated during the onset and continued persistence of salt stress [
18,
66,
67].
Excessive salinity causes stomata closure, which limits CO
2 uptake, contributing to the dysfunction of the photosynthetic apparatus and the decrease in growth [
30,
68,
69]. The effect of stomatal closure on the efficiency of photosynthesis is proportional to the amplitude of the reduction in leaf CO
2 partial pressure (reviewed by [
70]). Moreover, the significant amount of accumulated NaCl in chloroplasts induces the deterioration of PSII, which contributes to photodamage and affects photosynthetic efficiency [
71,
72,
73,
74].
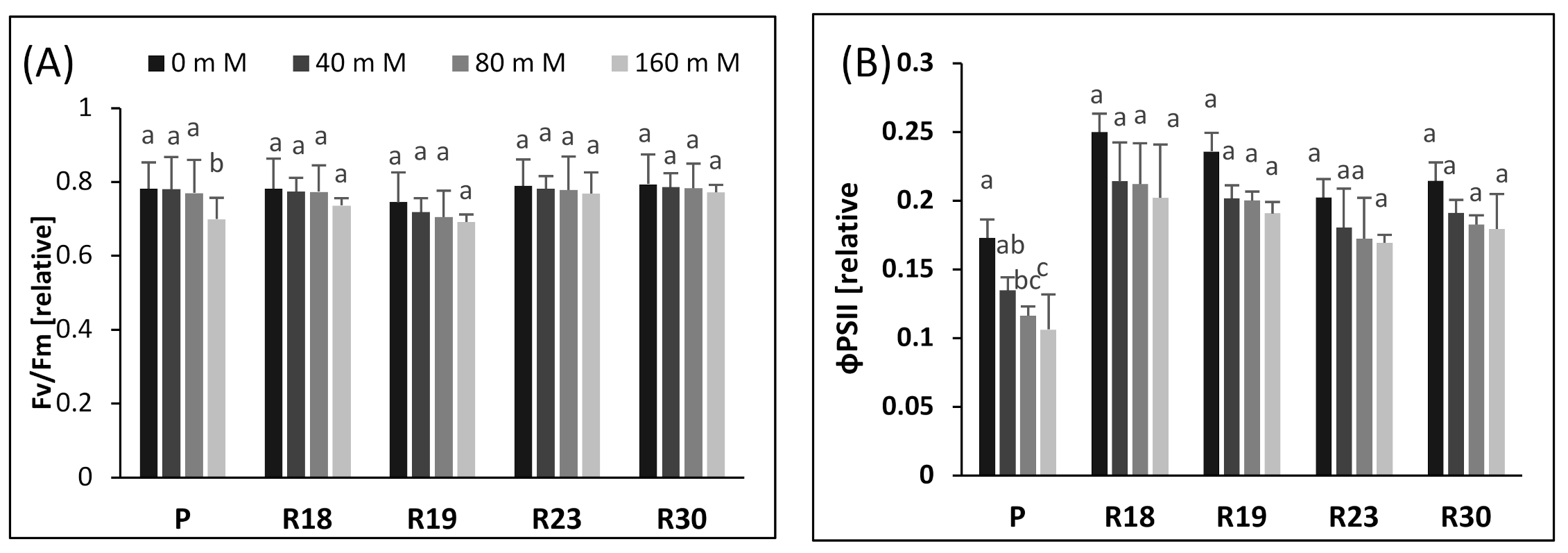
To verify the impact of increasing salinity on the photochemistry of PSII, we performed Chl a fluorescence measurement in salt-tolerant eggplant mutants selected in vitro. Our study shows that increasing salinity has no significant effect on Fv/Fm in all regenerants after 30 days of salt stress. Similar findings have been reported for salt-tolerant tomato [
36] and wheat [
75]. The small reduction in Fv/Fm could be caused by the downregulation of PSII rather than photodamage to PSII [
76,
77].
The study of ΦPSII reflects PSII activity and adaptation [
78]. It is clear that the decrease in growth is strongly related to the reduction in ΦPSII under saline conditions. Increasing salt concentration generated a significant decrease in control plants but a non-significant decrease in regenerants, due to the disruption of electron transport via PSII. These results are consistent with previous studies on salt-tolerant species reported by [
79] in oaks, [
36] in tomatoes, [
80] in soybeans, [
73] in roquettet and [
74] in gingko. It is clear that the reduction in electron transport rate was less pronounced in the regenerants, demonstrating that they tolerate higher salt content than the parental control.
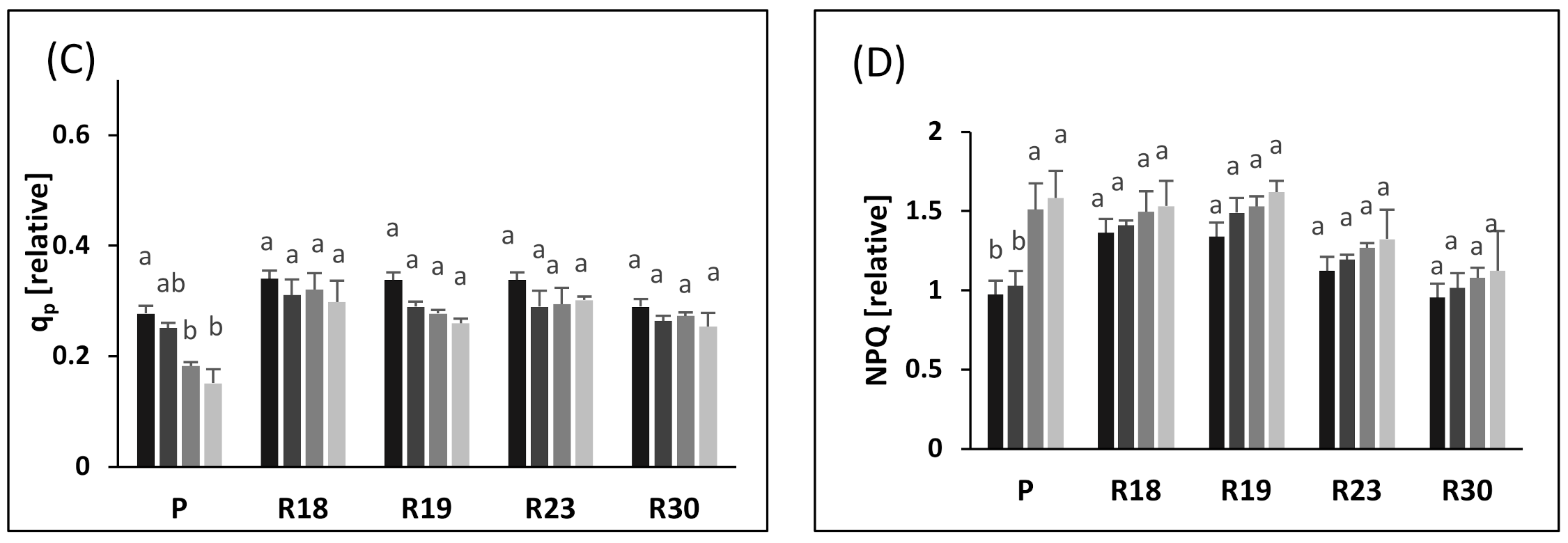
Similar to ΦPSII, increasing salt stress also contributed to lower qp in ‘P’, ‘R18’, ‘R19’, ‘R23’ and ‘R30’. However, the qp decrease was less pronounced in regenerants than in the parental control. As stated by [
81], qp reflects the capacity of PSII in reducing the primary electron acceptor QA under salt conditions and indicates the number of photons used by photochemical reactions per number of assimilated photons. Our results show that ‘R18’, ‘R19’, ‘R23’ and ‘R30’ showed higher PSII reaction center activity combined with better efficiency in converting light energy into chemical energy than ‘P’
As highlighted by [
77], photochemical quenching plays a major role in preserving the photosynthetic apparatus via shifting electrons to O
2 under abiotic stress. Stomata closure contributes to a limitation of transpiration rates [
36] and a reduction in assimilation efficiency [
82] with increasing salinity. Ribulose bisphosphate carboxylase/oxygenase (Rubisco) is the main plant enzyme involved in the first phase of the C3 photosynthetic carbon reduction cycle [
83]. During stomata closure, oxygenation of ribulose-1,5 bisphosphate in C3 plants replaces carboxylation [
84,
85,
86,
87].
Photosystem II is very vulnerable to salt stress. The absence of a significant effect of salinity on ΦPSII in all regenerated plants can be explained by the presence of an effective electron sink. Interestingly, the plants manage to maintain an effective conversion system of excitation energy via the downregulation of PSII RCs under salt condition [
88]. Photoreduction could occur on the acceptor side of PSI in the Mehler reaction [
89], which generates a pH gradient of the thylakoid membranes and produces a thermal loss of abundant excitation energy [
90].
The decline in the non-photochemical energy loss is likely the main mechanism used to cope with photodamage [
91]. The parental control exhibited a prominent rise in NPQ after 30 days of imposition of salt stress.
It is clear that the increase in the NPQ of ‘P’ contributed greatly to counteract the decrease in Fv/Fm. The increased NPQ dissipates some of the abundant excitation energy at the expense of photochemical utilization [
36,
92], leading to a downregulation of PSII to avoid over-reduction in the primary electron acceptor QA. This reaction indicates a protective process to prevent photodamage in the photosynthetic machinery [
76].
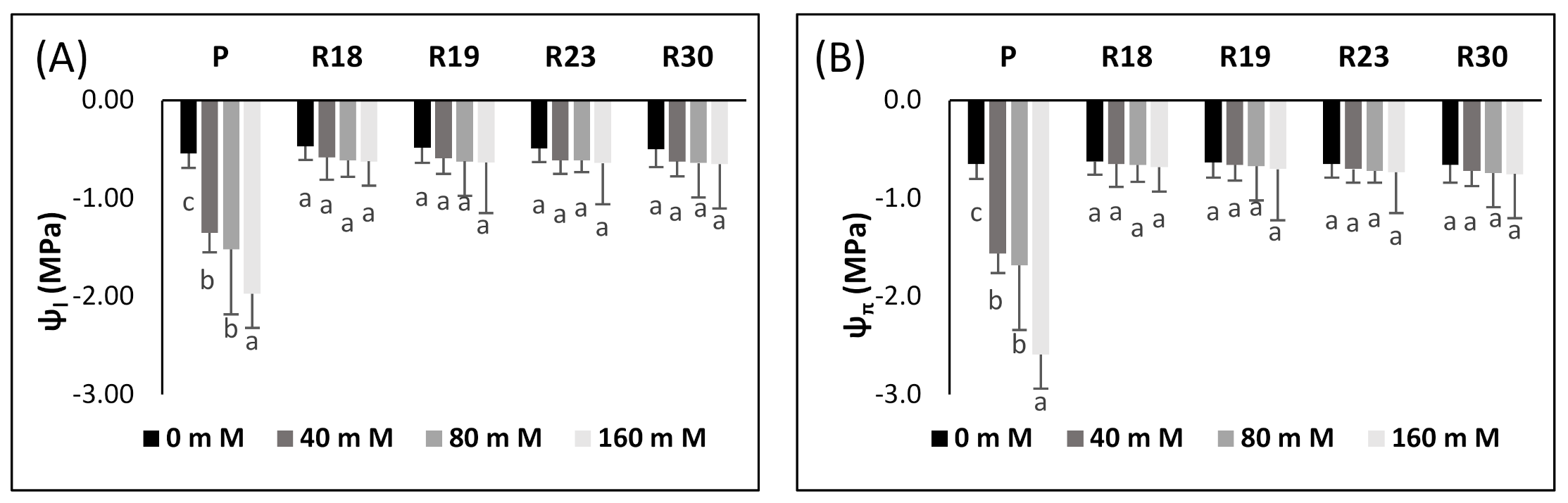
Under salinity conditions, foliar water potential and osmotic potential/EC changes are interrelated. It is obvious that the capacity of plants in maintaining a stable water potential difference between leaves and nutrient solution is of immense importance in order to impede desiccation. Salt stress decreased leaf turgor and atmospheric vapor pressure [
93].
In [
5], it was stated that the reduction in foliar water potential is associated with a linear decline in osmotic potential, thus leading to a fast diminishment in turgor potential.
The regenerated plant succeeded in maintaining ψ
l and ψ
π at relatively stable levels under increasing salinity levels, while the parental control failed to do so. It is evident that the decrease in ψ
l in ‘P’ occurred simultaneously with the reduction in ψ
π, thus keeping the same turgor status as the control. The aggravated decline in foliar ψ
π in parental controls might be explained by a substantial accumulation of Na
+ and Cl
− (
Table 7), considerably lowering water uptake, as reflected by the reduced TWC (
Table 4), which contributed to the amplification of internal vacuole concentration.
Many previous reports stated that salt stress reduced shoot and root growth, fresh and dry weight, plant height and yield characteristics [
94,
95].
The authors in [
30] emphasized that a decline in growth appears in two steps: a fast reaction to osmotic stress is followed by a slower reaction induced by Na accumulation in foliar tissue. In our experiment, the regenerants exhibited higher salt tolerance than the parental control. This was evident through higher shoot biomass in ‘R18’, ‘R19’, ‘R23’ and ‘R30’ when compared to ‘P’. Our results agree with earlier reports indicating that the ability of regenerants in maintaining stable shoot biomass might be due to their capacity to cope with salinity at the cellular level through a mechanism of exclusion/avoidance [
52,
96,
97]. While the excessive accumulation of salt in old foliar cell tissue continues, an attained toxic level decreases their ability to overcome salt influx (first phase). Then, an ionic specific response of the plant to salinity starts (second phase) [
30].
The foliar tissue water content (TWC) represents an efficient stress indicator. All regenerated plants succeeded in maintaining higher and quite stable TWC compared to the parental control regardless of the salt concentration. Our findings indicate the ability of ‘R18’, ‘R19’, ‘R23’ and ‘R30’ in maintaining enhanced turgor and cell expansion using a performant plasmolytic process in order to overcome salt stress [
97,
98]. To cope with salinity, plants adopt an osmoregulatory strategy through the accumulation of organic low molecular weight solutes, enabling plants to maintain their plant–water status and positive carbon balance [
18,
99,
100,
101,
102].
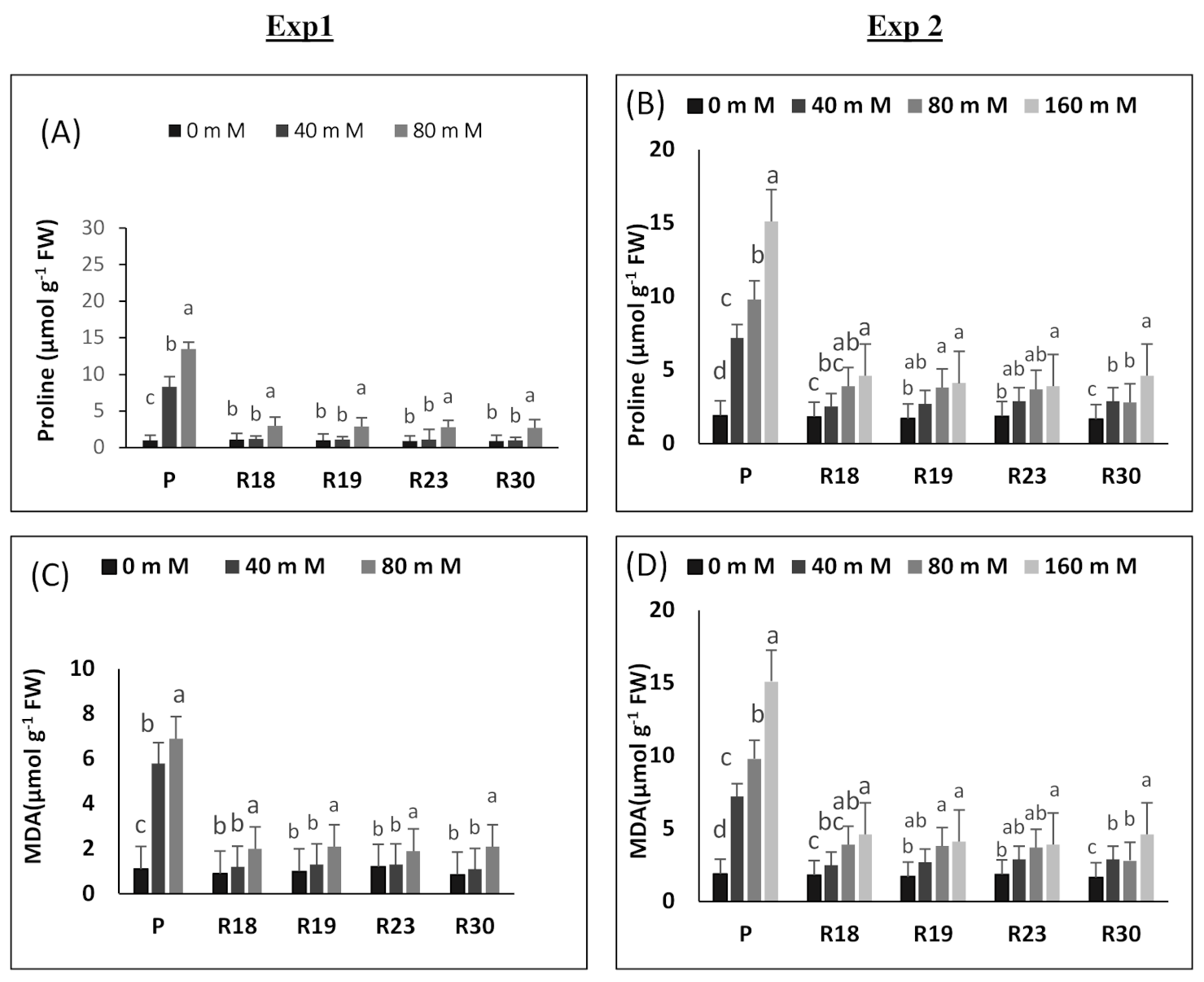
Proline catabolism is upregulated under increasing salinity concentrations by impeding its oxidation, which contributes to its substantial accumulation. Our results show that proline rose under salinity conditions in both parental control and regenerated plants. However, proline accumulation increase was more pronounced in ‘P’ than in ‘R18’, ‘R19’, ‘R23’ and ‘R30’. In the current work, it is noteworthy that the enhancement of proline accumulation seems to be induced by the metabolic damage generated by salt stress rather than appearing as a tolerance feature [
103,
104].
Saline conditions contribute to oxidative stress [
105]. A high accumulation of reactive oxygen species leads to oxidative damage and engenders the membrane lipid peroxidation, thus affecting membrane fluidity and selectivity [
101,
106]. Membranes represent the most vulnerable cell organelles to stress-induced damage, and the extent of damage is considered an efficient tool for evaluating salt tolerance [
105,
107,
108]. An apparent difference was detected between parental controls and regenerated plants in terms of lipid peroxidation under salt stress. ‘R18’, ‘R19’, ‘R23’ and ‘R30’ have the capacity of keeping lower levels of membrane peroxidation, indicating their superior capability for cellular protection to overcome the oxidative impairment caused by applied salt stress [
109,
110,
111].
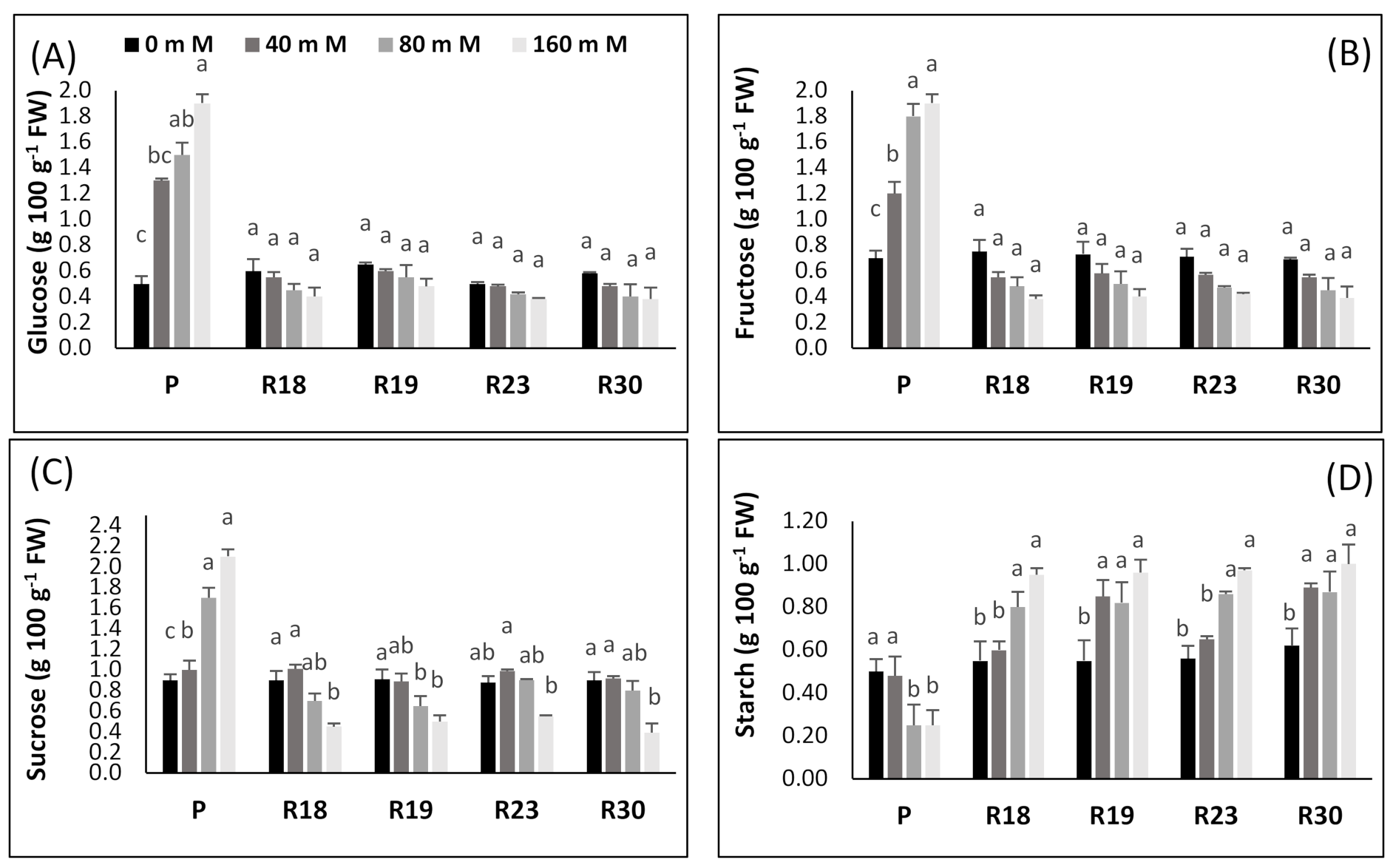
The increase in carbohydrates as a key factor involved in osmotic adjustment was stated previously by many authors [
101,
112]. These organic solutes provide the plant the ability to alleviate desiccation through improving its effectiveness in maintaining an osmotic balance in cell tissue [
113]. Carbohydrates also function as the buffer of cellular redox potential, preventing cell dehydration by preserving the membrane and protein structural stability and providing enough energy sources under severe stress [
114]. To overcome salinity-induced damage, plants adopt a process of compatible solutes accumulation, including amino acids, sugars and/or other composites [
115].
Our results indicate that regenerated plants exhibited the ability to overcome a long and increasing salt stress, while they were successful in maintaining better growth by maintaining a higher chlorophyll content under salt stress conditions. In contrast, a high salinity level led to a chlorophyll content reduction in the parental control line, which contributed to an early leaf senescence and plant withering.
As emphasized by [
116], salinity contributes to K deficiency, thus reducing chlorophyll content and affecting photosynthetic apparatus functioning. Furthermore, salt-induced oxidative stress can also decrease the chlorophyll content [
117,
118]. Our experiments showed that the significant decline in the chlorophyll content was only limited to parental control, whereas no significant change was found in all the regenerated plants.
This is in line with earlier reports [
52,
119,
120,
121] where rising salinity acted gravely on many processes; for example, water content, transpiration rate and leaf stomatal conductance in several crops. The negative effect of salt stress generated photosynthetic machinery damage through chloroplast dysfunction and chlorophyll depletion.
As stated by [
122], the availability of specific K
+ and Ca
2+ contents is crucially needed for preserving the integrity and functioning of cell membranes. The rise of Na
+ and Cl
− content is considered as an osmoregulation process adopted by plants under salinity condition, while a decline in K
+ and Ca
2+ occurred in stressed plants [
62].
In the present work, rising salinity level engendered an increase in foliar Na and Cl concentration associated with a reduction in K
+, Ca
2+, Mg
2+ and NO
3− in the parental control and in all regenerated plants. However, all the regenerated plants exhibited reduced leaf concentration of Na
+ and Cl
− and succeeded in keeping a more enhanced accumulation of K
+, Ca
2+ and Mg
2+ compared to the parental control. This is consistent with earlier reports obtained by [
17,
18] for salt-tolerant eggplant lines.
It is worth noting that the parental control line showed an accentuated reduction in K content. K deficiency is mainly due to the strong competition between the absorbed NaCl and specifically K uptake [
18,
66]. Therefore, the rise in Na/K and Na/Ca ratios was more pronounced in ‘P’ than in ‘R18’, ‘R19’, ‘R23’ and ‘R30’. Consequently, salt tolerance or sensitivity in plants is deeply related to Na/K and Na/Ca ratios. As emphasized by [
123,
124], tolerant lines exhibit lower Na/K and Na/Ca ratios.
To accomplish performant growth via normal cellular functioning, a reduced cytosol Na/K ratio is of immense importance in all plants. The imposition of salt stress and its continuity contributes to the inhibition of the K-specific transporters by Na, while competing with K uptake. Therefore, an excessive toxic concentration of Na is combined with a K deficiency [
125,
126]. Other salt-tolerant species are provided with an efficient osmotic potential regulation mechanism via preventing Na
+ and Cl
− uptake and enhancing the absorption of other crucially required ions, such as K
+ [
123].
Moreover, to overcome the substantial amount of foliar Na, many glycophytes are provided with the mechanism of excluding Na and Cl
- from the cytosol through vacuole compartmentalization [
127,
128].
The high salt tolerance in the regenerants is closely related to a lower amount of accumulated salt ions in leaves. This might be explained by a more efficient mechanism of salt exclusion in ‘R18’, ‘R19’, ‘R23’ and ‘R30’ leaves (more tolerant) compared to ‘P’ (susceptible) [
18,
128].
A substantial salt ion accumulation contributes to the alteration of osmoregulation, the disturbance of membrane potential equilibrium and the loss of turgor [
129]. Consequently, cell division and expansion are affected, thus causing deterioration in the plant biomass production process.
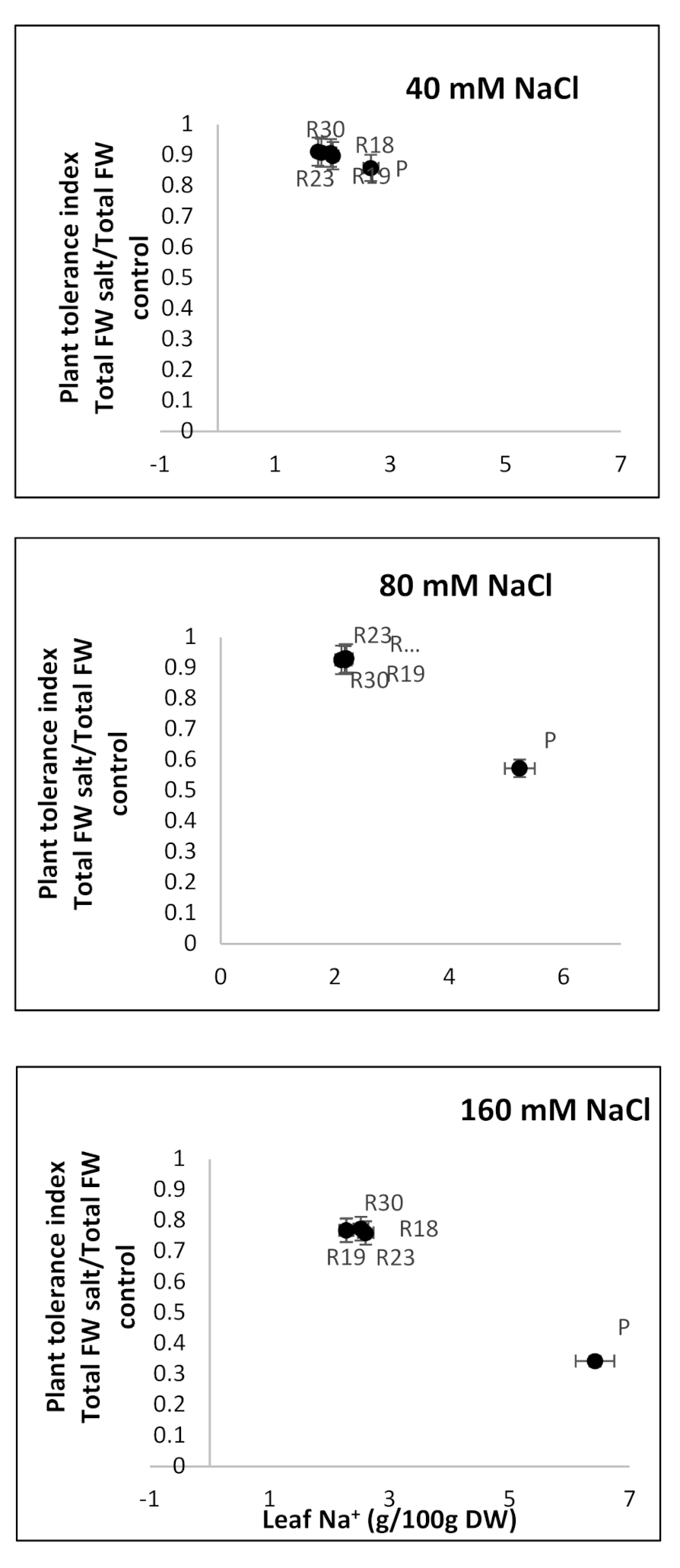
A positive correlation between PTI and leaf Na
+ concentration was evident. This result agrees with earlier findings. The authors of [
130], working on Arabidopsis, and of [
131], working on potato cultivars, emphasized that plants could be provided with mechanisms of Na
+ tissue tolerance, such as the high accumulation of osmoregulants and intracellular compartmentation. The same hypothesis was adopted by [
30] as an alternative tolerance process in glycophytes and obviously ‘R18’, ‘R19’, ‘R23’ and ‘R30’ acted by following the same strategy.
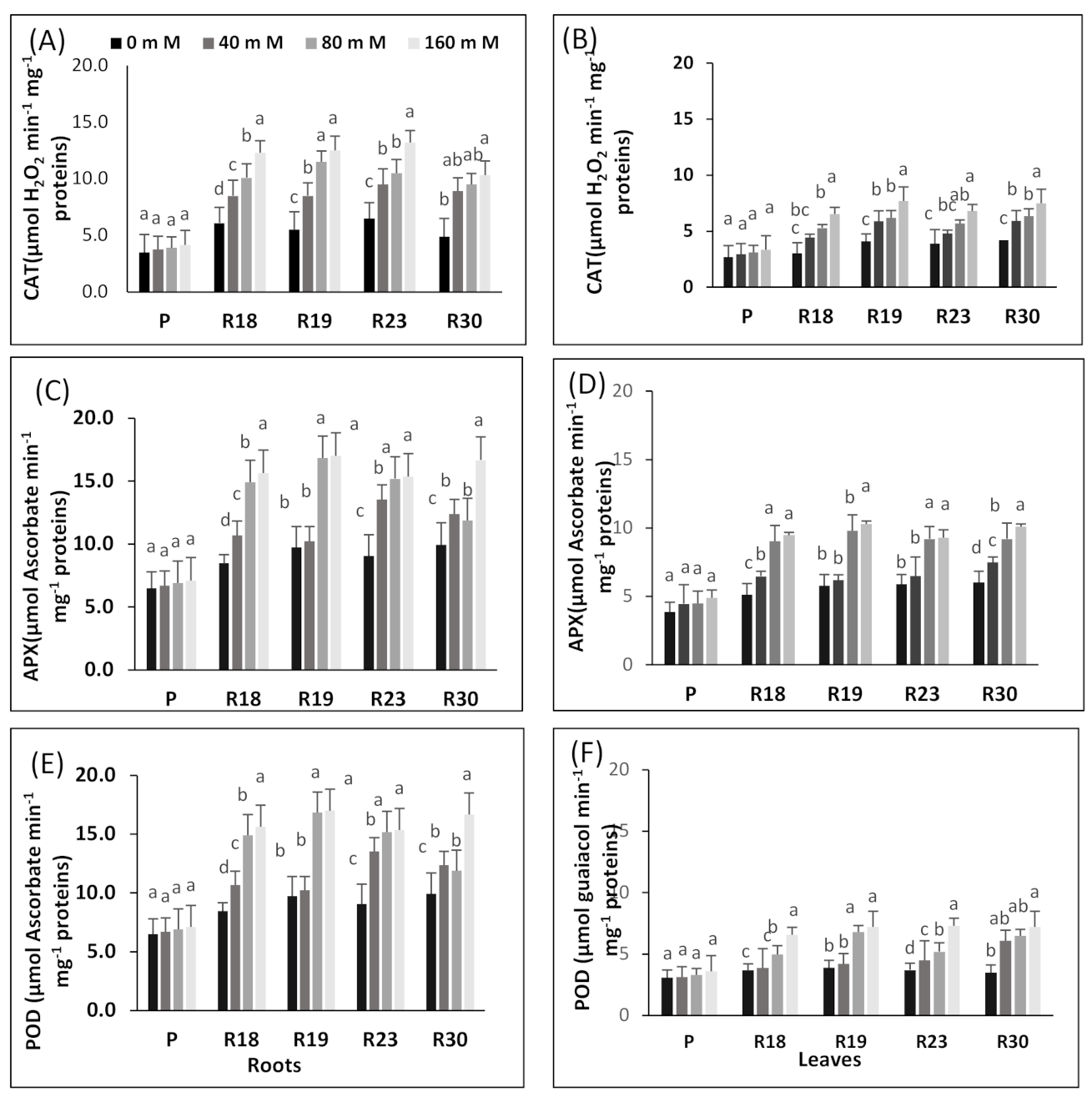
Under increasing salinity, the antioxidative machinery is upregulated in the regenerated plants. The effective protection developed by ‘R18’, ‘R19’, ‘R23’ and ‘R30’ against ROS in eggplant may be due to an efficient antioxidative system. These results are in line with many earlier reports [
132,
133].
As emphasized by [
134,
135], the strong oxidant H
2O
2 in peroxisomes is scavenged by CAT, and H
2O is transformed into water and molecular O
2. The enhanced activities of CAT and APX effectively decreased the rising amount of H
2O
2, thus leading to the amelioration of the cell membrane integrity and stability. In the parental control and the regenerated plants, CAT activity rose proportionally with the salt stress level. In ‘R18’, ‘R19’, ‘R23’ and ‘R30’, the most elevated salt concentration (160 mM NaCl) generated the highest CAT activity, thus leading to improved protection against ROS in all the regenerated plants. This result agrees with previous reports, stating that many salt-tolerant species exhibited higher CAT activity, such as maize [
136], sesame [
137], potato [
138], melon [
139] and wheat [
140]. Several earlier authors reported that salinity increased APX activity in many salt-tolerant cultivars [
106,
137,
141].
The susceptibility of the parental control could be due to the suppression of APX activity induced by rising salinity concentrations. This result is in concordance with previous reports dealing with Triticum aestivum [
106], potato [
138] and durum wheat [
140]. APX is required to keep an acceptable intracellular amount of H
2O
2 in higher plants [
142,
143].
Our findings show that POD enzyme activities in parental control and regenerated plants were more enhanced with increasing salinity levels and were more elevated in ‘R18’, ‘R19’, ‘R23’ and ‘R30’ (salt-tolerant) than in ‘P’ (salt-sensitive). It is noteworthy that several previous studies stated that POD enzymes are associated with salt tolerance in melon [
144], soybean [
145] and wheat [
140]. Consequently, POD enzymes contributed strongly to scavenging H
2O
2 in eggplant under salinity conditions. POD, which acts at both the intracellular and extracellular level, is an effective enzyme involved in the elimination of H
2O
2 [
146].
The salt tolerance response of the regenerated plants could be engendered by its enhanced capacity in downregulating ROS production or by its improved ability to overcome ROS compared to the parental control. Our findings indicate that antioxidant enzyme machinery may have a strong contribution in scavenging H2O2 in eggplant.
This is in line with earlier results reported by [
147] working on tomatoes, [
132] working on sugar beets, [
85] working on
Populus euphratica, [
148] working on rice and [
138] working on
Plantago.
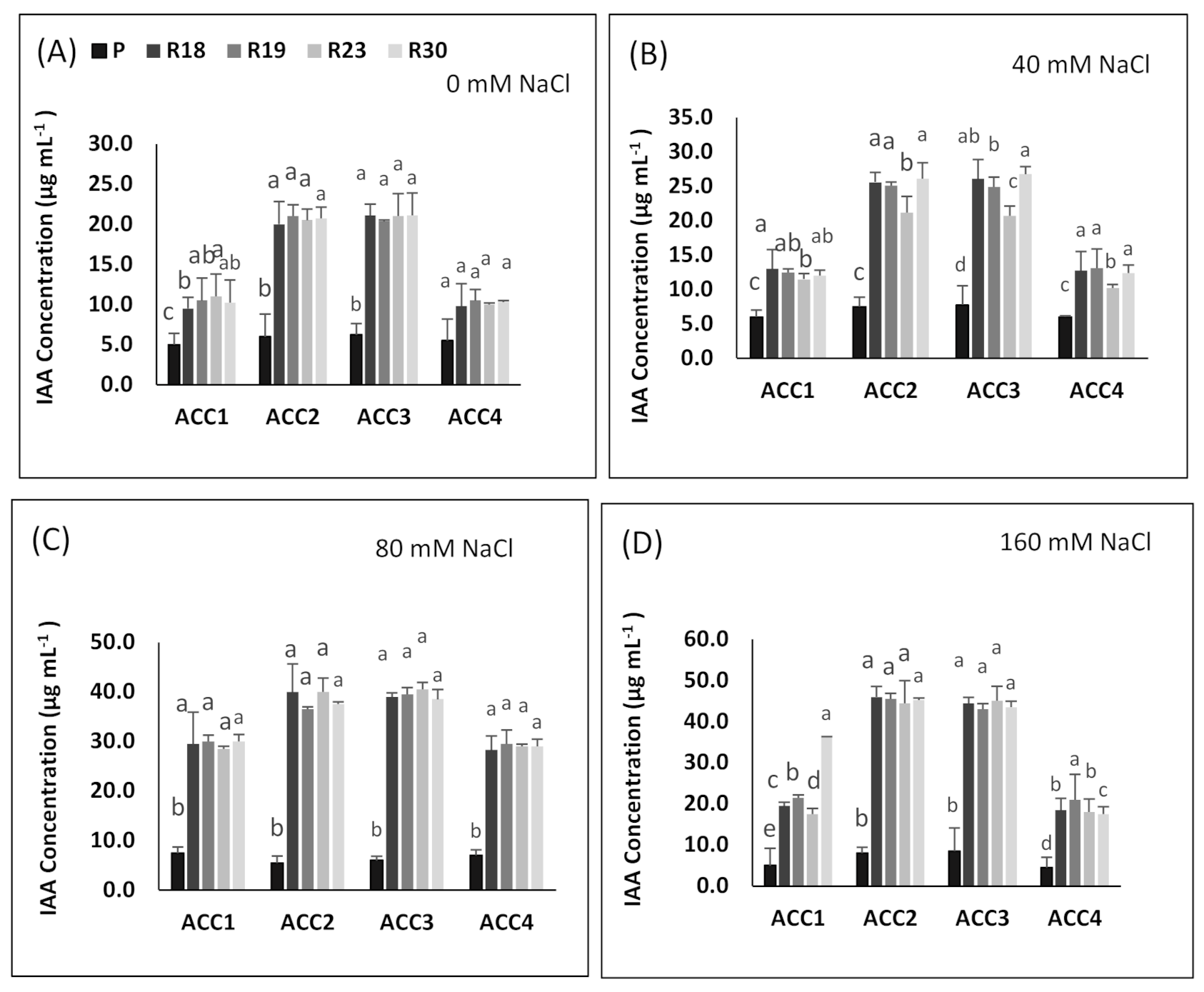
ACC deaminase activity exhibited by PGPR has been proved to play a key role in improving growth and stress tolerance in plants under stressed conditions [
149,
150]. ACC deaminase, as a microbial enzyme, contributes to dissociating stress-induced ACC into ammonia and α-ketobutyrate, which produces ethylene that causes serious damage to the physiology, growth and development of plants [
151,
152]. In the present work, the salt tolerance response of the regenerated plants was associated with higher ACC deaminase activity in all bacterial strains and specifically in ‘ACC2’ and ‘ACC3’ when compared to the parental line showing lower values. The results of the current study agree with previous published works, reporting that bacteria provided by ACC deaminase activity are able to counteract the toxic impact of salt stress [
153,
154].
Under salt stress, PGPB contribute in enhancing IAA additional production, which may be useful in stimulating root growth, counteracting the inhibition impact of salinity on root and shoot growth and ameliorating plant physiological features [
155].
In our study, all regenerants exhibited higher IAA accumulation in all isolates and specifically in ‘ACC2’ and ‘ACC3’ compared to the parental line in response to increasing salinity, which might be explained by their superior capacity to tolerate salt stress. This is consistent with previous work reported by [
156,
157,
158].
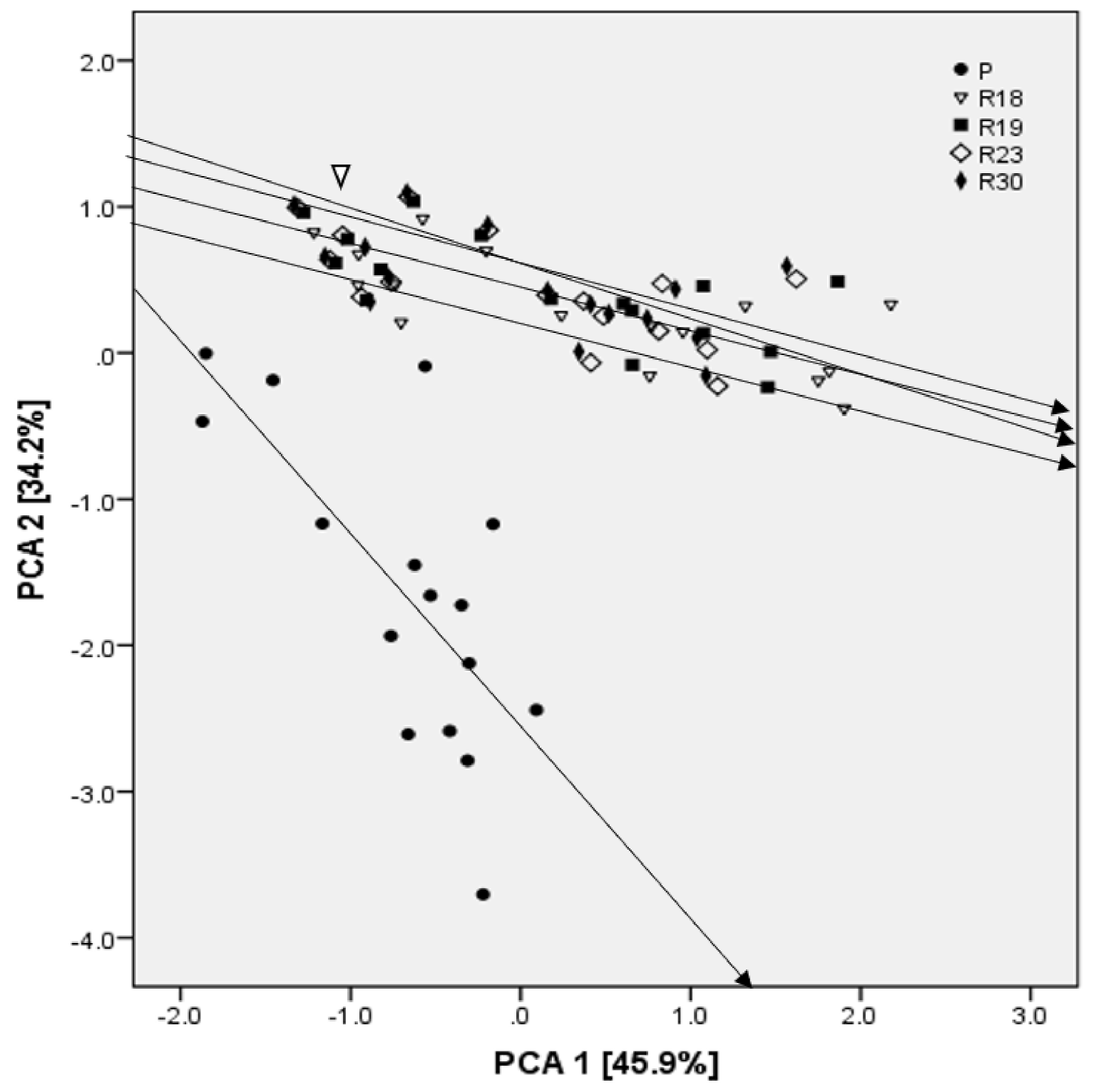
In addition, the PCA analysis showed that Na+ was one of the criteria explaining the majority of dissimilarities among parental and regenerants lines, as well as the characteristics related to the antioxidant machinery, proline, PSII, MDA and the leaf water potential.
In this study, the progeny of somaclonal variants showed significant dissimilarities for several characters when compared to the parental line, and a higher level of salt tolerance. The statistical analysis of the whole set of data included in the current study confirmed the change in genetic stability. Regenerants with consistent performance at advanced generations are assumed to bear gene or chromosome variations, which seem to be stable and heritable.
The success in applying somaclonal variation for salt tolerance depends on the rate and type of somaclonal variation obtained. Therefore, the low phenotypic variation among regenerants could be due to low physiological disturbance during tissue culture. Moreover, the identification of regenerants with putatively increased salt tolerance showed the positive impact of somaclonal variation in improving crop species for desired characters, besides the utility of culture-induced variation in genetic upgradation.