PAHs and PCBs Affect Functionally Intercorrelated Genes in the Sea Urchin Paracentrotus lividus Embryos
Abstract
:1. Introduction
2. Results
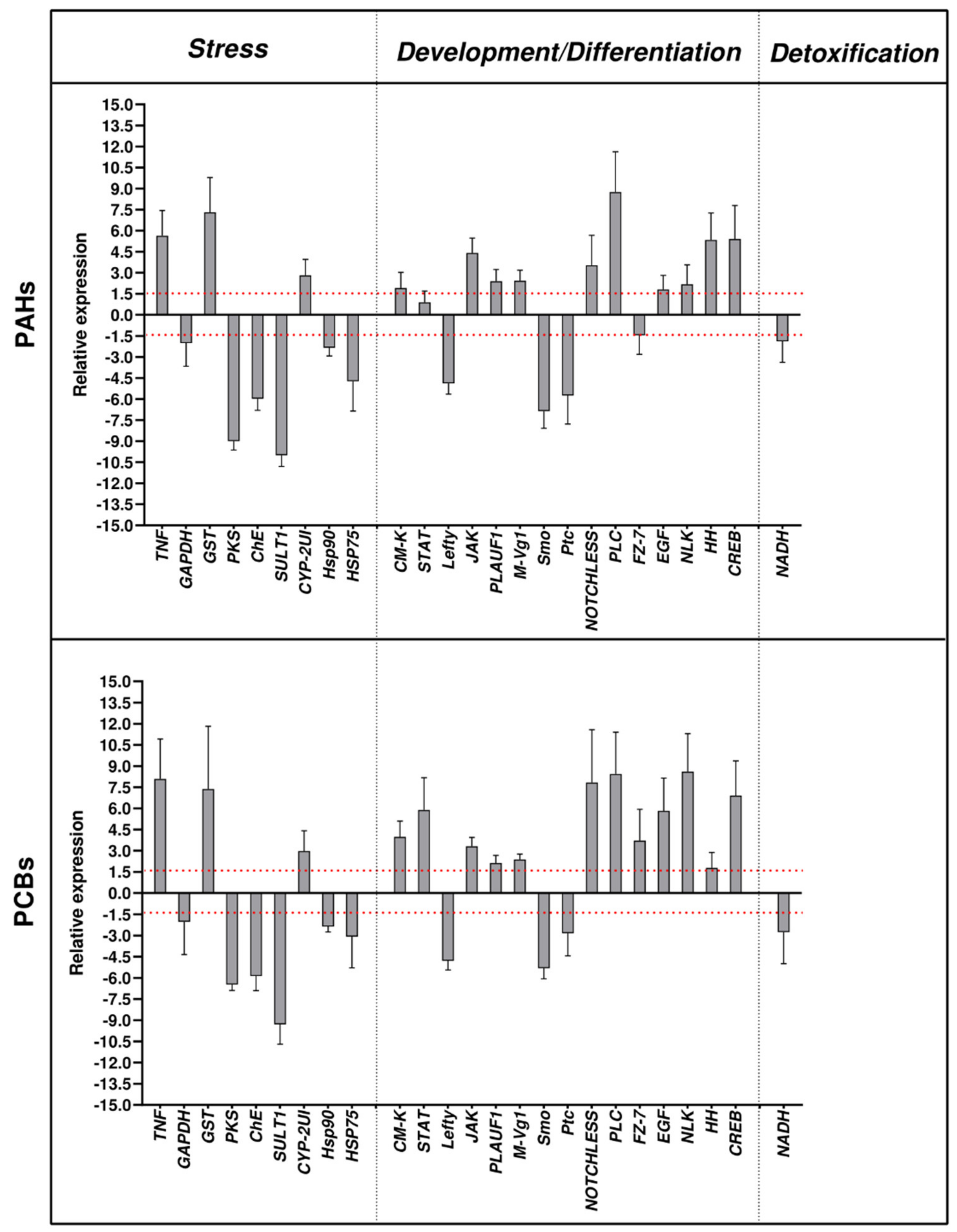
2.1. Effects of PAHs and PCBs on Gene Expression by Real Time qPCR
- -
- Stress genes:
- -
- Genes involved in development/differentiation:
- -
- Genes involved in detoxification:
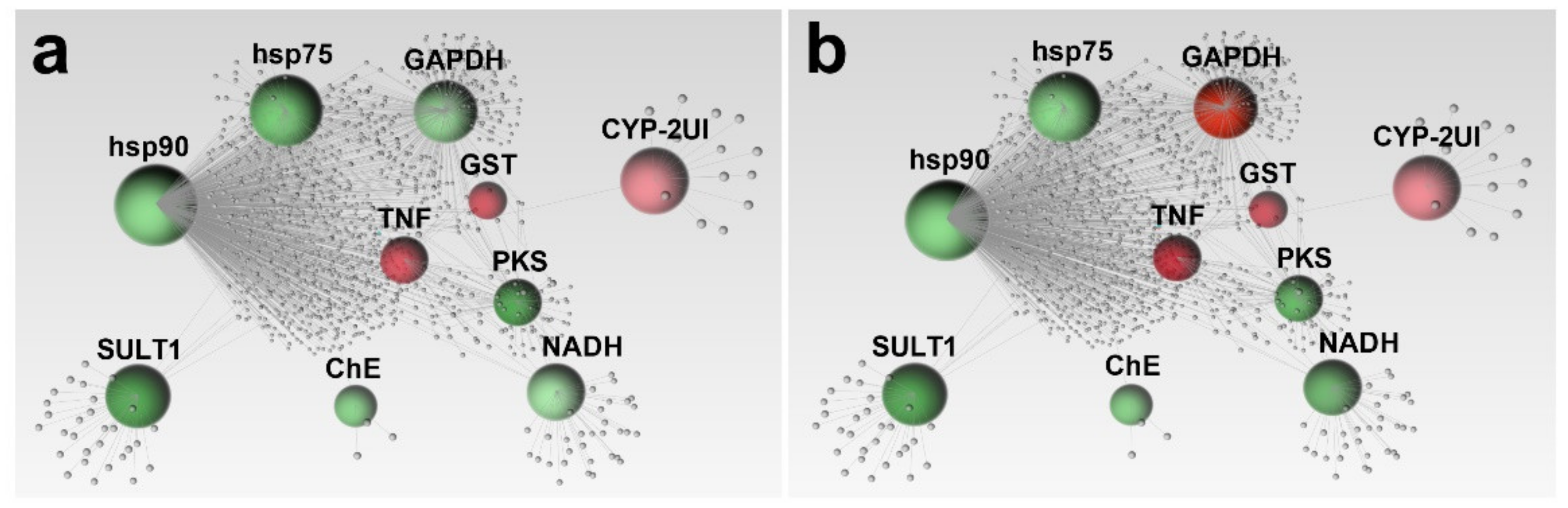
2.2. Network Analysis
3. Discussion
4. Materials and Methods
4.1. Experimental Conditions
4.2. RNA Extraction and cDNA Synthesis
4.3. Network Analysis
4.4. RT-qPCR Analysis
4.5. Gene Networks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doney, S.C.; Ruckelshaus, M.; Emmett Duffy, J.; Barry, J.P.; Chan, F.; English, C.A.; Galindo, H.M.; Grebmeier, J.M.; Hollowed, A.B.; Knowlton, N.; et al. Climate change impacts on marine ecosystems. Ann. Rev. Mar. Sci. 2012, 4, 11–37. [Google Scholar] [CrossRef] [Green Version]
- Gattuso, J.P.; Magnan, A.K.; Bopp, L.; Cheung, W.W.L.; Duarte, C.M.; Hinkel, J.; Mcleod, E.; Micheli, F.; Oschlies, A.; Williamson, P.; et al. Ocean solutions to address climate change and its effects on marine ecosystems. Front. Mar. Sci. 2018, 5, 337. [Google Scholar] [CrossRef] [Green Version]
- Kendall, M.A.; Burrows, M.T.; Southward, A.J.; Hawkins, S.J. Predicting the effects of marine climate change on the invertebrate prey of the birds of rocky shores. Ibis 2004, 146, 40–47. [Google Scholar] [CrossRef]
- Fangue, N.A.; O’Donnell, M.J.; Sewell, M.A.; Matson, P.G.; MacPherson, A.C.; Hofmann, G.E. A laboratory-based, experimental system for the study of ocean acidification effects on marine invertebrate larvae. Limnol. Oceanogr. Methods 2010, 8, 441–452. [Google Scholar] [CrossRef] [Green Version]
- Gattuso, J.P.; Magnan, A.; Billé, R.; Cheung, W.W.L.; Howes, E.L.; Joos, F.; Allemand, D.; Bopp, L.; Cooley, S.R.; Eakin, C.M.; et al. Contrasting futures for ocean and society from different anthropogenic CO2 emissions scenarios. Science 2015, 349. [Google Scholar] [CrossRef] [PubMed]
- Guinotte, J.M.; Fabry, V.J. Ocean acidification and its potential effects on marine ecosystems. Ann. N. Y. Acad. Sci. 2008, 1134, 320–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutignano, A.; Lamari, N.; D’ippolito, G.; Manzo, E.; Cimino, G.; Fontana, A. Lipoxygenase products in marine diatoms: A concise analytical method to explore the functional potential of oxylipins. J. Phycol. 2011, 47, 233–243. [Google Scholar] [CrossRef]
- D’Ippolito, G.; Lamari, N.; Montresor, M.; Romano, G.; Cutignano, A.; Gerecht, A.; Cimino, G.; Fontana, A. 15S-Lipoxygenase metabolism in the marine diatom Pseudo-nitzschia delicatissima. New Phytol. 2009, 183, 1064–1071. [Google Scholar] [CrossRef]
- Fontana, A.; D’Ippolito, G.; Cutignano, A.; Romano, G.; Lamari, N.; Gallucci, A.M.; Cimino, G.; Miralto, A.; Lanora, A. LOX-induced lipid peroxidation mechanism responsible for the detrimental effect of marine diatoms on zooplankton grazers. ChemBioChem 2007, 8, 1810–1818. [Google Scholar] [CrossRef]
- Fontana, A.; D’Ippolito, G.; Cutignano, A.; Miralto, A.; Ianora, A.; Romano, G.; Cimino, G. Chemistry of oxylipin pathways in marine diatoms. Pure Appl. Chem. 2007, 79, 481–490. [Google Scholar] [CrossRef]
- Miralto, A.; Barone, G.; Romano, G.; Poulet, S.A.; Ianora, A.; Russo, G.L.; Buttino, I.; Mazzarella, G.; Laablr, M.; Cabrini, M.; et al. The insidious effect of diatoms on copepod reproduction. Nature 1999, 402, 173–176. [Google Scholar] [CrossRef]
- Anselmo, H.M.R.; Koerting, L.; Devito, S.; van den Berg, J.H.J.; Dubbeldam, M.; Kwadijk, C.; Murk, A.J. Early life developmental effects of marine persistent organic pollutants on the sea urchin Psammechinus miliaris. Ecotoxicol. Environ. Saf. 2011, 74, 2182–2192. [Google Scholar] [CrossRef] [PubMed]
- Bellas, J.; Saco-Álvarez, L.; Nieto, Ó.; Beiras, R. Ecotoxicological evaluation of polycyclic aromatic hydrocarbons using marine invertebrate embryo-larval bioassays. Mar. Pollut. Bull. 2008, 57, 493–502. [Google Scholar] [CrossRef]
- Cunningham, V.L.; Buzby, M.; Hutchinson, T.; Mastrocco, F.; Parke, N.; Roden, N. Effects of HUMAN on Aquatic Life. Europe 2006, 40, 3456–3462. [Google Scholar]
- Lein, N.P.H.; Fujii, S.; Tanaka, S.; Nozoe, M.; Tanaka, H. Contamination of perfluorooctane sulfonate (PFOS) and perfluorooctanoate (PFOA) in surface water of the Yodo River basin (Japan). Desalination 2008, 226, 338–347. [Google Scholar] [CrossRef]
- Dutta, S.M.; Mustafi, S.B.; Raha, S.; Chakraborty, S.K. Biomonitoring role of some cellular markers during heat stress-induced changes in highly representative fresh water mollusc, Bellamya bengalensis: Implication in climate change and biological adaptation. Ecotoxicol. Environ. Saf. 2018, 157, 482–490. [Google Scholar] [CrossRef]
- Mizrahi, T.; Goldenberg, S.; Heller, J.; Arad, Z. Natural variation in resistance to desiccation and heat shock protein expression in the land snail Theba pisana along a climatic gradient. Physiol. Biochem. Zool. 2015, 88, 66–80. [Google Scholar] [CrossRef]
- Burton, G.A.; Johnston, E.L. Assessing contaminated sediments in the context of multiple stressors. Environ. Toxicol. Chem. 2010, 29, 2625–2643. [Google Scholar] [CrossRef] [Green Version]
- Yi, Y.; Yang, Z.; Zhang, S. Ecological risk assessment of heavy metals in sediment and human health risk assessment of heavy metals in fishes in the middle and lower reaches of the Yangtze River basin. Environ. Pollut. 2011, 159, 2575–2585. [Google Scholar] [CrossRef]
- Novelli, A.A.; Losso, C.; Libralato, G.; Tagliapietra, D.; Pantani, C.; Ghirardini, A.V. Is the 1:4 elutriation ratio reliable? Ecotoxicological comparison of four different sediment:water proportions. Ecotoxicol. Environ. Saf. 2006, 65, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Libralato, G.; Losso, C.; Novelli, A.A.; Citron, M.; Della Sala, S.; Zanotto, E.; Cepak, F.; Ghirardini, A.V. Ecotoxicological evaluation of industrial port of Venice (Italy) sediment samples after a decontamination treatment. Environ. Pollut. 2008, 156, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Lofrano, G.; Libralato, G.; Minetto, D.; De Gisi, S.; Todaro, F.; Conte, B.; Calabrò, D.; Quatraro, L.; Notarnicola, M. In situ remediation of contaminated marinesediment: An overview. Environ. Sci. Pollut. Res. 2016, 24, 5189–5206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pougnet, F.; Schäfer, J.; Dutruch, L.; Garnier, C.; Tessier, E.; Dang, D.H.; Lanceleur, L.; Mullot, J.U.; Lenoble, V.; Blanc, G. Sources and historical record of tin and butyl-tin species in a Mediterranean bay (Toulon Bay, France). Environ. Sci. Pollut. Res. 2014, 21, 6640–6651. [Google Scholar] [CrossRef]
- Mamindy-Pajany, Y.; Libralato, G.; Roméo, M.; Hurel, C.; Losso, C.; Ghirardini, A.V.; Marmier, N. Ecotoxicological evaluation of Mediterranean dredged sediment ports based on elutriates with oyster embryotoxicity tests after composting process. Water Res. 2010, 44, 1986–1994. [Google Scholar] [CrossRef]
- Nikolaou, A.; Kostopoulou, M.; Petsas, A.; Vagi, M.; Lofrano, G.; Meric, S. Levels and toxicity of polycyclic aromatic hydrocarbons in marine sediments. TrAC Trends Anal. Chem. 2009, 28, 653–664. [Google Scholar] [CrossRef]
- Liu, M.; Chen, L.; He, Y.; Baumann, Z.; Mason, R.P.; Shen, H.; Yu, C.; Zhang, W.; Zhang, Q.; Wang, X. Impacts of farmed fish consumption and food trade on methylmercury exposure in China. Environ. Int. 2018, 120, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Zupo, V.; Graber, G.; Kamel, S.; Plichta, V.; Granitzer, S.; Gundacker, C.; Wittmann, K.J. Mercury accumulation in freshwater and marine fish from the wild and from aquaculture ponds. Environ. Pollut. 2019, 255, 112975. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.G.; Chan, F.; Menge, B.A.; Hofmann, G.E. Transcriptomic responses to ocean acidification in larval sea urchins from a naturally variable pH environment. Mol. Ecol. 2013, 22, 1609–1625. [Google Scholar] [CrossRef]
- Hardy, N.A.; Lamare, M.; Uthicke, S.; Wolfe, K.; Doo, S.; Dworjanyn, S.; Byrne, M. Thermal tolerance of early development in tropical and temperate sea urchins: Inferences for the tropicalization of eastern Australia. Mar. Biol. 2014, 161, 395–409. [Google Scholar] [CrossRef]
- Sherman, E. Can sea urchins beat the heat? Sea urchins, thermal tolerance and climate change. PeerJ 2015, 3, e1006. [Google Scholar] [CrossRef]
- Au, D.W.T.; Lee, C.Y.; Chan, K.L.; Wu, R.S.S. Reproductive impairment of sea urchins upon chronic exposure to cadmium. Part I: Effects on gamete quality. Environ. Pollut. 2001, 111, 1–9. [Google Scholar] [CrossRef]
- Au, D.W.T.; Reunov, A.A.; Wu, R.S.S. Reproductive impairment of sea urchin upon chronic exposure to cadmium. Part II: Effects on sperm development. Environ. Pollut. 2001, 111, 11–20. [Google Scholar] [CrossRef]
- Albarano, L.; Zupo, V.; Caramiello, D.; Toscanesi, M.; Trifuoggi, M.; Guida, M.; Libralato, G.; Costantini, M. Sub-chronic effects of slight pah-and pcb-contaminated mesocosms in Paracentrotus lividus lmk: A multi-endpoint approach and de novo transcriptomic. Int. J. Mol. Sci. 2021, 22, 6674. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, Y.; Wang, Y.; Wang, Y. Assessment of toxic interactions of heavy metals in multi-component mixtures using sea urchin embryo-larval bioassay. Toxicol. Vitr. 2011, 25, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Bamforth, S.M.; Singleton, I. Bioremediation of polycyclic aromatic hydrocarbons: Current knowledge and future directions. J. Chem. Technol. Biotechnol. 2005, 80, 723–736. [Google Scholar] [CrossRef]
- Walker, C.H.; Hopkin, S.P.; Sibly, R.M.; Peakall, D.B. Principles of Ecotoxicology, 2nd ed.; Taylor & Francis: Abingdon-on-Thames, UK, 2001; ISBN 9780123859266. [Google Scholar]
- Perelo, L.W. Review: In situ and bioremediation of organic pollutants in aquatic sediments. J. Hazard. Mater. 2010, 177, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Richard B Meagher Phytoremediation of toxic elemental and organic pollutants. Curr. Opin. Plant Biol. 2000, 3, 153–162. [CrossRef]
- Pillai, M.C.; Vines, C.A.; Wikramanayake, A.H.; Cherr, G.N. Polycyclic aromatic hydrocarbons disrupt axial development in sea urchin embryos through a β-catenin dependent pathway. Toxicology 2003, 186, 93–108. [Google Scholar] [CrossRef]
- Steevens, J.A.; Slattery, M.; Schlenk, D.; Aryl, A.; Benson, W.H. Effects of ultraviolet-B light and polyaromatic hydrocarbon exposure on sea urchin development and bacterial bioluminescence. Mar. Environ. Res. 1999, 48, 439–457. [Google Scholar] [CrossRef]
- Suzuki, N.; Ogiso, S.; Yachiguchi, K.; Kawabe, K.; Makino, F.; Toriba, A.; Kiyomoto, M.; Sekiguchi, T.; Tabuchi, Y.; Kondo, T.; et al. Monohydroxylated polycyclic aromatic hydrocarbons influence spicule formation in the early development of sea urchins (Hemicentrotus pulcherrimus). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2015, 171, 55–60. [Google Scholar] [CrossRef]
- Gregorin, C.; Albarano, L.; Somma, E.; Costantini, M.; Zupo, V. Assessing the ecotoxicity of copper and polycyclic aromatic hydrocarbons: Comparison of effects on Paracentrotus lividus and Botryllus schlosseri, as alternative bioassay methods. Water 2021, 13, 711. [Google Scholar] [CrossRef]
- Carls, M.G.; Holland, L.; Larsen, M.; Collier, T.K.; Scholz, N.L.; Incardona, J.P. Fish embryos are damaged by dissolved PAHs, not oil particles. Aquat. Toxicol. 2008, 88, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Kiparissis, Y.; Akhtar, P.; Hodson, P.V.; Brown, R.S. Partition-controlled delivery of toxicants: A novel in vivo approach for embryo toxicity testing. Environ. Sci. Technol. 2003, 37, 2262–2266. [Google Scholar] [CrossRef]
- Di Toro, D.M.; Mcgrath, J.A.; Hansen, D.J. Technical basis for narcotic chemicals and polycyclic aromatic hydrocarbon criteria. I. Water and tissue. Environ. Toxicol. Chem. 2000, 19, 1951–1970. [Google Scholar] [CrossRef]
- Esposito, R.; Ruocco, N.; Albarano, L.; Ianora, A.; Manfra, L.; Libralato, G.; Costantini, M. Combined effects of diatom-derived oxylipins on the sea urchin Paracentrotus lividus. Int. J. Mol. Sci. 2020, 21, 719. [Google Scholar] [CrossRef] [Green Version]
- Ruocco, N.; Fedele, A.M.; Costantini, S.; Romano, G.; Ianora, A.; Costantini, M. New inter-correlated genes targeted by diatom-derived polyunsaturated aldehydes in the sea urchin Paracentrotus lividus. Ecotoxicol. Environ. Saf. 2017, 142, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Varrella, S.; Romano, G.; Ianora, A.; Bentley, M.G.; Ruocco, N.; Costantini, M. Molecular response to toxic diatom-derived aldehydes in the sea urchin Paracentrotus lividus. Mar. Drugs 2014, 12, 2089–2113. [Google Scholar] [CrossRef] [Green Version]
- Varrella, S.; Romano, G.; Costantini, S.; Ruocco, N.; Ianora, A.; Bentley, M.G.; Costantini, M. Toxic diatom aldehydes affect defence gene networks in sea urchins. PLoS ONE 2016, 11, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beeble, A.; Calestani, C. Expression pattern of polyketide synthase-2 during sea urchin development. Gene Expr. Patterns 2012, 12, 7–10. [Google Scholar] [CrossRef] [Green Version]
- Cunha, I.; García, L.M.; Guilhermino, L. Sea-urchin (Paracentrotus lividus) glutathione S-transferases and cholinesterase activities as biomarkers of environmental contamination. J. Environ. Monit. 2005, 7, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, C.; Aluigi, M.G.; Ferrando, S.; Gallus, L.; Ramoino, P.; Gatti, A.M.; Rottigni, M.; Falugi, C. Developmental abnormalities and changes in cholinesterase activity in sea urchin embryos and larvae from sperm exposed to engineered nanoparticles. Aquat. Toxicol. 2013, 130–131, 77–85. [Google Scholar] [CrossRef]
- Bonaventura, R.; Zito, F.; Chiaramonte, M.; Costa, C.; Russo, R. Nickel toxicity in P. lividus embryos: Dose dependent effects and gene expression analysis. Mar. Environ. Res. 2018, 139, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Geraci, F.; Pinsino, A.; Turturici, G.; Savona, R.; Giudice, G.; Sconzo, G. Nickel, lead, and cadmium induce differential cellular responses in sea urchin embryos by activating the synthesis of different HSP70s. Biochem. Biophys. Res. Commun. 2004, 322, 873–877. [Google Scholar] [CrossRef]
- Goldstone, J.V.; Hamdoun, A.; Cole, B.J.; Howard-Ashby, M.; Nebert, D.W.; Scally, M.; Dean, M.; Epel, D.; Hahn, M.E.; Stegeman, J.J. The chemical defensome: Environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev. Biol. 2006, 300, 366–384. [Google Scholar] [CrossRef] [Green Version]
- Sconzo, G.; Amore, G.; Capra, G.; Giudice, G.; Cascino, D.; Ghersi, G. Identification and characterization of a constitutive HSP75 in sea urchin embryos. Biochem. Biophys. Res. Commun. 1997, 234, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Chiaramonte, M.; Inguglia, L.; Vazzana, M.; Deidun, A.; Arizza, V. Stress and immune response to bacterial LPS in the sea urchin Paracentrous lividus (Lamarck, 1816). Fish Shellfish Immunol. 2019, 92, 384–394. [Google Scholar] [CrossRef]
- González, K.; Gaitán-Espitia, J.; Font, A.; Cárdenas, C.A.; González-Aravena, M. Expression pattern of heat shock proteins during acute thermal stress in the Antarctic sea urchin, Sterechinus neumayeri. Rev. Chil. Hist. Nat. 2016, 89, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Runcie, D.E.; Garfield, D.A.; Babbitt, C.C.; Wygoda, J.A.; Mukherjee, S.; Wray, G.A. Genetics of gene expression responses to temperature stress in a sea urchin gene network. Mol. Ecol. 2012, 21, 4547–4562. [Google Scholar] [CrossRef] [Green Version]
- Vergara-Amado, J.; Silva, A.X.; Manzi, C.; Nespolo, R.F.; Cárdenas, L. Differential expression of stress candidate genes for thermal tolerance in the sea urchin Loxechinus albus. J. Therm. Biol. 2017, 68, 104–109. [Google Scholar] [CrossRef]
- Nicholls, C.; Li, H.; Liu, J.P. GAPDH: A common enzyme with uncommon functions. Clin. Exp. Pharmacol. Physiol. 2012, 39, 674–679. [Google Scholar] [CrossRef]
- Barber, R.D.; Harmer, D.W.; Coleman, R.A.; Clark, B.J. GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genom. 2005, 21, 389–395. [Google Scholar] [CrossRef] [Green Version]
- Lesser, M.P.; Carleton, K.L.; Böttger, S.A.; Barry, T.M.; Walker, C.W. Sea urchin tube feet are photosensory organs that express a Rhabdomeric-like Opsin and PAX6. Proc. R. Soc. B Biol. Sci. 2011, 278, 3371–3379. [Google Scholar] [CrossRef] [Green Version]
- Voronina, E.; Marzluff, W.F.; Wessel, G.M. Cyclin B synthesis is required for sea urchin oocyte maturation. Dev. Biol. 2003, 256, 258–275. [Google Scholar] [CrossRef] [Green Version]
- Hansen, C.N.; Ketabi, Z.; Rosenstierne, M.W.; Palle, C.; Boesen, H.C.; Norrild, B. Expression of CPEB, GAPDH and U6snRNA in cervical and ovarian tissue during cancer development. Apmis 2009, 117, 53–59. [Google Scholar] [CrossRef]
- Merle, P.; De La Monte, S.; Kim, M.; Herrmann, M.; Tanaka, S.; Von Dem Bussche, A.; Kew, M.C.; Trepo, C.; Wands, J.R. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology 2004, 127, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.E.; Pandey, S.; Nedeljkovic-Kurepa, A.; Saxena, M.; Wang, A.; Pruitt, K. Frizzled 7 expression is positively regulated by SIRT1 and β-catenin in breast cancer cells. PLoS ONE 2014, 9, e98861. [Google Scholar]
- Vincan, E.; Darcy, P.K.; Farrelly, C.A.; Faux, M.C.; Brabletz, T.; Ramsay, R.G. Frizzled-7 dictates three-dimensional organization of colorectal cancer cell carcinoids. Oncogene 2007, 26, 2340–2352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasidharan Nair, V.; Toor, S.M.; Ali, B.R.; Elkord, E. Dual inhibition of STAT1 and STAT3 activation downregulates expression of PD-L1 in human breast cancer cells. Expert Opin. Ther. Targets 2018, 22, 547–557. [Google Scholar] [CrossRef]
- Guo, H.; Kuang, S.; Song, Q.L.; Liu, M.; Sun, X.X.; Yu, Q. Cucurbitacin I inhibits STAT3, but enhances STAT1 signaling in human cancer cells in vitro through disrupting actin filaments. Acta Pharmacol. Sin. 2018, 39, 425–437. [Google Scholar] [CrossRef] [Green Version]
- Choi, E.A.; Lei, H.; Maron, D.J.; Wilson, J.M.; Barsoum, J.; Fraker, D.L.; El-Deiry, W.S.; Spitz, F.R. Stat1-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand and the cell-surface death signaling pathway by interferon β in human cancer cells. Cancer Res. 2003, 63, 5299–5307. [Google Scholar]
- Carrier, T.J.; King, B.L.; Coffman, J.A. Gene expression changes associated with the developmental plasticity of sea urchin larvae in response to food availability. Biol. Bull. 2015, 228, 171–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, Y.; Li, J.; Sun, J.; Zhang, W.; Li, Y.; Cui, D.; Hu, W.; Chang, Y. The Impact of Chronic Heat Stress on the Growth, Survival, Feeding, and Differential Gene Expression in the Sea Urchin Strongylocentrotus intermedius. Front. Genet. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Coward, K.; Owen, H.; Poustka, A.J.; Hibbitt, O.; Tunwell, R.; Kubota, H.; Swann, K.; Parrington, J. Cloning of a novel phospholipase C-δ isoform from pacific purple sea urchin (Strongylocentrotus purpuratus) gametes and its expression during early embryonic development. Biochem. Biophys. Res. Commun. 2004, 313, 894–901. [Google Scholar] [CrossRef]
- Rongish, B.J.; Wu, W.; Kinsey, W.H. Fertilization-induced activation of phospholipase C in the sea urchin egg. Dev. Biol. 1999, 215, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Shearer, J.; De Nadai, C.; Emily-Fenouil, F.; Gache, C.; Whitaker, M.; Ciapa, B. Role of phospholipase Cγ at fertilization and during mitosis in sea urchin eggs and embryos. Development 1999, 126, 2273–2284. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, R.S.; Clough, R.R.; Bhullar, R.P. Regulation of phospholipase C-δ1 through direct interactions with the small GTPase Ral and calmodulin. J. Biol. Chem. 2005, 280, 21933–21941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duboc, V.; Röttinger, E.; Besnardeau, L.; Lepage, T. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev. Cell 2004, 6, 397–410. [Google Scholar] [CrossRef] [Green Version]
- Royet, J.; Bouwmeester, T.; Cohen, S.M. Notchless encodes a novel WD40-repeat-containing protein that modulates notch signaling activity. EMBO J. 1998, 17, 7351–7360. [Google Scholar] [CrossRef] [Green Version]
- Warner, J.F.; Mcclay, D.R. Left-right asymmetry in the Sea Urchin. Genesis 2014, 52, 481–487. [Google Scholar] [CrossRef]
- Materna, S.C.; Davidson, E.H. A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Dev. Biol. 2012, 364, 77–87. [Google Scholar] [CrossRef] [Green Version]
- Röttinger, E.; Croce, J.; Lhomond, G.; Besnardeau, L.; Gache, C.; Lepage, T. Nemo-like kinase (NLK) acts downstream of Notch/Delta signalling to downregulate TCF during mesoderm induction in the sea urchin embryo. Development 2006, 133, 4341–4353. [Google Scholar] [CrossRef] [Green Version]
- Ingham, P.W.; Taylor, A.M.; Nakano, Y. Role of the Drosophila patched gene in positional signalling. Nature 1991, 353, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Nachtergaele, S.; Whalen, D.M.; Mydock, L.K.; Zhao, Z.; Malinauskas, T.; Krishnan, K.; Ingham, P.W.; Covey, D.F.; Siebold, C.; Rohatgi, R. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. Elife 2013, 2, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Walton, K.D.; Warner, J.; Hertzler, P.H.; McClay, D.R. Hedgehog signaling patterns mesoderm in the sea urchin. Dev. Biol. 2009, 331, 26–37. [Google Scholar] [CrossRef] [Green Version]
- Lhomond, G.; McClay, D.R.; Gache, C.; Croce, J.C. Frizzled1/2/7 signaling directs β-catenin nuclearisation and initiates endoderm specification in macromeres during sea urchin embryogenesis. Development 2012, 139, 816–825. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.; Hu, N.; Hombría, J.C.G. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr. Biol. 2001, 11, 1700–1705. [Google Scholar] [CrossRef]
- Darnell, J.E. STATs and gene regulation. Science 1997, 277, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.X.; Zheng, Z.; Chen, X.; Perrimon, N. The JAK/STAT pathway in model organisms: Emerging roles in cell movement. Dev. Cell 2002, 3, 765–778. [Google Scholar] [CrossRef] [Green Version]
- Chiarelli, R.; Roccheri, M.C. Marine Invertebrates as Bioindicators of Heavy Metal Pollution. Open J. Met. 2014, 4, 93–106. [Google Scholar] [CrossRef] [Green Version]
- Marrone, V.; Piscopo, M.; Romano, G.; Ianora, A.; Palumbo, A.; Costantini, M. Defensome against toxic diatom aldehydes in the sea urchin Paracentrotus lividus. PLoS ONE 2012, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Trifuoggi, M.; Donadio, C.; Mangoni, O.; Ferrara, L.; Bolinesi, F.; Nastro, R.A.; Stanislao, C.; Toscanesi, M.; Di Natale, G.; Arienzo, M. Distribution and enrichment of trace metals in surface marine sediments in the Gulf of Pozzuoli and off the coast of the brownfield metallurgical site of Ilva of Bagnoli (Campania, Italy). Mar. Pollut. Bull. 2017, 124, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Ruocco, N.; Costantini, S.; Zupo, V.; Romano, G.; Ianora, A.; Fontana, A.; Costantini, M. High-quality RNA extraction from the sea urchin Paracentrotus lividus embryos. PLoS ONE 2017, 12, e0172171. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruocco, N.; Cavaccini, V.; Caramiello, D.; Ianora, A.; Fontana, A.; Zupo, V.; Costantini, M. Noxious effects of the benthic diatoms Cocconeis scutellum and Diploneis sp. on sea urchin development: Morphological and de novo transcriptomic analysis. Harmful Algae 2019, 86, 64–73. [Google Scholar] [CrossRef]
- Ruocco, N.; Costantini, S.; Zupo, V.; Lauritano, C.; Caramiello, D.; Ianora, A.; Budillon, A.; Romano, G.; Nuzzo, G.; D’Ippolito, G.; et al. Toxigenic effects of two benthic diatoms upon grazing activity of the sea urchin: Morphological, metabolomic and de novo transcriptomic analysis. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool ( REST © ) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT—PCR. Nucleic Acids Res. 2001, 29, 16–21. [Google Scholar] [CrossRef]
- Romano, G.; Costantini, M.; Buttino, I.; Ianora, A.; Palumbo, A. Nitric oxide mediates the stress response induced by diatom aldehydes in the sea urchin Paracentrotus lividus. PLoS ONE 2011, 6, e25980. [Google Scholar] [CrossRef] [Green Version]
- Pinsino, A.; Matranga, V.; Trinchella, F.; Roccheri, M.C. Sea urchin embryos as an in vivo model for the assessment of manganese toxicity: Developmental and stress response effects. Ecotoxicology 2010, 19, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.A.; Costa, S.; Gianguzza, M.; Roccheri, M.C.; Gianguzza, F. Effects of cadmium exposure on sea urchin development assessed by SSH and RT-qPCR: Metallothionein genes and their differential induction. Mol. Biol. Rep. 2013, 40, 2157–2167. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]


| Gene Names | Acronym | Accession Number | Primer | Sequence (5′→3′) | Amplicon Lenght (bp) |
|---|---|---|---|---|---|
| Calcium/calmodulin-dependent protein kinase type 1D | CM-K | PRJNA495004 | Pl_CM_F1 | GTTATCCTCCATTTTACGATGAG | 168 |
| Pl_CM_R1 | GCAGATATACGTGTGAGGAAG | ||||
| Camp-responsive element | CREB | PRJNA495004 | Pl_CREB_F2 | GTAACTAAAGCATCTGGGAGAC | 158 |
| Pl_CREB_R2 | GGTTCAGATATTAGTGGATGC | ||||
| Cholinesterase | ChE | SUB6701449 | Pl_ChE_F2 | CGAGATGGCGTATGTTTTGAG | 160 |
| Pl_ChE_R2 | GACTATGTTCCCGCTGACTG | ||||
| Citochrome P450 2UI isoform X2 | CYP-2UI | SUB6701449 | Pl_CYP-2UI_F1 | GCGCCTCTTCGTTCTATTCC | 174 |
| Pl_CUP-2UI_R1 | CGGCATAGTAGTAGACTAGC | ||||
| Epidermal growth factor | EGF | PRJNA495004 | Pl_EGF_F1 | CGGCGGTGTGTGTATCGATG | 189 |
| Pl_EGF_R1 | CAGTAGCCATCCTAGTGTTCC | ||||
| Frizzled7 | FZ-7 | PRJNA495004 | Pl_FZ_F1 | GATCGTGAGCGTAGCATATAC | 175 |
| Pl_FZ_R1 | CATGGTCTTTTTGGGCACTA | ||||
| Glutathione-S-transferase | GST | SUB6701449 | Pl_GST_F4 | GCCCGACTTACCTACTTTGC | 165 |
| Pl_GST_R4 | CTTGCAGCTCATCACTGATGG | ||||
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | PRJNA495004 | Pl_GAPDH_F1 | GTACTACTTCTCATTCACCTTG | 213 |
| Pl_GAPDH_R1 | CATAGCTCTGACACCGCCAC | ||||
| heat shock protein 75 | hsp75 | PRJNA495004 | Pl_hsp75_F2 | GGACTGGTGGAACAACTATATC | 173 |
| Pl_hsp75_R2 | CGATCACCACTCTCTGTCAC | ||||
| heat shock protein 90 | hsp90 | SUB6701449 | Pl_hsp90_F1 | GGGTGTGGTAGATTCTGATG | 148 |
| Pl_hsp90_R1 | GCTCTCCATGTATTCATCAG | ||||
| Hedgehog | HH | PRJNA495004 | Pl_HH_F1 | GGTACATGAGGCACAAGCTAG | 193 |
| Pl_HH_R1 | CCACTTCACATCACTTGACC | ||||
| Janus kinase | JAK | XM_030985987 | Pl_JAK_F2 | CACCTTACCCATACTAGACAG | 192 |
| Pl_JAK_R2 | CTTGCCAGAGCCTCCGCTGAC | ||||
| Lefty | Lefty | SUB6701449 | Pl_Lefty_F2 | CAGTCCAGACATGGGTGGCAG | 182 |
| Pl_Lefty_R2 | CATTTCGTCGACCACCTGCTG | ||||
| maternal Vg1 | M-Vg1 | SUB6701449 | Pl_M-Vg1_F1 | GCACCTGCACCTAGAGACTC | 145 |
| Pl_M-Vg1_R1 | GCATGACCTTTTCCGGCCTG | ||||
| NADH dehydrogenase | NADH | SUB2817153 | Pl_NADH_F1 | GTCTCCGTCGGATAAATCAAAG | 194 |
| Pl_NADH_R1 | CCGAAAAGGAAATAACGAAGC | ||||
| Nemo-like kinase protein | NLK | AY442297 | Pl_NLK_F1 | CCTCTACCAGATTCTCAGAG | 192 |
| Pl_NLK_R1 | GTGACACAGTACTACCGCGC | ||||
| Notchless protein | NOTCHLESS | PRJNA495004 | Pl_Notchless_F1 | GGGAAGCTAAGGCATCAGAC | 145 |
| Pl_Notchless_R1 | CGATCCTCTCAAGCACTTTAG | ||||
| Patched | Ptc | SUB6701449 | Pl_Ptc_F1 | CGGTCGTCAGTATCATCATG | 135 |
| Pl_Ptc_R1 | GCAACCACGACTCCGTAAGC | ||||
| Phospholipase C | PLC | AJ012336 | Pl_PLC_F1 | GAGACATTCACAGTGCCCAC | 139 |
| Pl_PLC_R1 | CTGACCGATACCAAGCTGTAC | ||||
| PLAUF 3 RNA-binding protein AUF1 mRNA | PLAUF 3 | AY682309.1 | Pl_PLAUF3_F2 | GGAGGATACGGCGGTGGCGG | 182 |
| Pl_PLAUF3_R2 | GTGTTGACTCCACAGGAGTG | ||||
| Polyketide synthase | PKS | SUB6701449 | Pl_PKS_F1 | GCTTCCTCGACCAGTCTGTC | 142 |
| Pl_PKS_R1 | CCTCCGAAGACAGTCATCTG | ||||
| signal transducer and activator of transcription | STAT1 | PRJNA495004 | Pl_STAT_F1 | GTGTGTCAATCAGCCAGTGC | 196 |
| Pl_STAT_R1 | GTACATCATGAGCTTACCATTTC | ||||
| Smoothened | Smo | SUB6701449 | Pl_Smo_F1 | CACGATCCATTACGGCGTTG | 217 |
| Pl_Smo_R1 | GCCCAACTCACACCCATGAC | ||||
| sulfotransferase 1C2-like | SULT1 | SUB6701449 | Pl_SULT1_F2 | CAGGCACTCACTGGCTCATG | 140 |
| Pl_SULT1_R2 | CTCTTCAGCTCTCGTCTTCG | ||||
| Tumor necrosis factor alpha | TNF | SUB2817153 | Pl_TNF_F1 | CCTGATGTGTATGCCTCTATC | 144 |
| Pl_TNF_R1 | CAAGATCCTCATGTCAGGAAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albarano, L.; Zupo, V.; Guida, M.; Libralato, G.; Caramiello, D.; Ruocco, N.; Costantini, M. PAHs and PCBs Affect Functionally Intercorrelated Genes in the Sea Urchin Paracentrotus lividus Embryos. Int. J. Mol. Sci. 2021, 22, 12498. https://doi.org/10.3390/ijms222212498
Albarano L, Zupo V, Guida M, Libralato G, Caramiello D, Ruocco N, Costantini M. PAHs and PCBs Affect Functionally Intercorrelated Genes in the Sea Urchin Paracentrotus lividus Embryos. International Journal of Molecular Sciences. 2021; 22(22):12498. https://doi.org/10.3390/ijms222212498
Chicago/Turabian StyleAlbarano, Luisa, Valerio Zupo, Marco Guida, Giovanni Libralato, Davide Caramiello, Nadia Ruocco, and Maria Costantini. 2021. "PAHs and PCBs Affect Functionally Intercorrelated Genes in the Sea Urchin Paracentrotus lividus Embryos" International Journal of Molecular Sciences 22, no. 22: 12498. https://doi.org/10.3390/ijms222212498
APA StyleAlbarano, L., Zupo, V., Guida, M., Libralato, G., Caramiello, D., Ruocco, N., & Costantini, M. (2021). PAHs and PCBs Affect Functionally Intercorrelated Genes in the Sea Urchin Paracentrotus lividus Embryos. International Journal of Molecular Sciences, 22(22), 12498. https://doi.org/10.3390/ijms222212498










