Optimization of Fermentation Conditions for Carrageenase Production by Cellulophaga Species: A Comparative Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Bacterial Strains
2.2.2. Optimization of Culture Medium by One-Factor-at-a-Time Experiment
2.2.3. Bacterial Growth Conditions
2.2.4. Determination of Carrageenase Activity
2.2.5. Effect of Temperature and pH on Enzymatic Degradation
2.2.6. Size-Exclusion Chromatography
2.2.7. Statistical Analysis
3. Results and Discussion
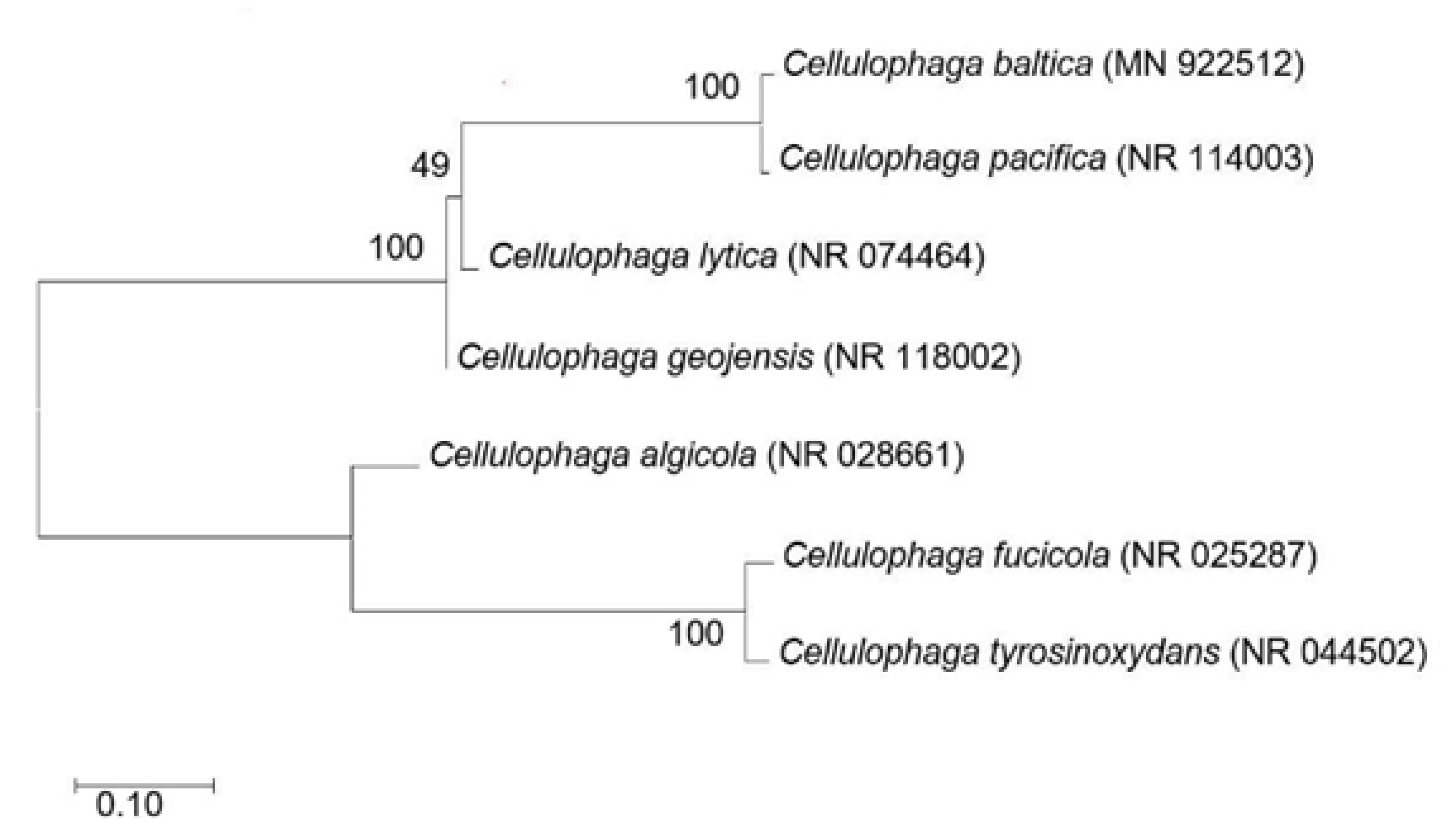
3.1. Phylogeny Analysis
3.2. Optimal Substrate Specificity
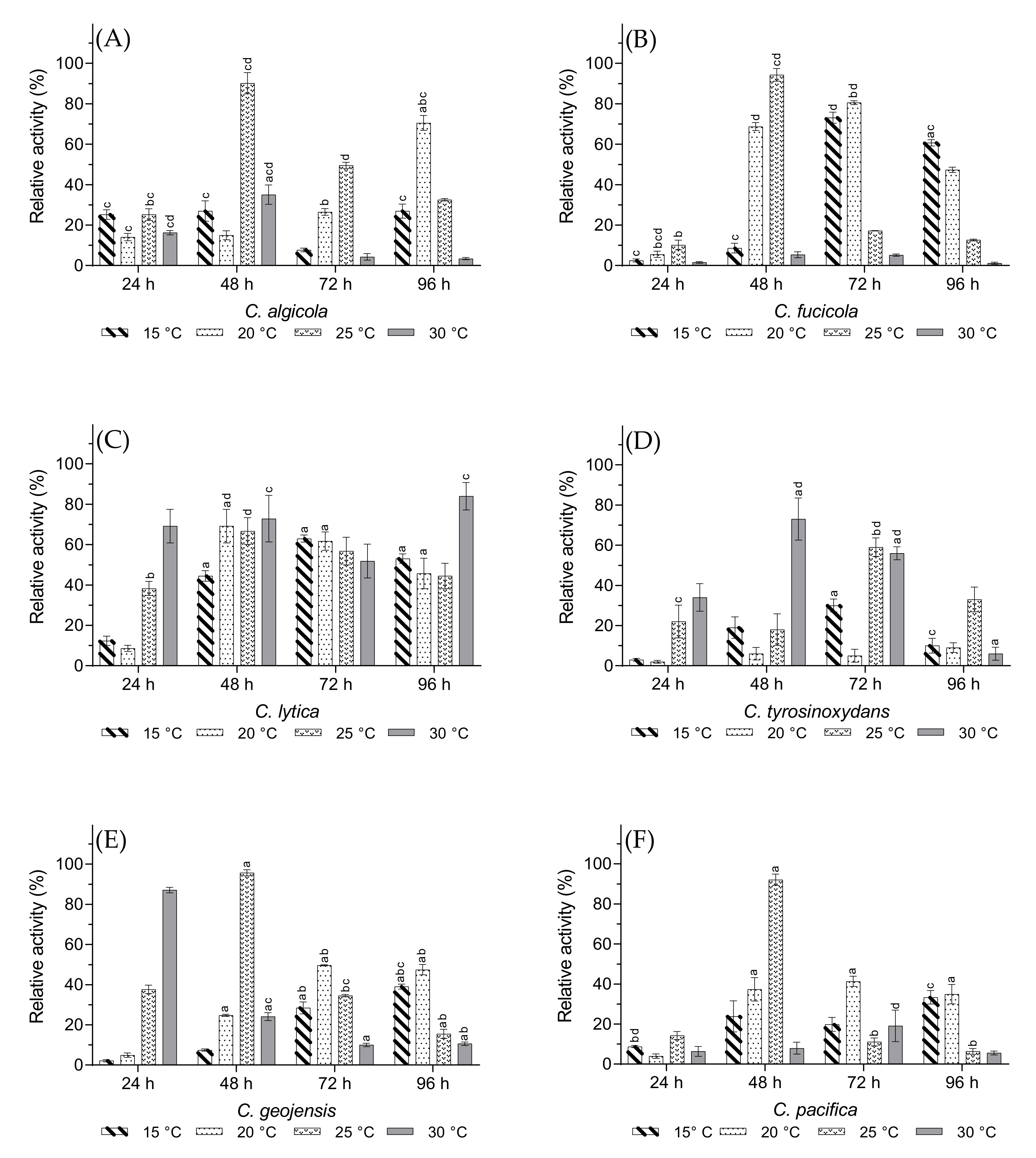
3.3. Optimal Growth Temperature and Incubation Time
3.4. Optimal Sea Salt Concentration
3.5. Optimal Nitrogen Source
3.6. Optimal Yeast Extract Concentration
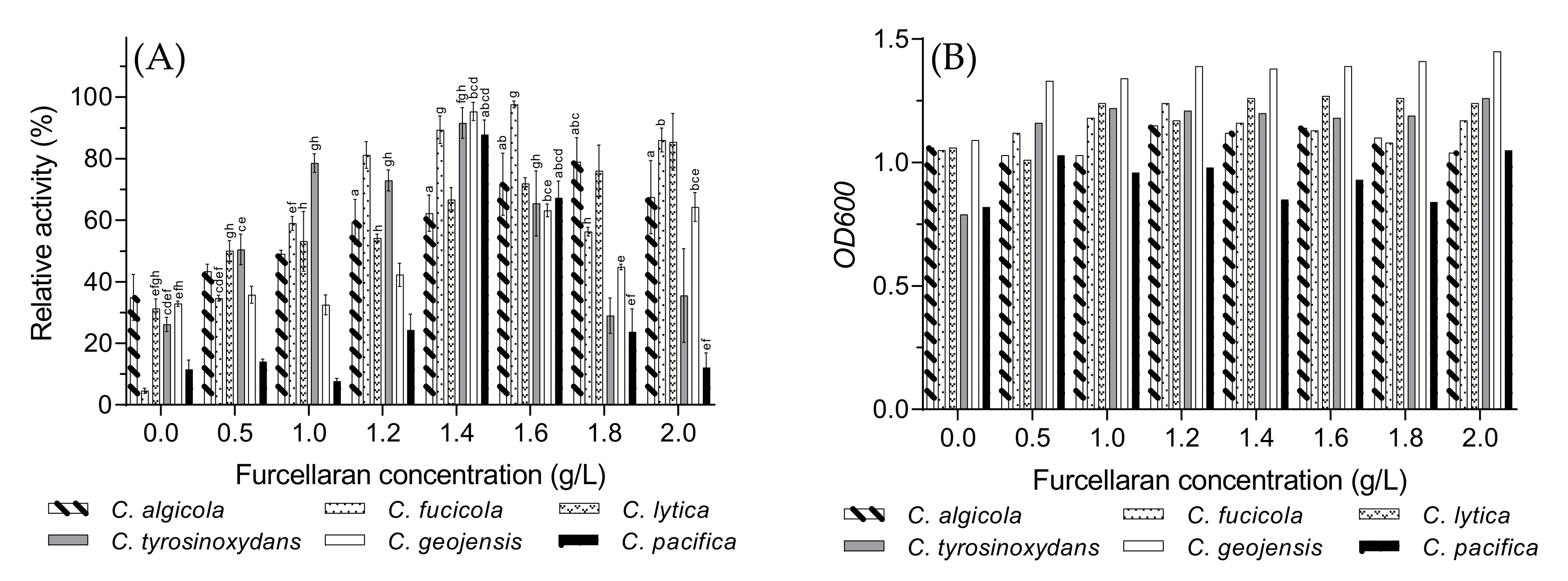
3.7. Optimal Furcellaran Concentration
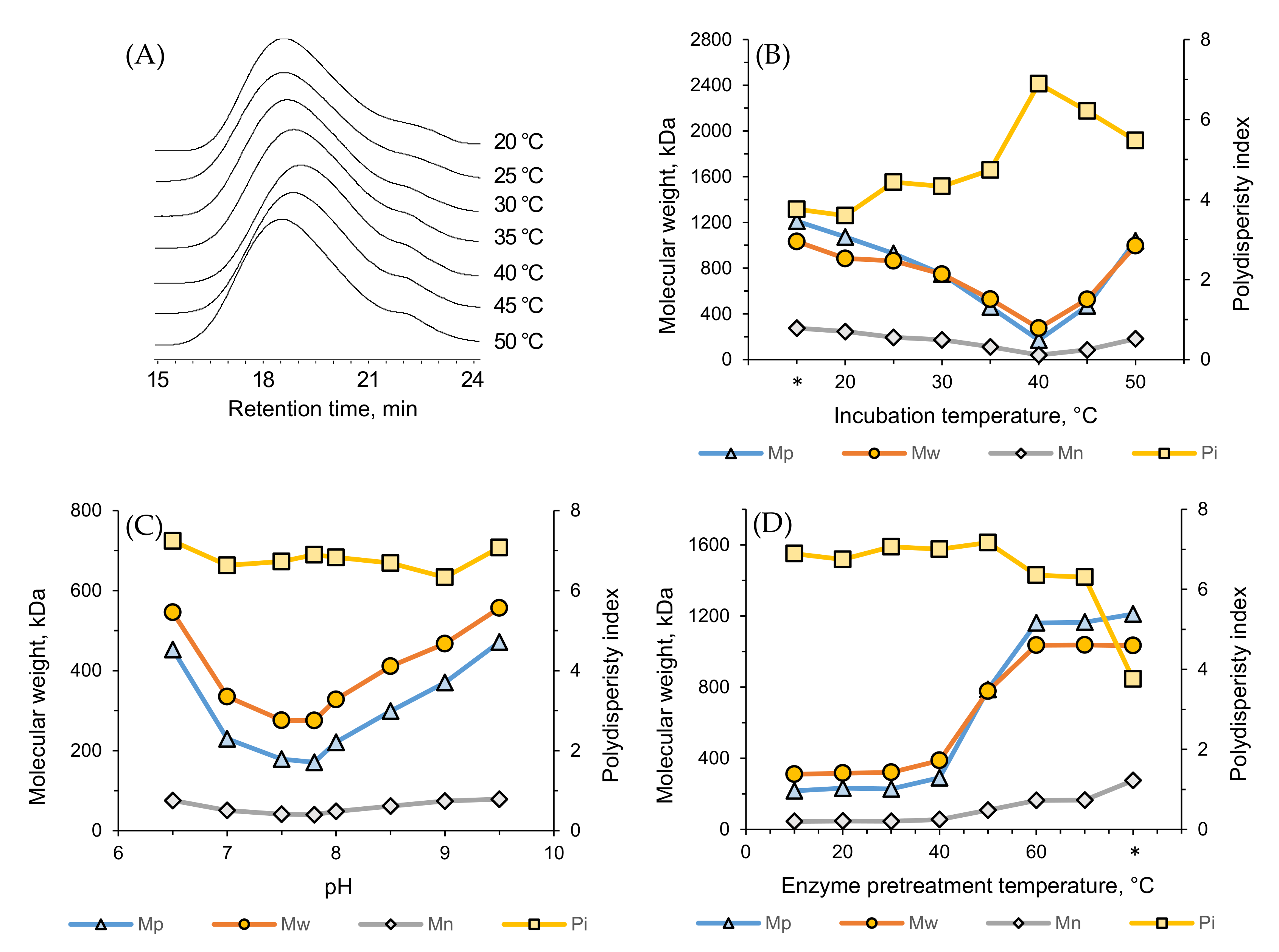
3.8. Effect of Temperature and pH on Enzymatic Degradation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, S.; Jianhua, H.; Mi, S. Cloning and characterization of a new cold-adapted and thermo-tolerant ι-carrageenase from marine bacterium. Int. J. Biol. Macromol. 2017, 102, 1059–1065. [Google Scholar] [CrossRef]
- Li, S.; He, N.; Han, Q.; Li, X.; Jung, S.; Suk Lee, B.; Kumar Mongre, R.; Wang, Z.P.; Wang, L.; Lee, M.S. Production of a thermo-tolerant κ-carrageenase via a food-grade host and anti-oxidant activity of its enzymatic hydrolysate. Food Chem. 2021, 339, 128027. [Google Scholar] [CrossRef]
- Li, J.; Hu, Q.; Seswita-Zilda, D. Purification and characterization of a thermostable λ-carrageenase from a hot spring bacterium, Bacillus sp. Biotechnol. Lett. 2014, 36, 1669–1674. [Google Scholar] [CrossRef]
- Mckim, J.M. Food additive carrageenan: Part I: A critical review of carrageenan in vitro studies, potential pitfalls, and implications for human health and safety. Crit. Rev. Toxicol. 2014, 8444, 1–33. [Google Scholar] [CrossRef]
- Singh, P.; Arunika, C. Bacterial carrageenases: An overview of production and biotechnological applications. 3 Biotech 2016, 6, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Shi, Q.; Wang, A.; Lu, Z.; Qin, C.; Hu, J.; Yin, J. Overview on the antiviral activities and mechanisms of marine polysaccharides from seaweeds. Carbohydr. Res. 2017, 453–454, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Sun, G.; Li, J.; Wang, L. A green strategy for maintaining intelligent response and improving antioxidant properties of κ-carrageenan-based film via cork bark extractive addition. Food Hydrocoll. 2020, 113, 106470. [Google Scholar] [CrossRef]
- Saluri, K.; Tuvikene, R. Anticoagulant and antioxidant activity of lambda- and theta-carrageenans of different molecular weights. Bioact. Carbohydr. Diet. Fibre 2020, 24, 100243. [Google Scholar] [CrossRef]
- Sousa, W.M.; Silva, R.O.; Bezerra, F.F.; Bingana, R.D.; Barros, F.C.N.; Costa, L.E.C.; Sombra, V.G.; Soares, P.M.G.; Feitosa, J.P.A.; de Paula, R.C.M.; et al. Sulfated polysaccharide fraction from marine algae Solieria filiformis: Structural characterization, gastroprotective and antioxidant effects. Carbohydr. Polym. 2016, 152, 140–148. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, P.; Yu, G.L.; Li, C.X.; Hao, C.; Qi, X.; Zhang, L.J.; Guan, H.S. Preparation and anti-influenza A virus activity of κ-carrageenan oligosaccharide and its sulphated derivatives. Food Chem. 2012, 133, 880–888. [Google Scholar] [CrossRef]
- Amin, M.L.; Mawad, D.; Dokos, S.; Koshy, P.; Martens, P.J.; Sorrell, C.C. Immunomodulatory properties of photopolymerizable fucoidan and carrageenans. Carbohydr. Polym. 2020, 230, 115691. [Google Scholar] [CrossRef]
- Guo, J.; Han, S.; Lu, X.; Guo, Z.; Zeng, S.; Zheng, X.; Zheng, B. κ-Carrageenan hexamer have significant anti-inflammatory activity and protect RAW264.7 Macrophages by inhibiting CD14. J. Funct. Foods 2019, 57, 335–344. [Google Scholar] [CrossRef]
- Sun, Y.; Yang, B.; Wu, Y.; Liu, Y.; Gu, X.; Zhang, H.; Wang, C. Structural characterization and antioxidant activities of κ-carrageenan oligosaccharides degraded by different methods. Food Chem. 2015, 178, 311–318. [Google Scholar] [CrossRef]
- Leiter, A.; Mailänder, J.; Wefers, D.; Bunzel, M.; Gaukel, V. Influence of acid hydrolysis and dialysis of κ-carrageenan on its ice recrystallization inhibition activity. J. Food Eng. 2017, 209, 26–35. [Google Scholar] [CrossRef]
- Muzyed, S.; Howlader, M.M.; Tuvikene, R. Fermentation optimization, purification and biochemical characterization of ι-carrageenase from marine bacterium Cellulophaga baltica. Int. J. Biol. Macromol. 2021, 166, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Abad, L.V.; Aurigue, F.B.; Montefalcon, D.R.V.; Manguiat, P.H.; Carandang, F.F.; Mabborang, S.A.; Hizon, M.G.S.; Abella, M.E.S. Effect of radiation-modified kappa-carrageenan as plant growth promoter on peanut (Arachis hypogaea L.). Radiat. Phys. Chem. 2018, 153, 239–244. [Google Scholar] [CrossRef]
- Kamińska-Dwórznicka, A.; Antczak, A.; Samborska, K.; Lenart, A. Acid hydrolysis of kappa-carrageenan as a way of gaining new substances for freezing process modification and protection from excessive recrystallisation of ice. Int. J. Food Sci. Technol. 2015, 50, 1799–1806. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, L.; Lu, X.; Zeng, S.; Zhang, Y.; Xu, H.; Zheng, B. Medium optimization and fermentation kinetics for κ-carrageenase production by Thalassospira sp. Fjfst-332. Molecules 2016, 21, 1479. [Google Scholar] [CrossRef]
- Yao, Z.; Wu, H.; Zhang, S.; Du, Y. Enzymatic preparation of κ-carrageenan oligosaccharides and their anti-angiogenic activity. Carbohydr. Polym. 2014, 101, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Dyrset, N.; Lystad, K.Q.; Levine, D.W. Development of a fermentation process for production of a κ-carrageenase from Pseudomonas carrageenovora. Enzyme Microb. Technol. 1997, 20, 418–423. [Google Scholar] [CrossRef]
- Tayco, C.C.; Tablizo, F.A.; Regalia, R.S.; Lluisma, A.O. Characterization of a κ-carrageenase-producing marine bacterium, Isolate ALAB-001. Philipp. J. Sci. 2013, 142, 45–54. [Google Scholar]
- Potin, P.; Sanseau, A.; Le Gall, Y.; Rochas, C.; Kloareg, B. Purification and characterization of a new κ-carrageenase from a marine Cytophaga-like bacterium. Eur. J. Biochem. 1991, 201, 241–247. [Google Scholar] [CrossRef]
- Cui, H.; Peng, Y.; Zhao, B.; Liu, Y.; Chen, F.; Wu, H.; Yao, Z. Cloning, identification and characterization of a novel κ-carrageenase from marine bacterium Cellulophaga lytica strain N5-2. Int. J. Biol. Macromol. 2017, 105, 509–515. [Google Scholar] [CrossRef]
- Perepelov, A.V.; Shashkov, A.S.; Tomshich, S.V.; Komandrova, N.A.; Nedashkovskaya, O.I. Structure of the O-specific polysaccharide from a marine bacterium Cellulophaga pacifica containing rarely occurred sugars, Fuc4NAc and ManNAcA. Carbohydr. Res. 2013, 372, 69–72. [Google Scholar] [CrossRef]
- Tomshich, S.V.; Kokoulin, M.S.; Kalinovsky, A.I.; Nedashkovskaya, O.I.; Komandrova, N.A. Structure of the O-specific polysaccharide from a marine bacterium Cellulophaga tyrosinoxydans. Carbohydr. Res. 2015, 413, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.E.; Nielsen, P.; Sjøholm, C. Description of Cellulophaga baltica gen. nov., sp. nov. and Cellulophaga fucicola gen. nov., sp. nov. and reclassification of Cytophaga lytica to Cellulophaga lytica gen. nov., comb. nov. Int. J. Syst. Bacteriol. 1999, 49, 1231–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Oh, K.H.; Lee, S.Y.; Oh, T.K.; Yoon, J.H. Cellulophaga geojensis sp. nov., a member of the family Flavobacteriaceae isolated from marine sand. Int. J. Syst. Evol. Microbiol. 2012, 62, 1354–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, J.P. Description of Cellulophaga algicola sp. nov., isolated from the surfaces of Antarctic algae, and reclassification of Cytophaga uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Cellulophaga uliginosa comb. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 1861–1868. [Google Scholar] [CrossRef] [Green Version]
- Kahng, H.Y.; Chung, B.S.; Lee, D.H.; Jung, J.S.; Park, J.H.; Jeon, C.O. Cellulophaga tyrosinoxydans sp. nov., a tyrosinase-producing bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 2009, 59, 654–657. [Google Scholar] [CrossRef] [Green Version]
- Holder, E.L.; Conmy, R.N.; Venosa, A.D. Comparative laboratory-scale testing of dispersant effectiveness of 23 crude oils using four different testing protocols. J. Environ. Prot. 2015, 6, 628–639. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Q.; Zhu, Y.; Li, J.; Wu, C.; Ni, H.; Xiao, A. Fermentation optimization and enzyme characterization of a new ι-carrageenase from Pseudoalteromonas carrageenovora ASY5. Electron. J. Biotechnol. 2018, 32, 26–34. [Google Scholar] [CrossRef]
- Guibet, M.; Barbeyron, T.; Genicot, S.; Kloareg, B.; Michel, G.; Helbert, W. Degradation of λ-carrageenan by Pseudoalteromonas carrageenovora λ-carrageenase: A new family of glycoside hydrolases unrelated to κ-and ι-carrageenases. Biochem. J. 2007, 404, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Araki, T.; Yoshifumi, H.; Tatsuo, M. Purification and characterization of κ-carrageenase from a marine bacterium, Vibrio sp. CA-1004. Fish. Sci. 1999, 65, 937–942. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Mao, W.; Gao, W.; Chi, Z.; Chi, Z.; Liu, G. Efficient production of a recombinant ι-carrageenase in Brevibacillus choshinensis using a new integrative vector for the preparation of ι-carrageenan oligosaccharides. Process Biochem. 2019, 76, 68–76. [Google Scholar] [CrossRef]
- Ma, S.; Tan, Y.; Yu, W.; Han, F. Cloning, expression and characterization of a new ℩-carrageenase from marine bacterium, Cellulophaga sp. Biotechnol. Lett. 2013, 35, 1617–1622. [Google Scholar] [CrossRef]
- Reimer, L.C.; Vetcininova, A.; Carbasse, J.S.; Söhngen, C.; Gleim, D.; Ebeling, C.; Overmann, J. BacDive in 2019: Bacterial phenotypic data for High-throughput biodiversity analysis. Nucleic Acids Res. 2019, 47, D631–D636. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.-C.; Kim, J.-T.; Kim, S.-H.; Rho, H.-K. Fluctuation characteristic of temperature and salinity in coastal waters around Jeju Island. Korean J. Fish. Aquat. Sci. 2003, 36, 306–316. [Google Scholar]
- Zhou, M.; Jian-she, M.; Jun, L.; Hai-ren, Y.; Ke-xin, H.; Zhao, X. A κ-carrageenase from a newly isolated Pseudoalteromonas-like bacterium, WZUC10. Biotechnol. Bioprocess Eng. 2008, 13, 545–551. [Google Scholar] [CrossRef]
- Senjyu, T. The Japan Sea Intermediate Water; its characteristics and circulation. J. Oceanogr. 1999, 55, 111–122. [Google Scholar] [CrossRef]
- Beltagy, E.A.; Youssef, A.S.; El-Shenawy, M.A.; El-Assar, S.A. Purification of kappa (k)-carrageenase from locally isolated Cellulosimicrobium cellulans. African J. Biotechnol. 2014, 11, 11438–11446. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, L.; Chen, Y.; Mou, H. Efficient extracellular production of κ-carrageenase in Escherichia coli: Effects of wild-type signal sequence and process conditions on extracellular secretion. J. Biotechnol. 2014, 185, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Jouanneau, D.; Boulenguer, P.; Mazoyer, J.; Helbert, W. Enzymatic degradation of hybrid ι-/ν-carrageenan by Alteromonas fortis ι-carrageenase. Carbohydr. Res. 2010, 345, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Hatada, Y.; Mizuno, M.; Li, Z.; Yukari, O. Hyper-production and characterization of the ι-carrageenase useful for ι-carrageenan oligosaccharide production from a deep-sea Bacterium, Microbulbifer thermotolerans JAMB-A94 T, and insight into the unusual catalytic mechanism. Mar. Biotechnol. 2011, 13, 411–422. [Google Scholar] [CrossRef]
- Shen, J.; Chang, Y.; Dong, S.; Chen, F. Cloning, expression and characterization of a ι-carrageenase from marine bacterium Wenyingzhuangia fucanilytica: A biocatalyst for producing ι-carrageenan oligosaccharides. J. Biotechnol. 2017, 259, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Rebuffet, E.; Barbeyron, T.; Jeudy, A.; Jam, M.; Czjzek, M.; Michel, G. Identification of catalytic residues and mechanistic analysis of family. Biochemistry 2010, 49, 7590–7599. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Howlader, M.M.; Molz, J.; Sachse, N.; Tuvikene, R. Optimization of Fermentation Conditions for Carrageenase Production by Cellulophaga Species: A Comparative Study. Biology 2021, 10, 971. https://doi.org/10.3390/biology10100971
Howlader MM, Molz J, Sachse N, Tuvikene R. Optimization of Fermentation Conditions for Carrageenase Production by Cellulophaga Species: A Comparative Study. Biology. 2021; 10(10):971. https://doi.org/10.3390/biology10100971
Chicago/Turabian StyleHowlader, Md Musa, Jana Molz, Nico Sachse, and Rando Tuvikene. 2021. "Optimization of Fermentation Conditions for Carrageenase Production by Cellulophaga Species: A Comparative Study" Biology 10, no. 10: 971. https://doi.org/10.3390/biology10100971
APA StyleHowlader, M. M., Molz, J., Sachse, N., & Tuvikene, R. (2021). Optimization of Fermentation Conditions for Carrageenase Production by Cellulophaga Species: A Comparative Study. Biology, 10(10), 971. https://doi.org/10.3390/biology10100971






