Fishing of α-Glucosidase’s Ligands from Aloe vera by α-Glucosidase Functionalized Magnetic Nanoparticles
Abstract
:1. Introduction
2. Results
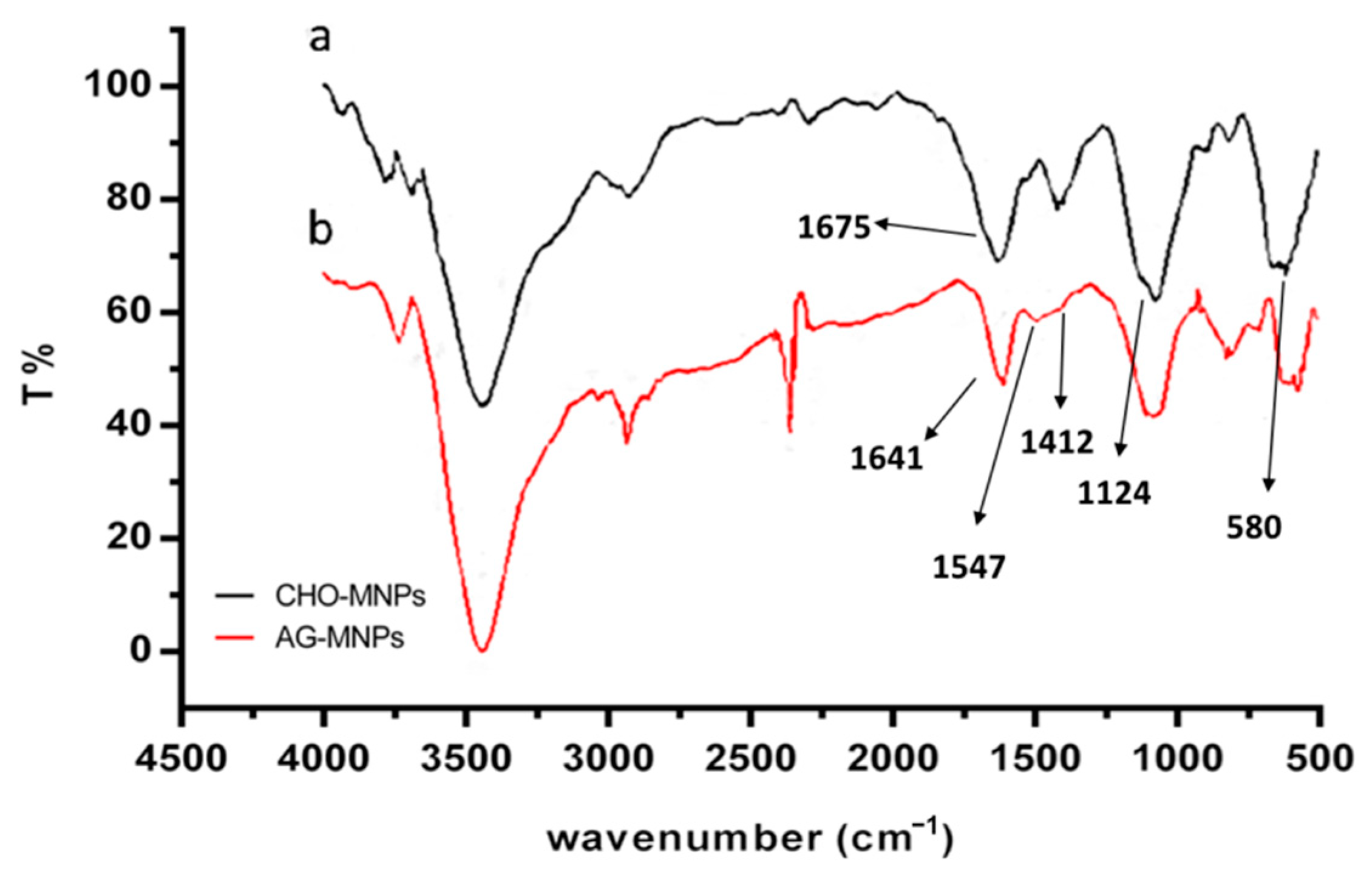
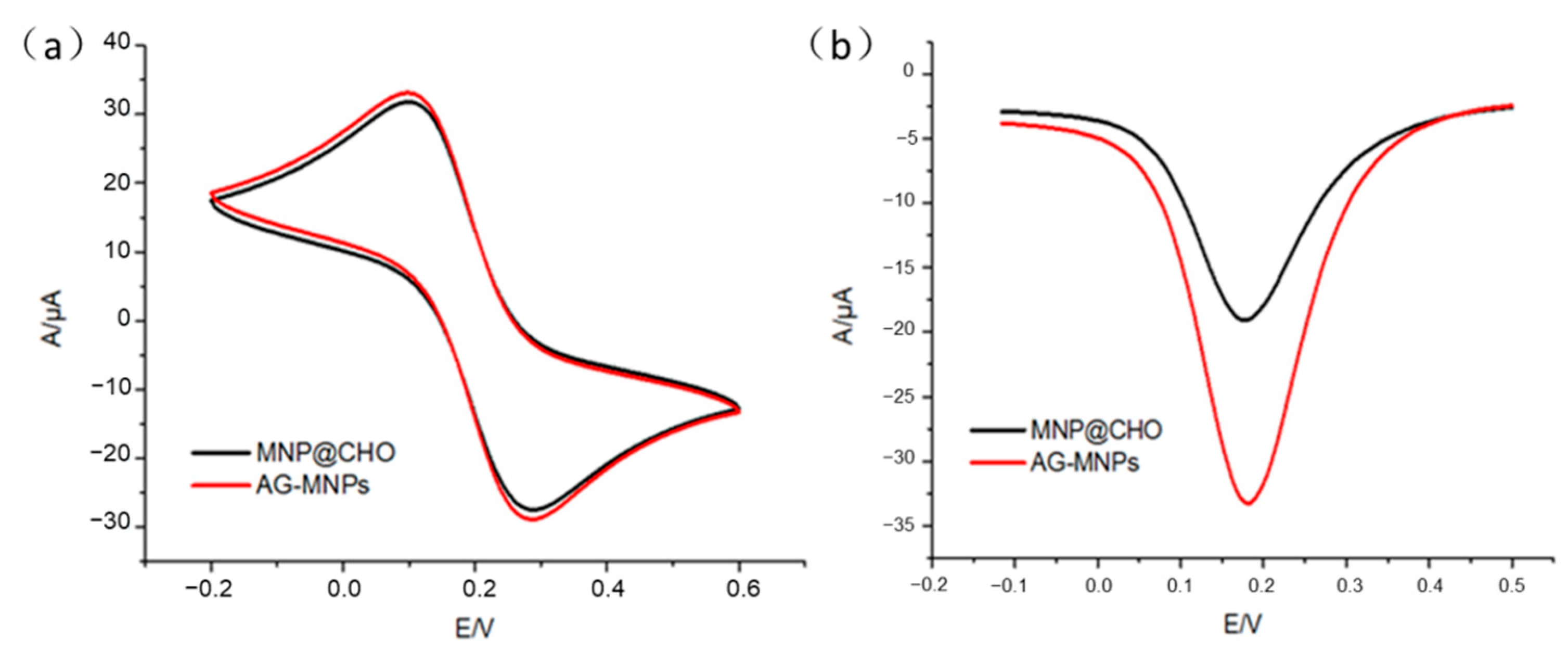
2.1. Characterization of AG-MNPs
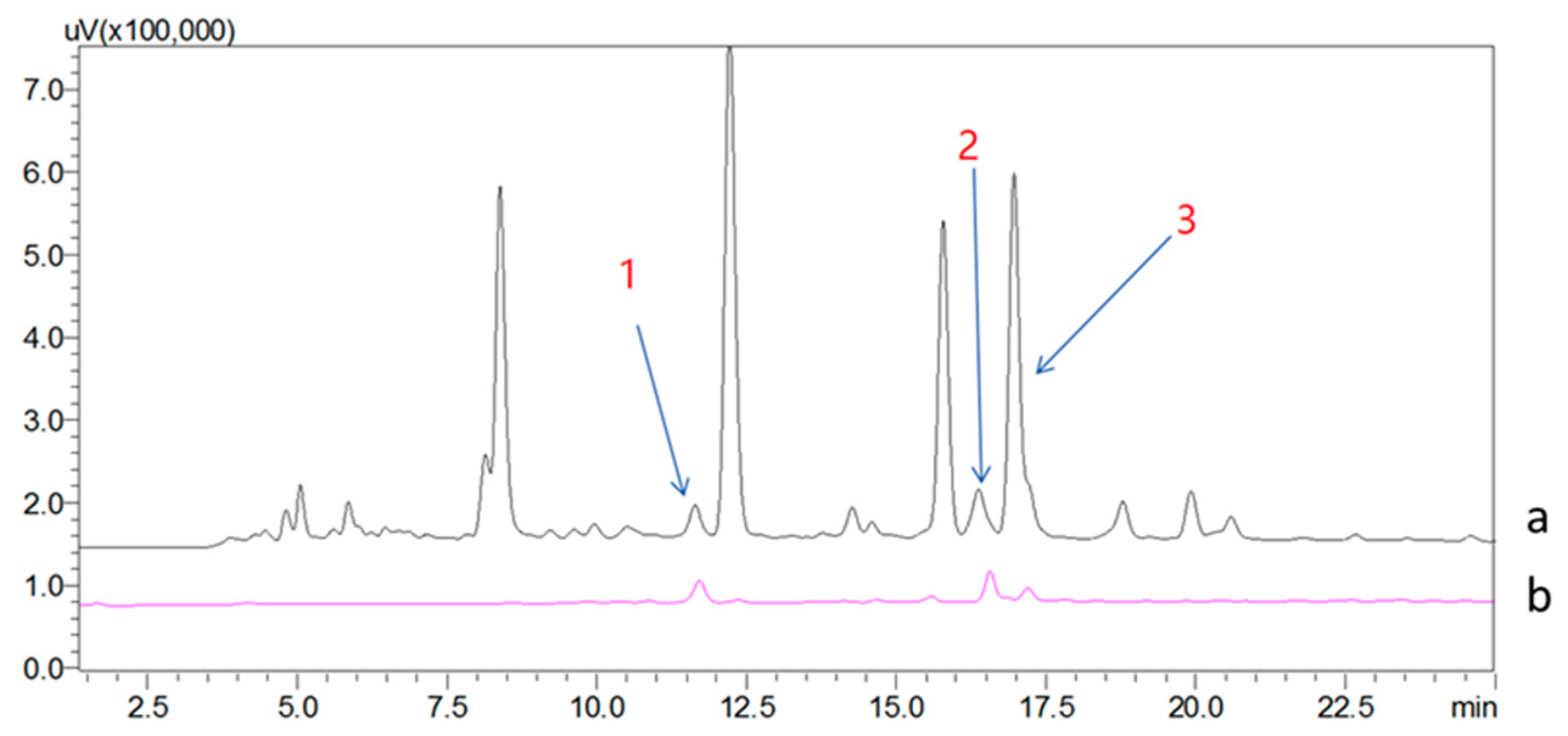
2.2. Screening and Structural Classification of α-Glucosidase Ligands from Extract of A. vera
2.3. Effects of the Ligands on the Enzymatic Activity of α-Glucosidase
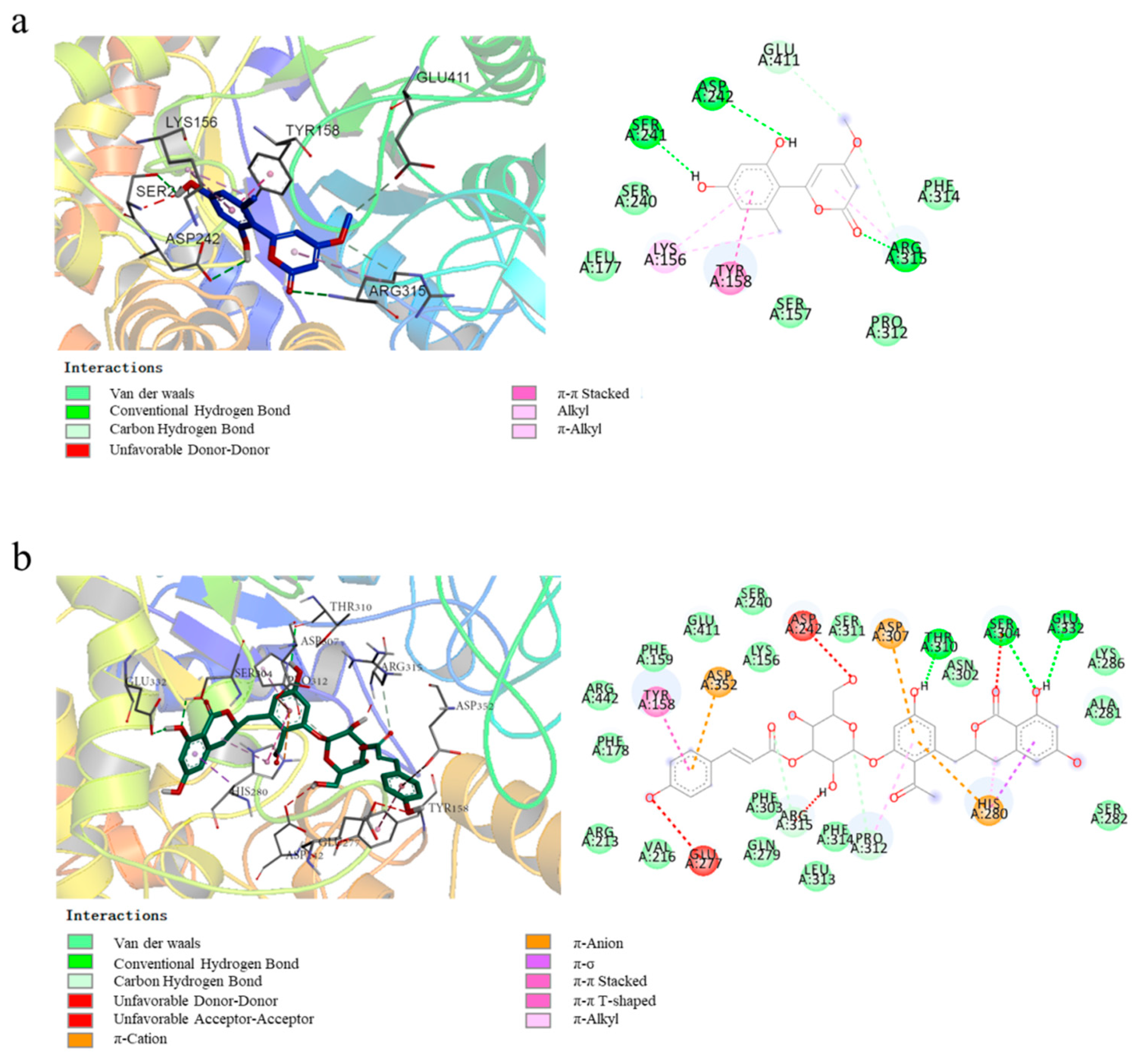
2.4. Molecular Docking Studies
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Apparatus and Instruments
3.3. Preparation of α-Glucosidase Functionalized MNPs
3.4. Characterization of α-Glucosidase Functionalized MNPs
3.5. Preparation of Extract of A. vera
3.6. Fishing of α-Glucosidase’s Ligands from A. vera
3.7. Enzymatic Activity Assay of the Enzyme’s Ligands
3.8. Molecular Docking Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Guariguata, L.; Whiting, D.R.; Hambleton, I.; Beagley, J.; Linnenkamp, U.; Shaw, J.E. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res. Clin. Pract. 2014, 103, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Ma, S.T. Recent Advances in Synthetic-Glucosidase Inhibitors. ChemMedChem 2017, 12, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Ghani, U. Re-exploring promising alpha-glucosidase inhibitors for potential development into oral anti-diabetic drugs: Finding needle in the haystack. Eur. J. Med. Chem. 2015, 103, 133–162. [Google Scholar] [CrossRef] [PubMed]
- He, J.H.; Chen, L.X.; Li, H. Progress in the discovery of naturally occurring anti-diabetic drugs and in the identification of their molecular targets. Fitoterapia 2019, 134, 270–289. [Google Scholar] [CrossRef] [PubMed]
- Ahlawat, K.S.; Khatkar, B.S. Processing, food applications and safety of Aloe vera products: A review. J. Food Sci. Technol-Mysore 2011, 48, 525–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yongchaiyudha, S.; Rungpitarangsi, V.; Bunyapraphatsara, N.; Chokechaijaroenporn, O. Antidiabetic activity of Aloe vera L juice. I. Clinical trial in new cases of diabetes mellitus. Phytomedicine 1996, 3, 241–243. [Google Scholar] [CrossRef]
- Huseini, H.F.; Kianbakht, S.; Hajiaghaee, R.; Dabaghian, F.H. Anti-hyperglycemic and Anti-hypercholesterolemic Effects of Aloe vera Leaf Gel in Hyperlipidemic Type 2 Diabetic Patients: A Randomized Double-Blind Placebo-Controlled Clinical Trial. Planta Med. 2012, 78, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Jong-Anurakkun, N.; Bhandari, M.R.; Hong, G.; Kawabata, J. Alpha-glucosidase inhibitor from Chinese aloes. Fitoterapia 2008, 79, 456–457. [Google Scholar] [CrossRef]
- Aldayel, T.S.; Grace, M.H.; Lila, M.A.; Yahya, M.A.; Omar, U.M.; Alshammary, G. LC-MS characterization of bioactive metabolites from two Yemeni Aloe spp. with antioxidant and antidiabetic properties. Arab. J. Chem. 2020, 13, 5040–5049. [Google Scholar] [CrossRef]
- Kim, J.H.; Cho, C.W.; Lee, J.I.; Vinh, L.; Kim, K.T.; Cho, I.S. An investigation of the inhibitory mechanism of alpha-glucosidase by chysalodin from Aloe vera. Int. J. Biol. Macromol. 2020, 147, 314–318. [Google Scholar] [CrossRef]
- Indrianingsih, A.W.; Tachibana, S.; Itoh, K. In vitro evaluation of antioxidant and alpha-Glucosidase inhibitory assay of several tropical and subtropical plants. In 5th Sustainable Future for Human Security; Trihartono, A., McLellan, B., Eds.; Elsevier Science Bv: Amsterdam, The Netherlands, 2015; Volume 28, pp. 639–648. [Google Scholar]
- Vuckovic, D.; Zhang, X.; Cudjoe, E.; Pawliszyn, J. Solid-phase microextraction in bioanalysis: New devices and directions. J. Chromatogr. A 2010, 1217, 4041–4060. [Google Scholar] [CrossRef]
- Kataoka, H. Current Developments and Future Trends in Solid-phase Microextraction Techniques for Pharmaceutical and Biomedical Analyses. Anal. Sci. 2011, 27, 893–905. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi, M.; Elmongy, H.; Madrakian, T.; Abdel-Rehim, M. Nanomaterials as sorbents for sample preparation in bioanalysis: A review. Anal. Chim. Acta 2017, 958, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.F.; Jiang, X.L.; Gong, Y.Z.; Hu, Y.D.; Bai, X.L.; Liao, X. Ligand fishing of anti-neurodegenerative components from Lonicera japonica using magnetic nanoparticles immobilised with monoamine oxidase B. J. Sep. Sci. 2019, 42, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.H.; Jiang, X.L.; Liu, Y.M.; Hu, J.J.; Liang, J.; Liao, X. Screening of lipase inhibitors from Scutellaria baicalensis extract using lipase immobilized on magnetic nanoparticles and study on the inhibitory mechanism. Anal. Bioanal. Chem. 2016, 408, 2275–2283. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.M.; Gray, A.I.; Reynolds, T.; Waterman, P.G. Anthraquinone, anthrone and phenylpyrone components of Aloe nyeriensis var. kedongensis leaf exudate. Phytochemistry 1987, 26, 2995–2997. [Google Scholar] [CrossRef]
- Speranza, G.; Dada, G.; Lunazzi, L.; Gramatica, P.; Manitto, P. Aloenin B, a New Diglucosylated 6-Phenyl-2-pyrone from Kenya Aloe. J. Nat. Prod. 1986, 49, 800–805. [Google Scholar] [CrossRef]
- Duri, L.; Morelli, C.F.; Crippa, S.; Speranza, G. 6-Phenylpyrones and 5-methylchromones from Kenya aloe. Fitoterapia 2004, 75, 520–522. [Google Scholar] [CrossRef]
- Kurizaki, A.; Watanabe, T.; Devkota, H.P. Chemical Constituents from the Flowers of Aloe arborescens. Nat. Prod. Commun. 2019, 14. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Xu, F.; Ji, T.F.; Li, J. A New Spermidine from the Fruits of Lycium ruthenicum. Chem. Nat. Compd. 2014, 50, 880–883. [Google Scholar] [CrossRef]
- Huang, S.Y.; Zou, X.Q. Advances and Challenges in Protein-Ligand Docking. Int. J. Mol. Sci. 2010, 11, 3016–3034. [Google Scholar] [CrossRef] [Green Version]
- Morris, C.J.; Della Corte, D. Using molecular docking and molecular dynamics to investigate protein-ligand interactions. Mod. Phys. Lett. B 2021, 35. [Google Scholar] [CrossRef]
- Zhu, Y.T.; Jia, Y.W.; Liu, Y.M.; Liang, J.; Ding, L.S.; Liao, X. Lipase Ligands in Nelumbo nucifera Leaves and Study of Their Binding Mechanism. J. Agric. Food Chem. 2014, 62, 10679–10686. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Xu, P.L.; Zeng, Q.; Liu, Y.M.; Ding, L.S.; Liao, X. Preparation of ds-DNA functionalized magnetic nanobaits for screening of bioactive compounds from medicinal plant. Mater. Sci. Eng. C-Mater. Biol. Appl. 2015, 56, 401–408. [Google Scholar] [CrossRef]
- Feng, J.; Yang, X.W.; Wang, R.F. Bio-assay guided isolation and identification of alpha-glucosidase inhibitors from the leaves of Aquilaria sinensis. Phytochemistry 2011, 72, 242–247. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Zeng, L.; Zhang, G.W.; Lin, S.Y.; Gong, D.M. Inhibitory Mechanism of Apigenin on alpha-Glucosidase and Synergy Analysis of Flavonoids. J. Agric. Food Chem. 2016, 64, 6939–6949. [Google Scholar] [CrossRef]
- Shen, Y.P.; Wang, M.; Zhou, J.W.; Chen, Y.F.; Wu, M.R.; Yang, Z.Z.; Yang, C.Y.; Xia, G.H.; Tam, J.P.; Zhou, C.S.; et al. Construction of Fe3O4@alpha-glucosidase magnetic nanoparticles for ligand fishing of alpha-glucosidase inhibitors from a natural tonic Epimedii Folium. Int. J. Biol. Macromol. 2020, 165, 1361–1372. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
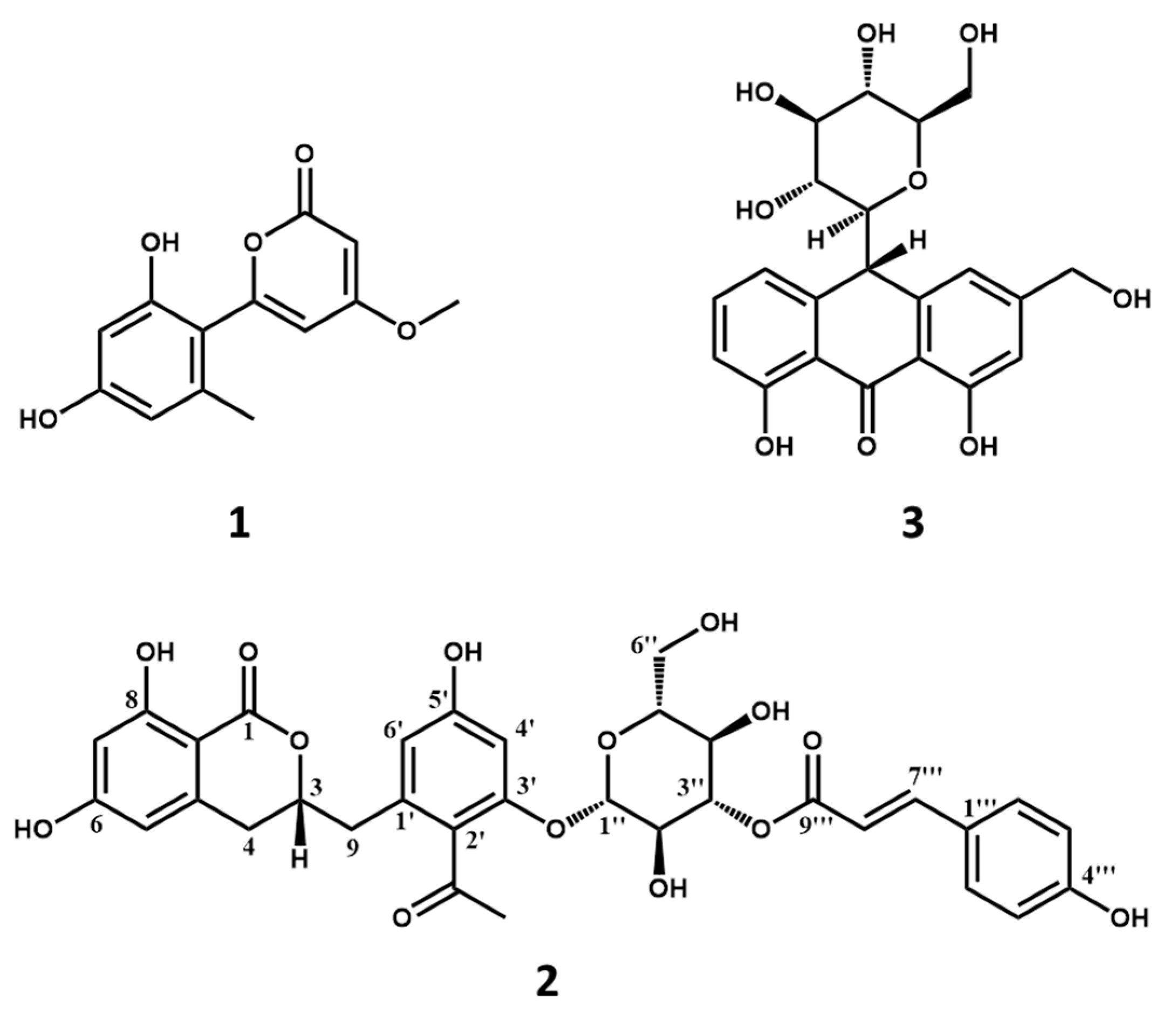
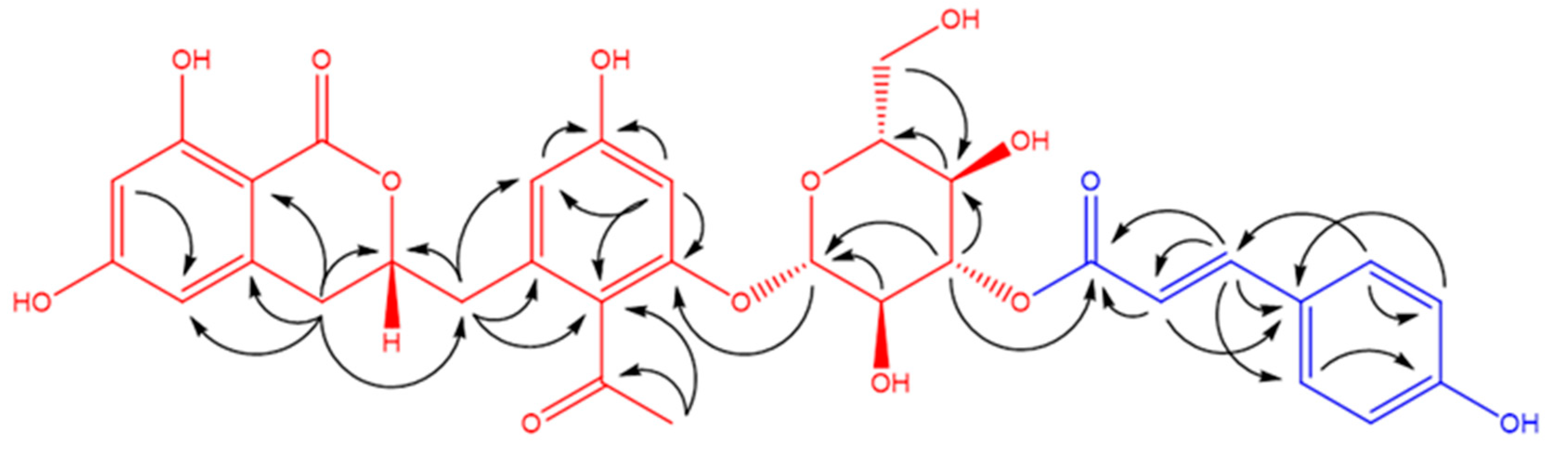

| 2 | |||||
|---|---|---|---|---|---|
| No. | δH | δC | No. | δH | δC |
| 1 | 166.8 | 1″ | 5.19, d, 8.0 | 98.9 | |
| 3 | 4.65, m | 79.8 | 2″ | 3.71, m | 74.8 |
| 4 | 2.76, m | 32.1 | 3″ | 5.13, m | 73.2 |
| 4a | 141.7 | 4″ | 3.55, m | 70.0 | |
| 5 | 6.16, m | 106.6 | 5″ | 3.55, m | 77.1 |
| 6 | 164.2 | 6″ | 3.97, dd, 12.0, 2.0 | 61.0 | |
| 7 | 6.17, m | 100.8 | 3.78, dd, 12.0, 5.4 | ||
| 8 | 164.8 | 1‴ | 125.9 | ||
| 8a | 100.1 | 2‴ | 7.43, d, 8.6 | 129.8 | |
| 9 | 2.98, dd, 13.8, 7.2 | 37.6 | 3‴ | 6.76, d, 8.6 | 115.4 |
| 2.84, dd, 13.8, 5.0 | 4‴ | 159.9 | |||
| 1′ | 136.8 | 5‴ | 6.76, d, 8.6 | 115.4 | |
| 2′ | 123.8 | 6‴ | 7.43, d, 8.6 | 129.8 | |
| 3′ | 156.3 | 7‴ | 7.62, d, 15.9 | 145.8 | |
| 4′ | 6.63, d, 2.0 | 101.1 | 8‴ | 6.31, d, 15.9 | 113.6 |
| 5′ | 159.5 | 9‴ | 166.8 | ||
| 6′ | 6.47, d, 2.0 | 112.0 | |||
| COCH3 | 205.5 | ||||
| COCH3 | 2.45, s | 31.9 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, H.; Wan, H.; Hu, Y.-K.; Ayeni, E.A.; Chang, Q.; Ma, C.; Liao, X. Fishing of α-Glucosidase’s Ligands from Aloe vera by α-Glucosidase Functionalized Magnetic Nanoparticles. Molecules 2021, 26, 5840. https://doi.org/10.3390/molecules26195840
Yuan H, Wan H, Hu Y-K, Ayeni EA, Chang Q, Ma C, Liao X. Fishing of α-Glucosidase’s Ligands from Aloe vera by α-Glucosidase Functionalized Magnetic Nanoparticles. Molecules. 2021; 26(19):5840. https://doi.org/10.3390/molecules26195840
Chicago/Turabian StyleYuan, Hao, Hong Wan, Yi-Kao Hu, Emmanuel Ayodeji Ayeni, Qiang Chang, Chao Ma, and Xun Liao. 2021. "Fishing of α-Glucosidase’s Ligands from Aloe vera by α-Glucosidase Functionalized Magnetic Nanoparticles" Molecules 26, no. 19: 5840. https://doi.org/10.3390/molecules26195840
APA StyleYuan, H., Wan, H., Hu, Y.-K., Ayeni, E. A., Chang, Q., Ma, C., & Liao, X. (2021). Fishing of α-Glucosidase’s Ligands from Aloe vera by α-Glucosidase Functionalized Magnetic Nanoparticles. Molecules, 26(19), 5840. https://doi.org/10.3390/molecules26195840








