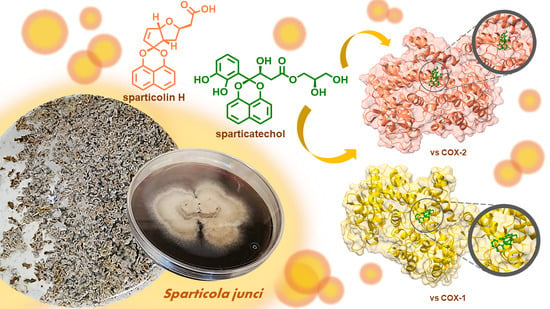
COX Inhibitory and Cytotoxic Naphthoketal-Bearing Polyketides from Sparticola junci
Abstract
:1. Introduction
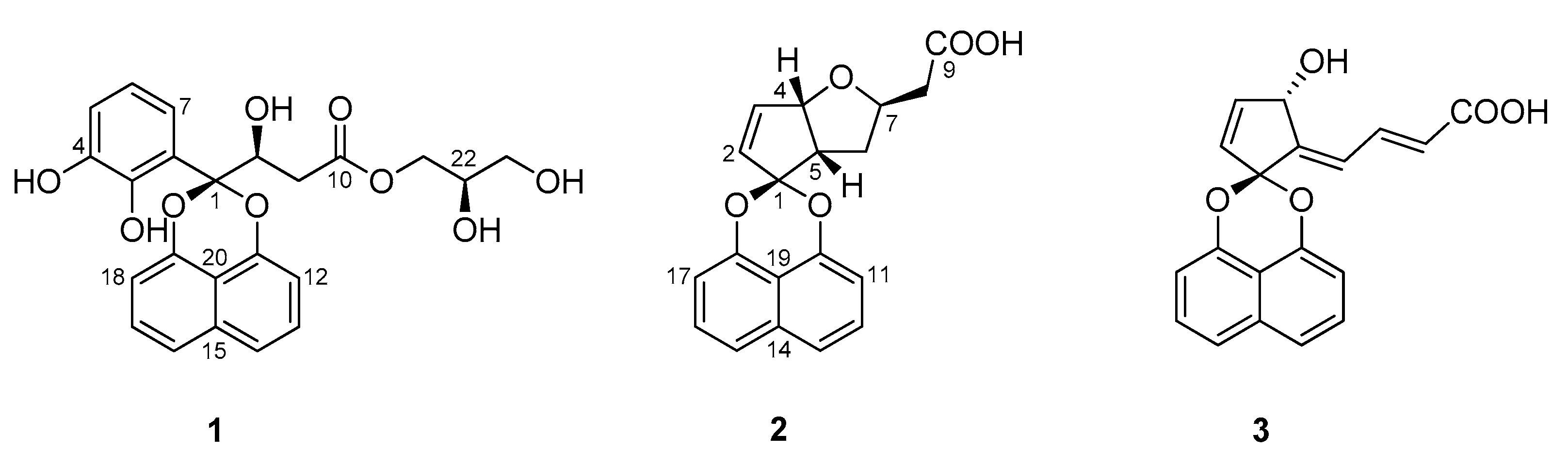
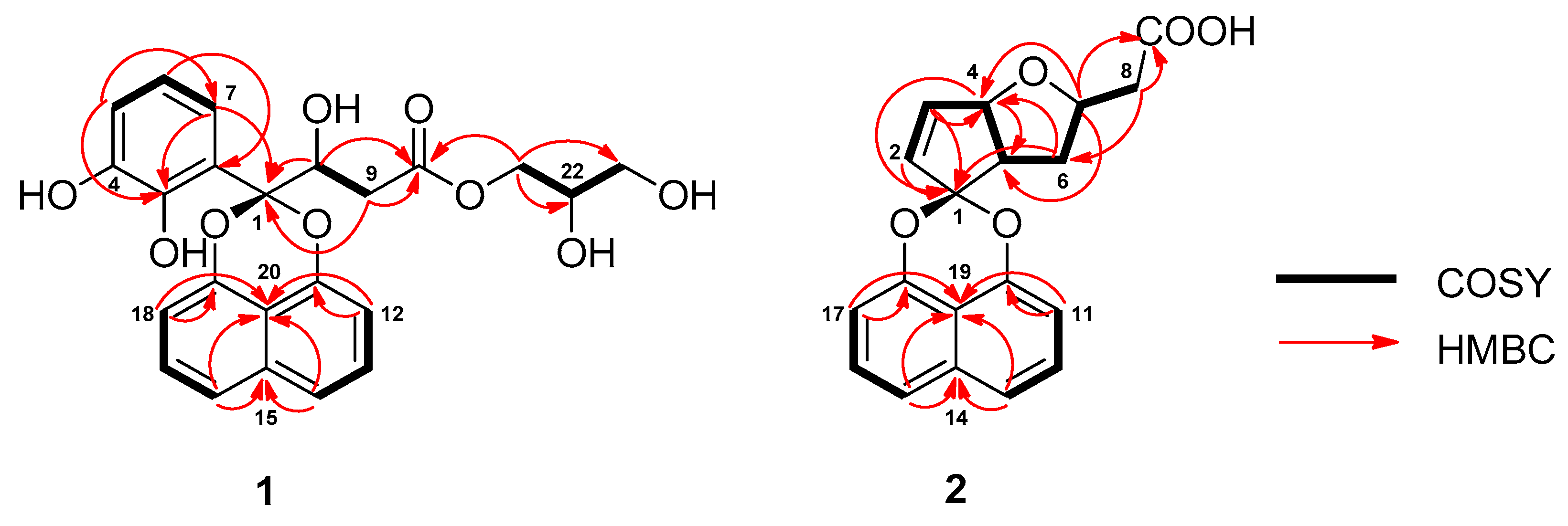
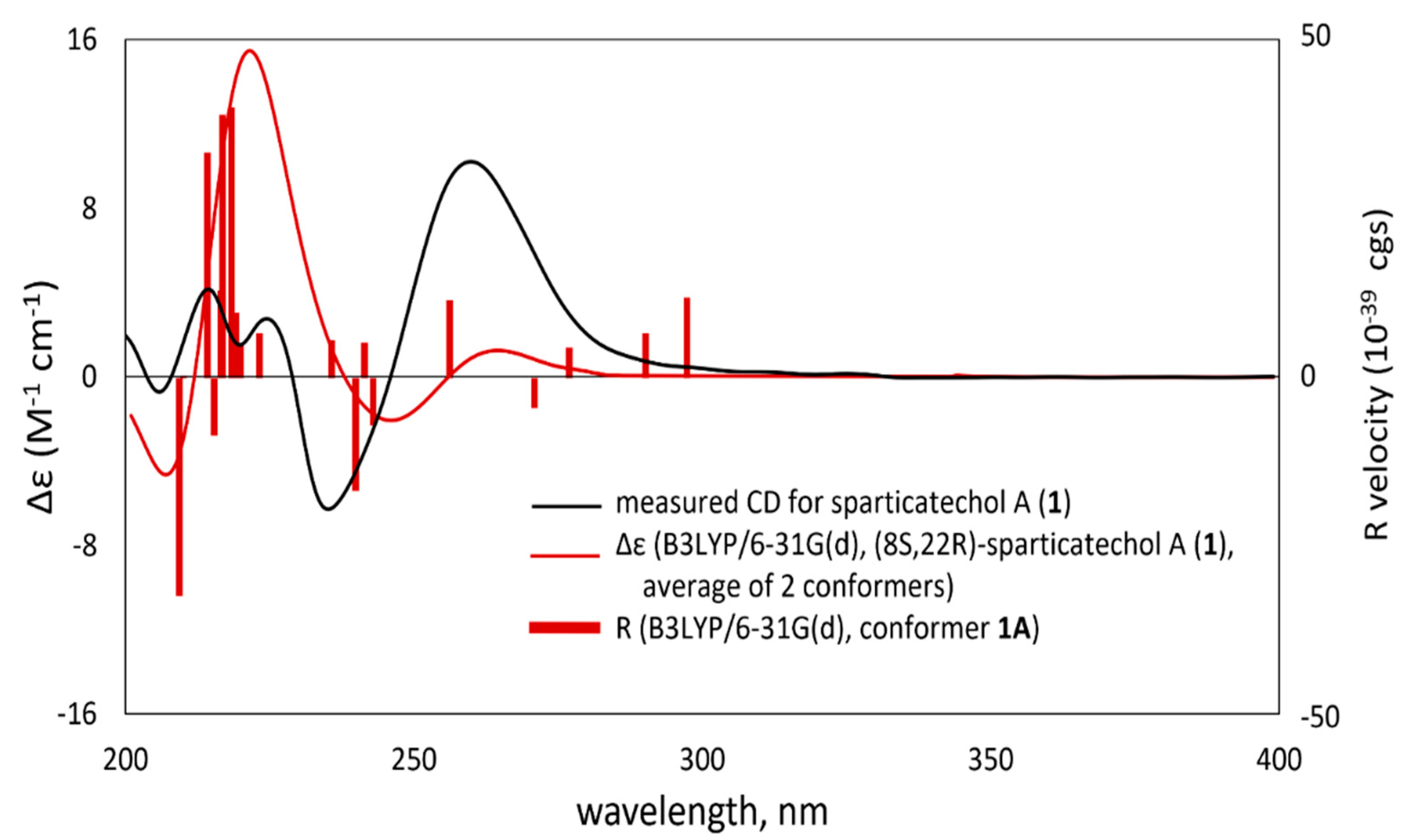
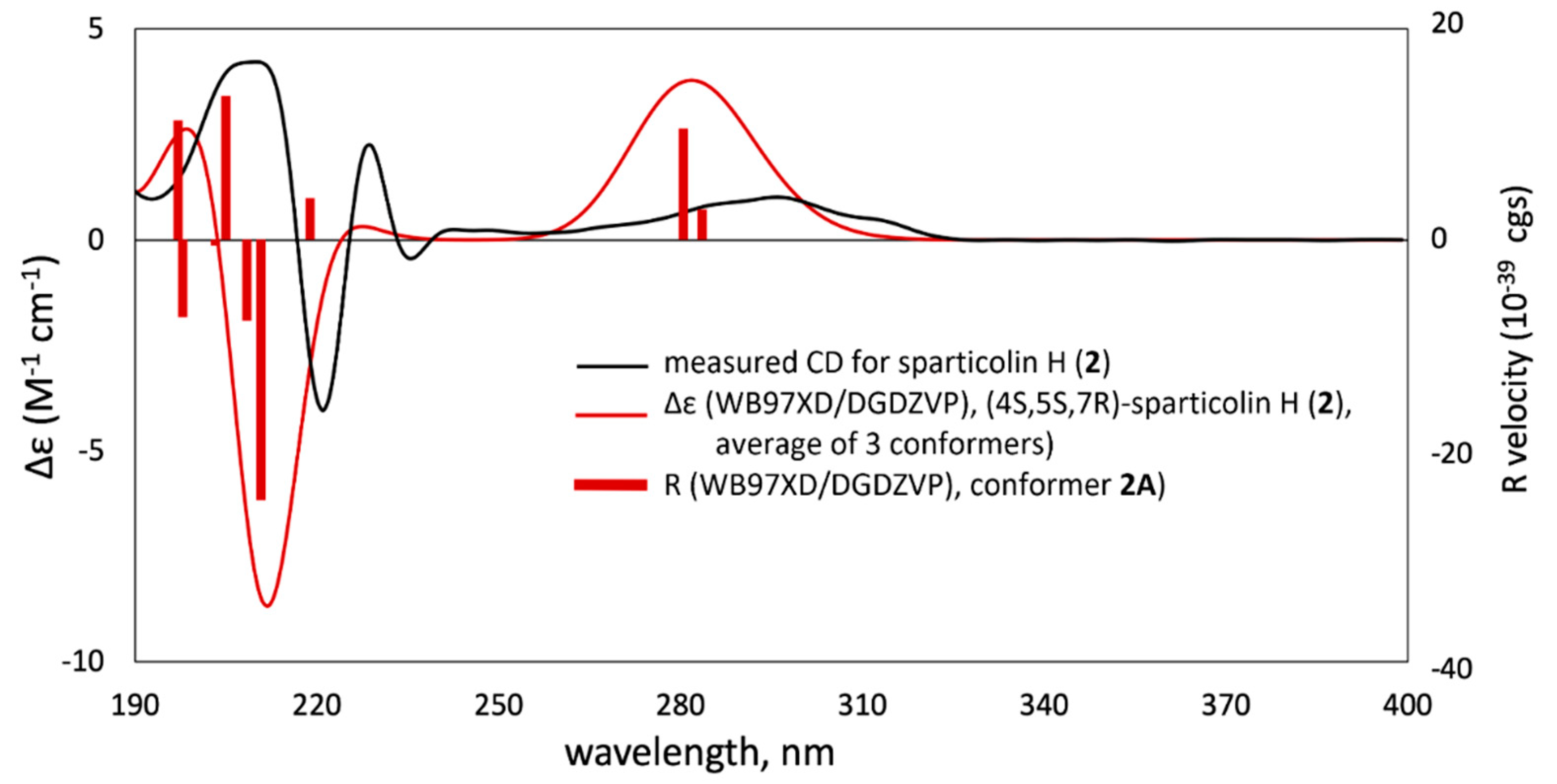
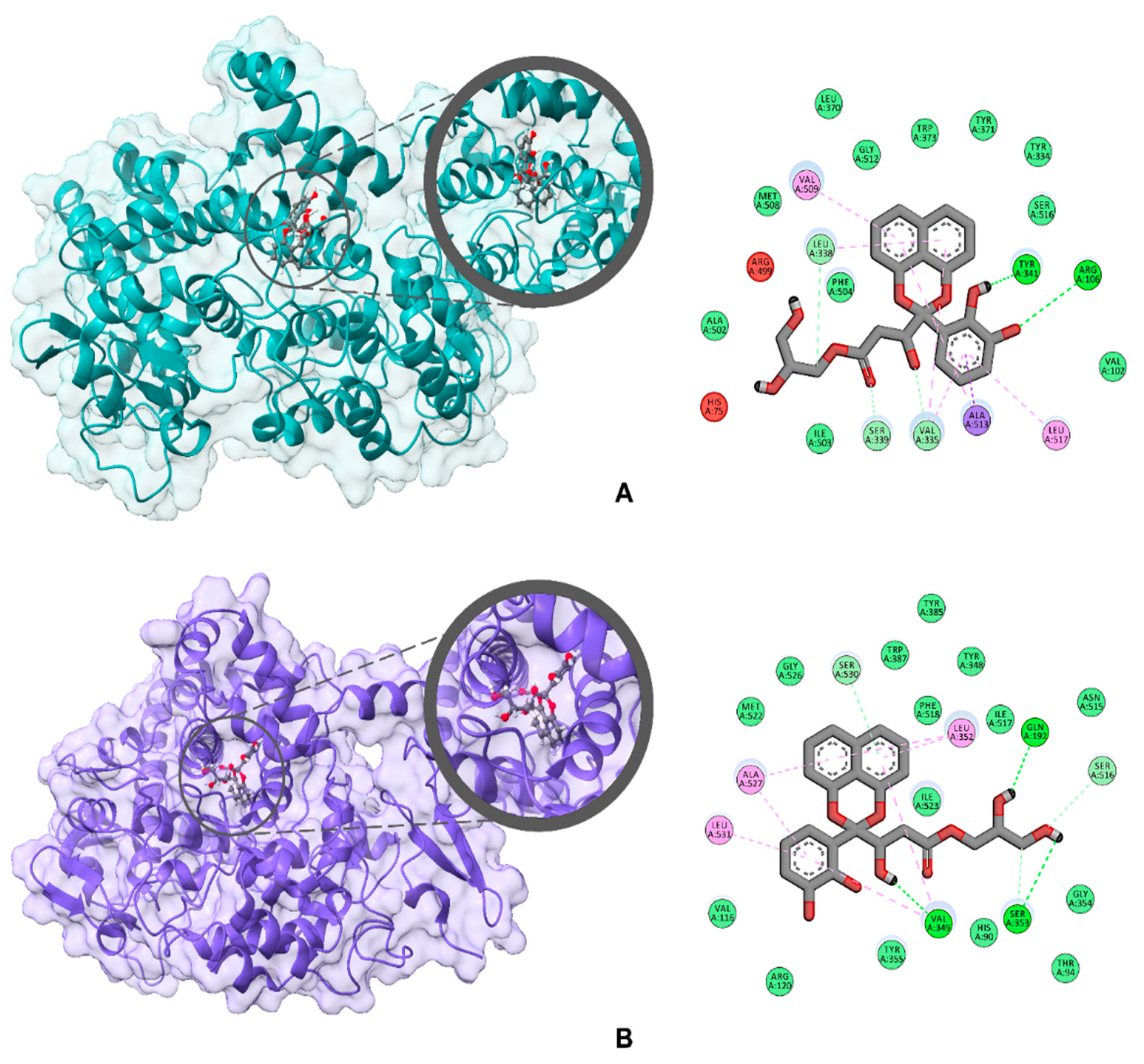
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material and Fermentation
3.3. Extraction and Isolation
3.4. Biological Assays
3.5. Computational Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuephadungphan, W.; Macabeo, A.P.G.; Luangsa-ard, J.J.; Stadler, M. Discovery of novel biologically active secondary metabolites from Thai mycodiversity with anti-infective potential. CRBIOT 2004, 3, 160–172. [Google Scholar] [CrossRef]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar]
- Bills, G.F.; Gloer, J.B. Biologically active secondary metabolites from the fungi. Microbiol. Spectr. 2016, 4, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.-S.; Guo, Y.-W.; Krohn, K. Structure, bioactivities, biosynthetic relationships and chemical synthesis of the spirodioxynaphthalenes. Nat. Prod. Rep. 2010, 27, 1840–1870. [Google Scholar] [CrossRef]
- Ogishi, H.; Chiba, N.; Mikawa, T.; Sasaki, T.; Miyaji, S.; Sezaki, M. Mitsubishi Kasei Corp., JP 01294686. Chem. Abstr. 1990, 113, 38906q. [Google Scholar]
- Petersen, F.; Moerker, T.; Vanzanella, F.; Peter, H.H. Production of cladospirone bisepoxide, a new fungal metabolite. J. Antibiot. 1994, 47, 1098–1103. [Google Scholar] [CrossRef]
- Thiergardt, R.; Rihs, G.; Hug, P.; Peter, H.H. Cladospirone bisepoxide: Definite structure assignment including absolute configuration and selective chemical transformations. Tetrahedron 1995, 51, 733–742. [Google Scholar] [CrossRef]
- Krohn, K.; Michel, A.; Flörke, U.; Aust, H.-J.; Draeger, S.; Schulz, B. Palmarumycins C1–C16 from Coniothyrium sp.: Isolation, structure elucidation and biological activity. Liebigs Ann. Chem. 1994, 1994, 1099–1108. [Google Scholar] [CrossRef]
- Jiao, P.; Swenson, D.C.; Gloer, J.B.; Campbell, J.; Shearer, C.A. Decaspirones A–E, Bioactive Spirodioxynaphthalenes from the freshwater aquatic fungus Decaisnella thyridioides. J. Nat. Prod. 2006, 69, 1667–1671. [Google Scholar] [CrossRef] [Green Version]
- Dai, J.; Krohn, K.; Elsässer, B.; Flörke, U.; Draeger, S.; Schulz, B.; Pescitelli, G.; Salvadori, P.; Antus, S.; Kurtán, T. Metabolic products of the endophytic fungus Microsphaeropsis sp. from Larix decidua. Eur. J. Org. Chem. 2007, 29, 4845–4954. [Google Scholar] [CrossRef]
- Chu, M.; Truumees, I.; Patel, M.G.; Gullo, V.P.; Pai, J.-K.; Das, P.R.; Puar, M.S. Two new phospholipase d inhibitors, Sch 49211 and Sch 49212, produced by the fungus Nattrasia mangiferae. Bioorg. Med. Chem. Lett. 1994, 4, 1539–1542. [Google Scholar] [CrossRef]
- Van der Sar, S.A.; Lang, G.; Mitova, M.; Blunt, J.; Cole, A.L.J.; Cummings, N.; Ellis, G.; Munro, M.H.G. Biosynthesis of spiro-mamakone A, a structurally unprecedented fungal metabolite. J. Org. Chem. 2008, 73, 8635–8638. [Google Scholar] [CrossRef] [PubMed]
- Phukhamsakda, C.; Macabeo, A.P.G.; Huch, V.; Cheng, T.; Hyde, K.D.; Stadler, M. Sparticolins A–G, Biologically active oxidized pirodioxynaphthalene derivatives from the ascomycete Sparticola junci. J. Nat. Prod. 2019, 82, 2878–2885. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.Y.M.; Phukhamsakda, C.; Quimque, M.T.J.; Hyde, K.D.; Stadler, M.; Macabeo, A.P.G. Catechol-bearing polyketide derivatives from Sparticola junci. J. Nat. Prod. 2021, 84, 2053–2058. [Google Scholar] [CrossRef]
- Macabeo, A.P.G.; Pilapil, L.A.E.; Garcia, K.Y.M.; Quimque, M.T.J.; Phukhamsakda, C.; Cruz, A.J.C.; Hyde, K.D.; Stadler, M. Alpha-Glucosidase- and Lipase-Inhibitory Phenalenones from a new species of Pseudolophiostoma originating from Thailand. Molecules 2020, 25, 965. [Google Scholar] [CrossRef] [Green Version]
- Phukhamsakda, C.; Macabeo, A.P.G.; Yuyama, K.T.; Hyde, K.D.; Stadler, M. Biofilm inhibitory abscisic acid derivatives from the plant-associated Dothideomycete fungus, Roussoella sp. Molecules 2018, 23, 2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walton, L.J.; Corre, C.; Challis, G.L. Mechanisms for incorporation of glycerol-derived precursors into polyketide metabolites. J. Ind. Microbiol. 2005, 33, 105–120. [Google Scholar] [CrossRef]
- Yan, Y.; Zang, X.; Jamieson, C.S.; Lin, H.-C.; Houk, K.N.; Zhou, J.; Tang, Y. Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance. Chem. Sci. 2020, 11, 9554–9562. [Google Scholar] [CrossRef]
- Quimque, M.T.; Notarte, K.I.; Letada, A.; Fernandez, R.A.; Pilapil, D.Y.; Pueblos, K.R.; Agbay, J.C.; Dahse, H.-M.; Wenzel-Storjohann, A.; Tasdemir, D.; et al. Potential cancer- and Alzheimer’s Disease-targeting phosphodiesterase inhibitors from Uvaria alba: Insights from in vitro and consensus virtual screening. ACS Omega 2021, 6, 8403–8417. [Google Scholar] [CrossRef] [PubMed]
- Acharya, R.; Chacko, S.; Bose, P.; Lapenna, A.; Pattanayak, S.P. Structure based multitargeted molecular docking analysis of selected furanocoumarins against breast cancer. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Quimque, M.T.J.; Liman, R.A.; Agbay, J.C.; Macabeo, A.P.G.; Corpuz, M.J.-A.; Wang, Y.-M.; Lu, T.-T.; Lin, C.H.; Villaflores, O.B. Computational and experimental assessments of magnolol as a neuroprotective agent and utilization of UiO-66(Zr) as Its drug delivery system. ACS Omega 2021, 6, 24382–24396. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karuth, F.; Dahse, H.-M.; Rüttinger, H.-H.; Frohberg, P. Synthesis and characterization of novel 1,2,4-triazine derivatives with antiproliferative activity. Bioorg. Med. Chem. 2010, 18, 1816–1821. [Google Scholar] [CrossRef]
- Becker, K.; Wessel, A.C.; Luangsa-ard, J.J.; Stadler, M. Viridistratins A–C, antimicrobial and cytotoxic benzo[j]fluoranthenes from stromata of Annulohypoxylon viridistratum (Hypoxylaceae, Ascomycota). Biomolecules 2020, 10, 805. [Google Scholar] [CrossRef] [PubMed]
- Kuephadungphan, W.; Macabeo, A.P.G.; Luangsa-ard, J.J.; Tasanathai, K.; Thanakitpipattana, D.; Phongpaichit, S.; Yuyama, K.; Stadler, M. Studies on the biologically active secondary metabolites of the new spider parasitic fungus Gibellula gamsii. Mycol. Prog. 2019, 18, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Collins, L.; Franzblau, S.G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997, 41, 1004–1009. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.M.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision A. 03; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Modell. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Zoete, V. A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [Green Version]

| Position | 1 | 2 | ||
|---|---|---|---|---|
| δHa (Mult., J in Hz) | δC a, Type | δH b (Mult., J in Hz) | δC c, Type | |
| 1 | - | 105.1, C | - | 111.0, C |
| 2 | - | 123.5, C | 5.88 (ddd, 5.7, 1.0, 0.4) | 130.1, CH |
| 3 | - | 145.1, C | 6.25 (dd, 5.7, 1.7) | 139.3, CH |
| 4 | - | 147.3, C | 5.07 (ddd, 7.0, 1.8, 1.0) | 85.7, CH |
| 5 | 6.59 (dd, 7.9, 1.5) | 117.0, CH | 3.14 (td, 8.5, 7.1) | 51.1, CH |
| 6 | 6.42 (dd, 7.9, 1.5) | 119.8, CH | 1.86–1.73 (m) | 32.7, CH2 |
| 7 | 6.80 (dd, 7.9, 1.5) | 121.6, CH | 4.24 (dtd, 12.1, 6.1, 5.6) | 78.7, CH |
| 8 | 4.81 (dd, 14.0, 3.1) | 73.9, CH | 2.52 (d, 6.3) | 39.9, CH2 |
| 9a | 2.68 (dd, 15.7, 9.4) | 38.0, CH2 | - | 173.5, C |
| 9b | 2.97 (dd, 15.7, 3.2) | |||
| 10 | - | 173.1, C | - | 148.2, C |
| 11 | - | 148.7, C | 6.87 (dd, 8.9, 7.4) | 108.6, CH |
| 12 | 7.02 (dd, 8.3, 1.3) | 110.2, CH | 7.40 (dd, 8.9, 7.4) | 127.1, CH |
| 13 | 7.37 (dd, 8.3) | 121.6, CH | 7.47 (dd, 8.9, 7.4) | 120.1, CH |
| 14 | 7.39 (dd, 8.3, 1.3) | 128.2, CH | - | 134.5, C |
| 15 | - | 135.5, C | 7.47 (dd, 8.9, 7.4) | 120.1, CH |
| 16 | 7.39 (dd, 8.3, 1.3) | 128.2, CH | 7.40 (dd, 8.9, 7.4) | 127.1, CH |
| 17 | 7.37 (dd, 8.3) | 121.6, CH | 6.87 (dd, 8.9, 7.4) | 108.6, CH |
| 18 | 7.02 (dd, 8.3, 1.3) | 110.2, CH | - | 148.4, C |
| 19 | - | 148.9, C | - | 114.1, C |
| 20 | - | 115.5, C | - | - |
| 21a | 4.10 (td, 11.4, 6.3) | 66.9, CH2 | - | - |
| 21b | 4.19 (td, 11.4, 4.3) | - | - | |
| 22 | 3.80 (dt, 15.2, 6.7) | 71.1, CH | - | - |
| 23 | 3.56 (ddd, 7.3, 5.7, 2.3) | 64.0, CH2 | - | - |
| Compound | Antiproliferative Effect GI50 (µM) | Cytotoxicity CC50 (µM) | Cytotoxicity IC50 (µM) | ||
|---|---|---|---|---|---|
| HUVEC | K-562 | HeLa | L929 | KB3.1 | |
| 1 | >50 | 97.5 | 85.3 | − | − |
| 2 | >50 | 80.8 | 124.7 | 22.9 | 21.8 |
| 3 | >50 | 91.3 | 119.8 | − | − |
| Imatinib | 18.5 | 0.17 | 65.8 | N.D. | N.D. |
| Epothilone B | N.D. | N.D. | N.D. | 1.57 × 10−3 | 3.9 × 10−3 |
| Compound | COX-1 IC50 (µM) | COX-2 IC50 (µM) |
|---|---|---|
| 1 | 8.8 × 10−3 | 0.3 |
| 2 | 1.8 | 1.5 |
| 3 | 1.4 | 2.3 |
| Celecoxib | 18.09 | 0.656 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia, K.Y.M.; Quimque, M.T.J.; Primahana, G.; Ratzenböck, A.; Cano, M.J.B.; Llaguno, J.F.A.; Dahse, H.-M.; Phukhamsakda, C.; Surup, F.; Stadler, M.; et al. COX Inhibitory and Cytotoxic Naphthoketal-Bearing Polyketides from Sparticola junci. Int. J. Mol. Sci. 2021, 22, 12379. https://doi.org/10.3390/ijms222212379
Garcia KYM, Quimque MTJ, Primahana G, Ratzenböck A, Cano MJB, Llaguno JFA, Dahse H-M, Phukhamsakda C, Surup F, Stadler M, et al. COX Inhibitory and Cytotoxic Naphthoketal-Bearing Polyketides from Sparticola junci. International Journal of Molecular Sciences. 2021; 22(22):12379. https://doi.org/10.3390/ijms222212379
Chicago/Turabian StyleGarcia, Katherine Yasmin M., Mark Tristan J. Quimque, Gian Primahana, Andreas Ratzenböck, Mark Joseph B. Cano, Jeremiah Francis A. Llaguno, Hans-Martin Dahse, Chayanard Phukhamsakda, Frank Surup, Marc Stadler, and et al. 2021. "COX Inhibitory and Cytotoxic Naphthoketal-Bearing Polyketides from Sparticola junci" International Journal of Molecular Sciences 22, no. 22: 12379. https://doi.org/10.3390/ijms222212379