Anthracene and Pyrene Biodegradation Performance of Marine Sponge Symbiont Bacteria Consortium
Abstract
1. Introduction
2. Results
2.1. Characteristics of Seawater at Sampling Location
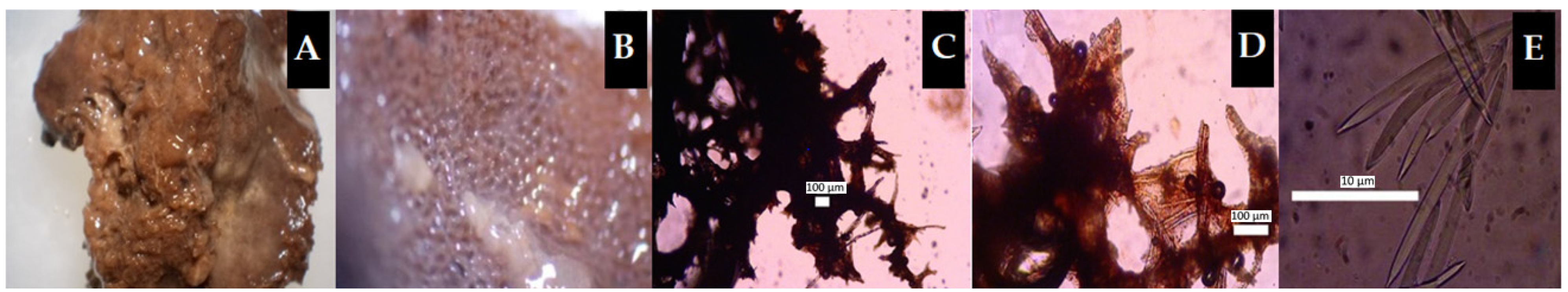
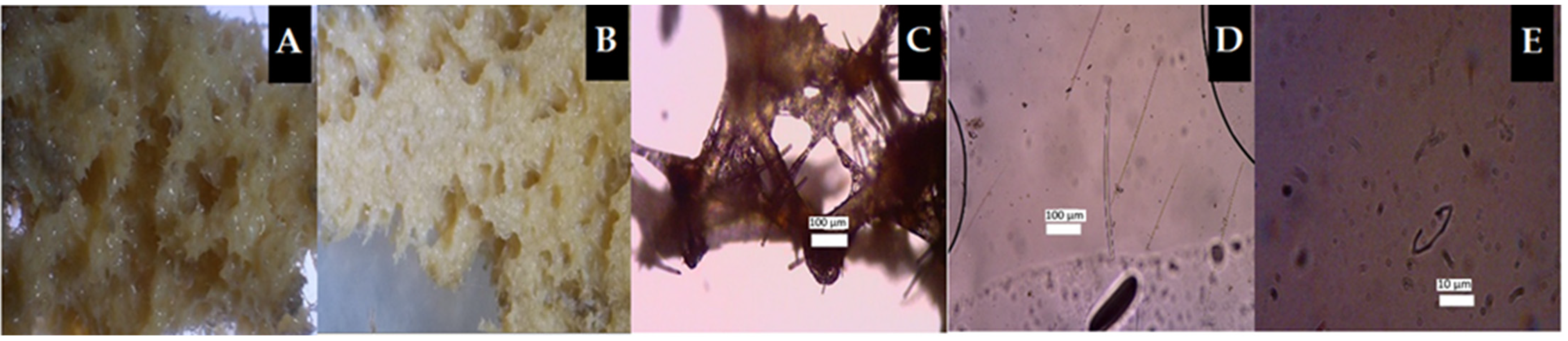
2.2. Marine Sponge Morphology
2.3. Isolation and Phenotypic Characteristics of Marine Sponge Symbiont Bacteria
2.4. Genotypic Analysis of Marine Sponge Symbiont Bacteria
2.5. Biodegradation of PAH Components
2.5.1. Evaluating PAH Biodegradation Performance of the Marine Sponge Symbiont Bacterial Consortium
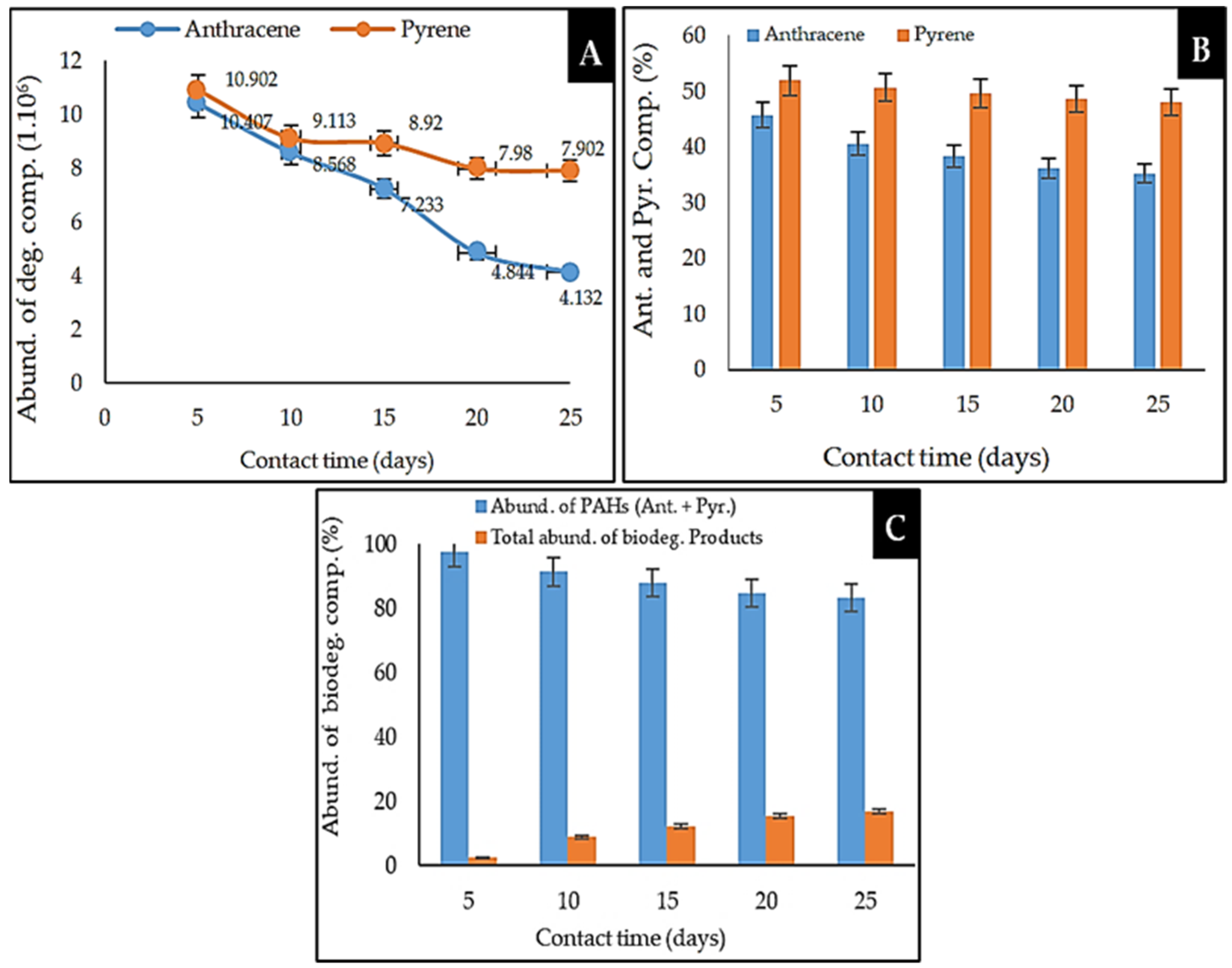
2.5.2. Analysis of the Biodegradation Performance of Anthracene and Pyrene PAHs by Marine Sponge Symbiont Consortium
2.5.3. Functional Groups of Biodegradation Products
2.6. Comparison of the Biodegradation of Anthracene and Pyrene by the Marine Sponge Symbiont Consortium Bacteria
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Sampling Point Characterization
4.3. Marine Sponge Morphology
4.4. Isolation and Phenotypic Analysis of Marine Sponge Symbiont Bacteria
4.5. Genotypic Analysis of the Bacterial Isolate
4.6. Biodegradation Interactions and Evaluation of the Processes
4.6.1. Interaction of Biodegradation Components
4.6.2. Parameters of Biodegradation of PAHs by Bacteria
4.6.3. Biodegradation Performance of PAHs by the Bacterial Consortium
4.6.4. Detection of Functional Groups for Identification of Biodegradation Products of PAHs by Bacteria Consortium
4.7. Comparison of Biodegradation Rates of Anthracene and Pyrene
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bendouz, M.; Dionne, D.; Tran, L.H.; Coudert, L.; Mercier, G.; Blais, J.F. Polycyclic Aromatic Hydrocarbon Oxidation from Concentrates Issued from an Attrition Process of Polluted Soil Using the Fenton Reagent and Permanganate. Water Air Soil Pollut. 2017, 228, 115. [Google Scholar] [CrossRef]
- Bezza, F.A.; Chirwa, E.M.N. Biosurfactant-enhanced bioremediation of aged polycyclic aromatic hydrocarbons (PAHs) in creosote contaminated soil Fisseha. Chemosphere 2016, 144, 635–644. [Google Scholar] [CrossRef]
- Dadrasnia, A.; Usman, M.M.; Lim, K.T.; Farahiyah, F.H.; Rodzhan, N.S.M.; Karim, S.H.A.; Ismail, S. Bio-Enhancement of Petroleum Hydrocarbon Polluted Soil Using Newly Isolated Bacteria. Polycycl. Aromat. Compd. 2020, 40, 484–493. [Google Scholar] [CrossRef]
- Fang, H.; Shi, Y.; Zhou, M.; Niu, Q. Influence of n-Hexadecane and Naphthalene on Anaerobic Digestion: Kinetic Simulation, DOM Variation and Microbial Community Assessment. IOP Conf. Ser. Earth Environ. Sci. 2020, 555, 012038. [Google Scholar] [CrossRef]
- Ja’nczuk, B.; Szymczyk, K.; Zdziennicka, A. Adsorption Properties of Hydrocarbon and Fluorocarbon Surfactants Ternary Mixture at the Water-Air Interface. Molecules 2021, 26, 4313. [Google Scholar] [CrossRef] [PubMed]
- Khabouchi, I.; Khadhar, S.; Chaouachi, D.; Chekirbene, A.; Doumenq, P. Study of organic pollution in superficial sediments of Meliane river catchment area: Aliphatic and polycyclic aro-matic hydrocarbons Study of organic pollution in superficial sediments of Meliane river catchment area: Aliphatic and polycyclic aromatic hydrocarbons. Enviroment Monit. Assess. 2020, 192, 282–290. [Google Scholar] [CrossRef]
- Lu, C.; Hong, Y.; Liu, J.; Gao, Y.; Ma, Z.; Yang, B.; Ling, W.; Waigi, M.G. A PAH-degrading bacterial community enriched with contaminated agricultural soil and its utility for microbial bioremediation. Environ. Pollut. 2019, 251, 773–782. [Google Scholar] [CrossRef]
- Medić, A.; Lješević, M.; Inui, H.; Beškoski, V.; Kojić, I.; Stojanović, K.; Karadžić, I. Efficient biodegradation of petroleum: N -alkanes and polycyclic aromatic hydrocarbons by polyextremophilic Pseudomonas aeruginosa san ai with multidegradative capacity. RSC Adv. 2020, 10, 14060–14070. [Google Scholar] [CrossRef]
- Parhamfar, M.; Abtahia, H.; Godinib, K.; Saeedi, R.; Sartaje, M.; Villaseñorf, J.; Coulong, F.; Kumarg, V.; Soltanighiash, T.; Radi, E.G.; et al. Biodegradation of heavy oily sludge by a two-step inoculation composting process using synergistic effect of indigenous isolated bacteria. Process Biochem. 2020, 91, 223–230. [Google Scholar] [CrossRef]
- Laothamteep, N.; Kawano, H.; Vejarano, F.; Minakuchi, C.S.; Shintani, M.; Nojiri, H.; Pinyakong, O. Effects of environmental factors and coexisting substrates on PAH degradation and transcriptomic responses of the defined bacterial consortium OPK. Environ. Pollut. 2021, 277, 116769. [Google Scholar] [CrossRef] [PubMed]
- Marzuki, I.; Kamaruddin, M.; Ahmad, R. Identification of marine sponges-symbiotic bacteria and their application in degrading polycyclic aromatic hydrocarbons. Bioivers. J. Biol. Divers. 2021, 22, 1481–1488. [Google Scholar] [CrossRef]
- Abass, O.K.; Zhuo, M.; Zhang, K. Concomitant degradation of complex organics and metals recovery from fracking wastewater: Roles of nano zerovalent iron initiated oxidation and adsorption. Chem. Eng. J. 2017, 328, 159–171. [Google Scholar] [CrossRef]
- Nursid, M.; Marraskuranto, E.; Atmojo, K.B.; Hartono, T.M.P.; Meinita, M.D.N. Investigation on antioxidant compounds from Marine Algae Extracts collected from Binuangeun Coast, Banten, Indonesia. Squalen Bull. Mar. Fish. Postharvest Biotechnol. 2016, 11, 59–67. [Google Scholar] [CrossRef][Green Version]
- Onwosia, C.O.; Odibob, F.-J.K.; Enebechia, C.K.; Nwankwegub, A.S.; Ikeleb, A.I.; Okehc, O.C. Soil and Sediment Contamination: An International Bioremediation of Diesel-contaminated Soil by Composting with Locally Generated Bulking Agents Bioremediation of Diesel-contaminated. Soil Sediment. Contam. 2017, 26, 438–456. [Google Scholar] [CrossRef]
- Košnář, Z.; Částková, T.; Wiesnerová, L.; Praus, L.; Jablonský, I.; Koudela, M.; Tlustoš, P. Comparing the removal of polycyclic aromatic hydrocarbons in soil after different bioremediation approaches in relation to the extracellular enzyme activities. J. Environ. Sci. 2019, 76, 249–258. [Google Scholar] [CrossRef]
- Yetti, E.; Thontowi, A.; Yopi, Y.; Lisdiyanti, P. Screening of Marine bacteria capable of degrading various polyaromatic hydrocarbons. Squalen Bull. Mar. Fish. Postharvest Biotechnol. 2015, 10, 121–127. [Google Scholar] [CrossRef][Green Version]
- Obire, O.; Aleruchi, O.; Wemedo, S. Fungi in Biodegradation of Polycyclic Aromatic Hydrocarbons in Oilfield Wastewater. Acta Sci. Microbiol. 2020, 3, 220–224. [Google Scholar] [CrossRef]
- Sinsona, M.-J.; Juinio, M.M.A. Effects of sediment enrichment with macroalgae, Sargassum spp., on the behavior, growth, and survival of juvenile sandfish, Holothuria scabra. Aquac. Rep. 2018, 12, 56–63. [Google Scholar] [CrossRef]
- Araújo, S.C.D.S.; Silva-Portela, R.C.B.; De Lima, D.C.; Da Fonsêca, M.M.B.; Araújo, W.J.; Da Silva, U.B.; Napp, A.; Pereira, E.; Vainstein, M.H.; Agnez-Lima, L.F. MBSP1: A biosurfactant protein derived from a metagenomic library with activity in oil degradation. Sci. Rep. 2020, 10, 1340–1352. [Google Scholar] [CrossRef]
- Campana, S.; Hudspith, M.; Lankes, D.; de Kluijver, A.; Demey, C.; Schoorl, J.; Absalahh, S.; der Meer, M.T.J.; Mueller, B.; de Goeij, J.-M. Processing of Naturally Sourced Macroalgal- and Coral-Dissolved Organic Matter (DOM) by High and Low Microbial Abundance Encrusting Sponges. Front. Mar. Sci. 2021, 8, 640583. [Google Scholar] [CrossRef]
- Gu, B.; Li, F.; Liu, Y.; Mao, L.; Tao, H. Effect of Vegetable Growth on Content and Composition of Antibiotics in Litopenaeus vannamei Pond Sediments in Crop/Aquacultural Rotation Process. Sustainability 2021, 13, 8400. [Google Scholar] [CrossRef]
- Maldonado, M.; López-Acosta, M.; Busch, K.; Slaby, B.M.; Bayer, K.; Beazley, L.; Hentschel, U.; Kenchington, E.; Rapp, H.T. A Microbial Nitrogen Engine Modulated by Bacteriosyncytia in Hexactinellid Sponges: Ecological Implications for Deep-Sea Communities. Front. Mar. Sci. 2021, 8, 638505. [Google Scholar] [CrossRef]
- Morganti, T.M.; Ribes, M.; Yahel, G.; Coma, R. Size Is the Major Determinant of Pumping Rates in Marine Sponges. Front. Physiol. 2019, 10, 1474. [Google Scholar] [CrossRef]
- Orani, A.M.; Barats, A.; Vassileva, E.; Thomas, O.P. Marine sponges as a powerful tool for trace elements biomonitoring studies in coastal environment. Mar. Pollut. Bull. 2018, 131, 633–645. [Google Scholar] [CrossRef]
- Page, H.N.; Hewett, C.; Tompkins, H.; Hall, E.R. Ocean acidification and direct interactions affect coral, macroalga, and sponge growth in the florida keys. J. Mar. Sci. Eng. 2021, 9, 739. [Google Scholar] [CrossRef]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Turon, M.; Cáliz, J.; Garate, L.; Casamayor, E.O.; Uriz, M.J. Showcasing the role of seawater in bacteria recruitment and microbiome stability in sponges. Sci. Rep. 2018, 8, 15201. [Google Scholar] [CrossRef] [PubMed]
- Campana, S.; Busch, K.; Hentschel, U.; Muyzer, G.; de Goeij, J.M. DNA-stable isotope probing (DNA-SIP) identifies marine sponge-associated bacteria actively utilizing dissolved organic matter (DOM). Environ. Microbiol. 2021, 23, 4489–4504. [Google Scholar] [CrossRef]
- Marzuki, I.; Sinardi, S.; Pratama, I.; Chaerul, M.; Paserangi, I.; Kamaruddin, M.; Asaf, R. Performance of sea sponges micro symbionts as a biomaterial in biodegradation naphthalene waste of modified. In Proceedings of the 5th International Seminar on Sustainable Urban Development, Jakarta, Indonesia, 5 August 2020; Volume 737, p. 012016. [Google Scholar] [CrossRef]
- Costa, G.; Violi, B.; Bavestrello, G.; Pansini, M.; Bertolino, M. Aplysina aerophoba (Nardo, 1833) (Porifera, Demospongiae): An unexpected miniaturised growth form the tidal zone of Mediterranean caves: Morphology and DNA barcoding. Eur. Zool. J. 2020, 87, 73–81. [Google Scholar] [CrossRef]
- Baquiran, J.I.P.; Nada, M.A.L.; Posadas, N.; Manogan, D.P.; Cabaitan, P.C.; Conaco, C. Population structure and microbial community diversity of two common tetillid sponges in a tropical reef lagoon. PeerJ 2020, 8, e9017. [Google Scholar] [CrossRef] [PubMed]
- Marzuki, I.; Daris, L.; Nisaa, K.; Emelda, A. The power of biodegradation and bio-adsorption of bacteria symbiont sponges sea on waste contaminated of polycyclic aromatic hydrocarbons and heavy metals. In Proceedings of the International Conference on Fisheries and Marine, Ternate, Maluku, Indonesia, 13 July 2020; Volume 584, p. 012013. [Google Scholar] [CrossRef]
- Yang, Q.; Franco, C.M.M.; Lin, H.W.; Zhang, W. Untapped sponge microbiomes: Structure specificity at host order and family levels. FEMS Microbiol. Ecol. 2019, 95, fiz136. [Google Scholar] [CrossRef] [PubMed]
- Lavy, A.; Keren, R.; Haber, M.; Schwartz, I.; Ilan, M. Implementing sponge physiological and genomic information to enhance the diversity of its culturable associated bacteria. FEMS Microbiol. Ecol. 2014, 87, 486–502. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Wang, H.; Bai, Y.; Xue, J.; Gao, Y.; Hu, S.; Wu, T.; Sum, J. Systematic degradation mechanism and pathways analysis of the immobilized bacteria: Permeability and biodegradation, kinetic and molecular simulation. Environ. Sci. Ecotechnol. 2020, 2, 100028. [Google Scholar] [CrossRef]
- Su, X.M.; Bamba, A.M.; Zhang, S.; Zhang, Y.G.; Hashmi, M.Z.; Lin, H.J.; Ding, L.X. Revealing potential functions of VBNC bacteria in polycyclic aromatic hydrocarbons biodegradation. Lett. Appl. Microbiol. 2018, 66, 277–283. [Google Scholar] [CrossRef]
- Marzuki, I.; Daris, L.; Yunus, S.; Riana, A.D. Selection and characterization of potential bacteria for polycyclic aromatic biodegradation of hydrocarbons in sea sponges from Spermonde Islands, Indonesia. Aquac. Aquar. Conserv. Legis. 2020, 13, 3493–3506. [Google Scholar]
- Rua, C.P.J.; de Oliveira, L.S.; Froes, A.; Tschoeke, D.A.; Soares, A.C.; Leomil, L.; Gregoracci, G.B.; Coutinho, R.; Hajdu, E.; Thompson, C.C.; et al. Microbial and Functional Biodiversity Patterns in Sponges that Accumulate Bromopyrrole Alkaloids Suggest Horizontal Gene Transfer of Halogenase Genes. Microb. Ecol. 2018, 76, 825–838. [Google Scholar] [CrossRef]
- Al-Dhabaan, F.A. Morphological, biochemical and molecular identification of petroleum hydrocarbons biodegradation bacteria isolated from oil polluted soil in Dhahran, Saud Arabia. Saudi J. Biol. Sci. 2019, 26, 1247–1252. [Google Scholar] [CrossRef]
- Miao, L.L.; Qu, J.; Liu, Z.P. Hydroxylation at Multiple Positions Initiated the Biodegradation of Indeno[1,2,3-cd]Pyrene in Rhodococcus aetherivorans IcdP1. Front. Microbiol. 2020, 11, 568381. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Tian, J.; Yu, Y.; Chand, A.; Zhang, S.; Meng, Q.; Li, X.; Wang, S. Multifunctional graphene-based composite sponge. Sensors 2020, 20, 329. [Google Scholar] [CrossRef] [PubMed]
- Marzuki, I.; Ali, M.Y.; Syarif, H.U.; Gusty, S.; Ritnawati, R.; Daris, L.; Nisaa, K. Investigation of Biodegradable Bacteria as Bio indicators of the Presence of PAHs Contaminants in Marine Waters in the Marine Tourism Area of Makassar City. In Proceedings of 6th International Conference on Tropical Coastal Region Eco-Development, Semarang, Indonesia. IOP Conf. Ser. Earth Environ. Sci. 2020, 750, 012006. [Google Scholar] [CrossRef]
- Khan, M.N.; Ullah, H.; Naeem, S.; Uddin, J.; Hamid, Y.; Ahmad, W.; Ding, J. Remediation of Emerging Heavy Metals from Water Using Natural Adsorbent: Adsorption Performance and Mechanistic Insights. Sustainability 2021, 13, 8817. [Google Scholar] [CrossRef]
- Siahaya, N.; Noor, A.; Sukamto, N.; de Voogd, N. A preliminary effort to assign sponge (Callispongia sp.) as trace metal biomonitor for Pb, Cd, Zn, and Cr, an environmental perspective in Hative gulf waters Ambon. Adv. Biol. Chem. 2013, 3, 549–552. [Google Scholar] [CrossRef][Green Version]
- Melawaty, L.; Noor, A.; Harlim, T.; de Voogd, N. Essential metal Zn in sponge Callyspongia aerizusa from Spermonde Archipelago. Adv. Biol. Chem. 2014, 4, 86–90. [Google Scholar] [CrossRef][Green Version]
- Karimpour, M.; Ashrafi, S.D.; Taghavi, K.; Mojtahedi, A.; Roohbakhsh, E.; Naghipur, D. Adsorption of cadmium and lead onto live and dead cell mass of Pseudomonas aeruginosa: A dataset. Data Brief 2018, 18, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Schmittmann, L.; Jahn, M.T.; Pita, L.; Hentschel, U. Decoding cellular dialogues between sponges, bacteria, and phages. In Cellular Dialogues in the Holobiont, 1st ed.; CRC Press: Boca Raton, FL, USA, 2020; pp. 49–63. [Google Scholar] [CrossRef]
- Bart, M.C.; Hudspith, M.; Rapp, H.T.; Verdonschot, P.F.M.; de Goeij, J.M. A Deep-Sea Sponge Loop? Sponges Transfer Dissolved and Particulate Organic Carbon and Nitrogen to Associated Fauna. Front. Mar. Sci. 2021, 8, 604879. [Google Scholar] [CrossRef]
- Arjuna, A.; Olson, M.T.; Tokman, S.; Walia, R.; Mohanakumar, T.; Hashimi, A. Samad.; Smith, M.A.; Bremner, R.M.; Omar, A.; Norton, N. Antibody-Mediated Rejection and Sponge Effect in a Redo Lung Transplant Recipient. Case Rep. Transplant. 2021, 2021, ID6637154. [Google Scholar] [CrossRef]
- Weigel, B.L.; Erwin, P.M. Effects of reciprocal transplantation on the microbiome and putative nitrogen cycling functions of the intertidal sponge, Hymeniacidon heliophila. Sci. Rep. 2017, 7, 43247. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Hu, J.; Lu, H. Histological observation of a gelatin sponge transplant loaded with bone marrow-derived mesenchymal stem cells combined with platelet-rich plasma in repairing an annulus defect. PLoS ONE 2017, 12, e0171500. [Google Scholar] [CrossRef]
- Guo, J.; Wen, X. Performance and kinetics of benzo(a)pyrene biodegradation in contaminated water and soil and improvement of soil properties by biosurfactant amendment. Ecotoxicol. Environ. Saf. 2021, 207, 111292. [Google Scholar] [CrossRef]
- Ray, M.; Kumar, V.; Banerjee, C.; Gupta, P.; Singh, S.; Singh, A. Investigation of biosurfactants produced by three indigenous bacterial strains, their growth kinetics and their anthracene and fluorene tolerance. Ecotoxicol. Environ. Saf. 2021, 208, 111621. [Google Scholar] [CrossRef]
- Smułek, W.; Sydow, M.; Matejuk, Z.J.; Kaczorek, E. Bacteria involved in biodegradation of creosote PAH—A case study of long-term contaminated industrial area. Ecotoxicol. Environ. Saf. 2020, 187, 109843. [Google Scholar] [CrossRef] [PubMed]
- Tziveleka, L.-A.; Ioannou, E.; Tsiourvas, D.; Berillis, P.; Foufa, E.; Roussis, V. Collagen from the marine sponges Axinella cannabina and Suberites carnosus: Isolation and morphological, biochemical, and biophysical characterization. Mar. Drugs 2017, 15, 152. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, A.; Provoste, F.; Rodríguez, M.; Prieto, A.L. A mechanistic model to assess the fate of naphthalene and benzo(A)pyrene in a chilean wwtp. Processes 2021, 9, 1313. [Google Scholar] [CrossRef]
- Scheuch, G.A.; Zuñiga, J.R.; Fuentes, E.; Bravo, D.; Donoso, J.M.P. Effect of co-contamination by PAHs and heavy metals on bacterial communities of diesel contaminated soils of south shetland islands, antarctica. Microorganisms 2020, 8, 1749. [Google Scholar] [CrossRef] [PubMed]
- Marzuki, I.; Pratama, I.; Ismail, H.E.; Paserangi, I.; Kamaruddin, M.; Chaerul, M.; Ahmad, R. The Identification and Distribution Components of Polycyclic Aromatic Hydrocarbon Contaminants at the Port of Paotere, Makassar, South Sulawesi. In Proceedings of the the 1st International Conference on Biotechnology and Food Sciences, Surabaya, Indonesia, 11 September 2021; Volume 679, p. 012017. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Sherman, N. Microbiology: A Laboratory Manual, 10th ed.; Rockland Community College, Inc., Publishing as Benjamin Cummings: San Fransisco, CA, USA, 2014; pp. 110–168. [Google Scholar]



| Sample Code | Coordinate | Salinity (‰) | Temperature ((°C) | pH | TDS (mg/L) | EC (ds/m) | Depth MSL (m) | Distance from the Beach (m) |
|---|---|---|---|---|---|---|---|---|
| Sp1 | 5°6′ 38, 12376″ S | 28.3 | 29.4 | 7.47 | 7.41 | 14.46 | 3.20 | 200 |
| 119° 17′70, 76544″ E | ||||||||
| Sp2 | 5°6′ 11, 62476″ S | 28.1 | 30.9 | 7.69 | 7.21 | 14.20 | 3.74 | 250 |
| 119° 17′60, 06228″ E | ||||||||
| Sp3 | 5°6′ 23, 55372″ S | 27.3 | 30.3 | 7.70 | 7.50 | 12.87 | 4.25 | 370 |
| 190° 20′27, 62376″ E |
| Parameters | Marine Sponge Symbiont Bacteria Isolate | ||
|---|---|---|---|
| Sp1 (Niphates sp.) | Sp2 (Hyrtios erectus) | Sp3 (Clathria (Thalysias) reinwardtii) | |
| Morphology | jagged stem shape, cream color, clustered distribution, endospore is less clear | jagged stem shape, brown color, separate distribution with endospores | round shape, bluish-cream color, clustered distribution, endospores are less clear |
| Gram staining | reaction (−) with safranin reagent and (−) with 1% KOH alkaline solvent, Gram (+) | fixed color with safranin reagent and (−) with 1% KOH alkaline solvent, Gram (+) | reaction (−) with safranin and (−) with 1% KOH alkaline solvent, Gram (+) |
| Indole test | − | − | + |
| Triple sugar iron agar (TSIA) | + | + | − |
| Nitrate test | + | + | − |
| Simmons citrate test | − | + | + |
| Methyl red (Mr) test | − | + | + |
| Voges–Proskauer (VP) test | + | − | + |
| Urease test | + | + | + |
| Provisional guess | Bacillus group | Pseudomonas group | Acinetobacter group |
| Bacterial Isolate | Sequence Samples | Range Sequence Gen-Bank | Identities Quality (%) | Gaps (%) | Species Type |
|---|---|---|---|---|---|
| Sp1-Bc | 15–967 (952) | 710,748–711,700 (952) | 922/956 (96.44) | 6/956 (0.63) | Bacillus pumilus strain GLB197 |
| Sp2-Ps | 14–975 (961) | 3,666,632–3,667,587 (955) | 890/974 (91.14) | 30/974 (3.08) | Pseudomonas stutzeri strain SLG510A3-8 |
| Sp3-Ac | 9–961 (952) | 12–964 (952) | 951/954 (99.69) | 2/954 (0.21) | Acinetobacter calcoaceticus strain SLCDA976 |
| Biodegradation Parameters | Interaction Period (Days) | |||||
|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 15 | 20 | 25 | |
| Turbidity of interaction media (NTU) | 1.01 | 6.56 | 12.82 | 17.62 | 24.83 | 26.43 |
| Temperature (°C) | 29 | 29 | 30 | 30 | 30 | 29 |
| pH | 6.67 | 6.68 | 6.13 | 6.10 | 6.06 | 6.43 |
| Abundance of gas bubbles | nt | nt | + | ++ | ++ | + |
| Fermentation smell | nt | nt | √ | √√ | √√ | √√ |
| Peak Number | Retention Time | Comp. Peak Height (106) | Quality (%) | Total Conc. (%) | Group of Comp. | Approximate Comp. on the Library/ ID |
|---|---|---|---|---|---|---|
| Interaction time 5 days | ||||||
| 1 | 10.287 | 0.182 | 95 | 1.057 | Aromatic | Naphthalene |
| 2 | 17.468 | 0.120 | 47 | 0.424 | Aldehyde | --- |
| 3 | 18.566 | 10.407 | 95 | 45.693 | Aromatic ** | Anthracene |
| 4 | 21.819 | 10.902 | 96 | 51.914 | Aromatic ** | Pyrene |
| 5 | 24.947 | 0.223 | 90 | 0.912 | Alcohol | Phenol |
| Interaction time 10 days | ||||||
| 1 | 6.083 | 0.132 | 91 | 0.959 | Methyl | Cyclotetrasiloxane |
| 2 | 17.471 | 0.794 | 53 | 3.752 | Aldehyde | --- |
| 3 | 18.559 | 8.568 | 95 | 40.589 | Aromatic ** | Anthracene |
| 4 | 21.309 | 0.239 | 78 | 1.401 | Aldehyde | --- |
| 5 | 21.809 | 9.113 | 96 | 50.686 | Aromatic ** | Pyrene |
| 6 | 24.944 | 0.185 | 90 | 1.070 | Alcohol | Phenol |
| 7 | 26.824 | 0.240 | 62 | 1.543 | Carboxylic acid | --- |
| Interaction time 15 days | ||||||
| 1 | 6.088 | 0.109 | 91 | 0.558 | Organosilicon | Cyclotetrasiloxane |
| 2 | 17.471 | 0.471 | 53 | 1.663 | Carboxylate | --- |
| 3 | 18.564 | 7.233 | 96 | 38.311 | Aromatic ** | Anthracene |
| 4 | 21.311 | 0.139 | 72 | 0.561 | Indole, aldehyde | --- |
| 5 | 21.818 | 8.920 | 96 | 49.619 | Aromatic ** | Pyrene |
| 6 | 22.085 | 0.080 | 93 | 0.384 | Amide | Hexadecanamide |
| 7 | 23.637 | 0.187 | 91 | 0.760 | Amide | 9-octadecetamide |
| 8 | 23.685 | 0.123 | 93 | 1.084 | Amide | 9-octadecenamide |
| 9 | 24.944 | 0.506 | 93 | 1.824 | Alcohol aromatic | Phenol |
| 10 | 26.823 | 0,700 | 81 | 3.236 | Carboxylate | --- |
| Interaction time 20 days | ||||||
| 1 | 16.728 | 0.131 | 98 | 1.858 | Alcohol | Benzenemethanol |
| 2 | 17.467 | 0.139 | 53 | 0.982 | Carboxylate | --- |
| 3 | 18.553 | 4.844 | 95 | 36.105 | Aromatic ** | Anthracene |
| 4 | 20.329 | 0.141 | 46 | 0.946 | Aliphatic | --- |
| 5 | 20.776 | 0.199 | 43 | 1.323 | Aliphatic | --- |
| 6 | 21.803 | 7.980 | 96 | 48.603 | Aromatic ** | Pyrene |
| 7 | 24.944 | 0.163 | 93 | 1.168 | Alcohol aromatic | Phenol |
| 8 | 26.823 | 0.219 | 49 | 2.016 | Carboxylate | Tetraphtelic acid |
| Interaction time 25 days | ||||||
| 1 | 17.470 | 0.242 | 64 | 1.112 | Methylene | --- |
| 2 | 18.560 | 4.132 | 95 | 35.297 | Aromatic ** | Anthracene |
| 3 | 20.332 | 0.148 | 47 | 0.697 | Sulfurous acid | --- |
| 4 | 20.778 | 0.332 | 47 | 1.508 | Aliphatic | --- |
| 5 | 21.140 | 0.178 | 47 | 1.253 | Alcohol aromatic | --- |
| 6 | 21.811 | 7.902 | 96 | 47.959 | Aromatic ** | Pyrene |
| 7 | 24.944 | 0.126 | 78 | 0.681 | Alcohol aromatic | --- |
| 8 | 26.823 | 0.234 | 91 | 1.504 | Carboxylate | Tetraphtelic acid |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzuki, I.; Asaf, R.; Paena, M.; Athirah, A.; Nisaa, K.; Ahmad, R.; Kamaruddin, M. Anthracene and Pyrene Biodegradation Performance of Marine Sponge Symbiont Bacteria Consortium. Molecules 2021, 26, 6851. https://doi.org/10.3390/molecules26226851
Marzuki I, Asaf R, Paena M, Athirah A, Nisaa K, Ahmad R, Kamaruddin M. Anthracene and Pyrene Biodegradation Performance of Marine Sponge Symbiont Bacteria Consortium. Molecules. 2021; 26(22):6851. https://doi.org/10.3390/molecules26226851
Chicago/Turabian StyleMarzuki, Ismail, Ruzkiah Asaf, Mudian Paena, Admi Athirah, Khairun Nisaa, Rasheed Ahmad, and Mudyawati Kamaruddin. 2021. "Anthracene and Pyrene Biodegradation Performance of Marine Sponge Symbiont Bacteria Consortium" Molecules 26, no. 22: 6851. https://doi.org/10.3390/molecules26226851
APA StyleMarzuki, I., Asaf, R., Paena, M., Athirah, A., Nisaa, K., Ahmad, R., & Kamaruddin, M. (2021). Anthracene and Pyrene Biodegradation Performance of Marine Sponge Symbiont Bacteria Consortium. Molecules, 26(22), 6851. https://doi.org/10.3390/molecules26226851












