Microarray-Based Screening of Differentially Expressed Genes of E. coli O157:H7 Sakai during Preharvest Survival on Butterhead Lettuce
Abstract
:1. Introduction
2. Experimental Section
2.1. Bacterial Strains and Culture Conditions
2.2. Plant Growth Conditions
2.3. Inoculation of the Lettuce Plants and Measurement of Total Pathogen Populations on Lettuce Leaves
2.4. RNA Extraction for Microarray Experiment
2.5. Microarray Labeling Procedure
2.6. Preparation of Prokaryotic Hybridization Samples for Agilent 8 × 15K Arrays
2.7. Uninoculated Lettuce Control
2.8. Data Analysis
3. Results and Discussion
| Kyle et al., 2010 [3] | Fink et al., 2012 [4] | This Study | |
|---|---|---|---|
| Strain | E. coli O157:H7 EDL933 | E. coli K12 MG1655 E. coli O157:H7 EDL933 | E. coli O157:H7 Sakai |
| Growth medium | M9-glucose a | LB b | MOPS-enriched c |
| Growth temperature of E. coli | 28 °C | 37 °C | 18 °C |
| Time points | 15 min; 30 min | One and three days | 1 h, 2 days |
| Lettuce type | romaine lettuce | green leaf lettuce | butterhead lettuce |
| Type of interaction | leaf lysate supernatant | surface sterilized leaves sodium hypochlorite | growing non-sterilized plants |
| Inoculum level | 108 CFU/mL | 107 CFU/cm2 lettuce | 108 CFU/g lettuce |
| Plant growth conditions | Not applicable | 100% RH, 25 °C photoperiod 16 h for three days. | Growth chamber, 18 °C, 75% RH, photoperiod 16 h for two days |
| Reference control for expression analysis | Cells grown in M9-glucose at 28 °C | Cells grown in LB at 37 °C | Cells were re-suspended and kept for 1 h in MgSO4 buffer after initial growth in MOPS-enriched medium, both at 18 °C |
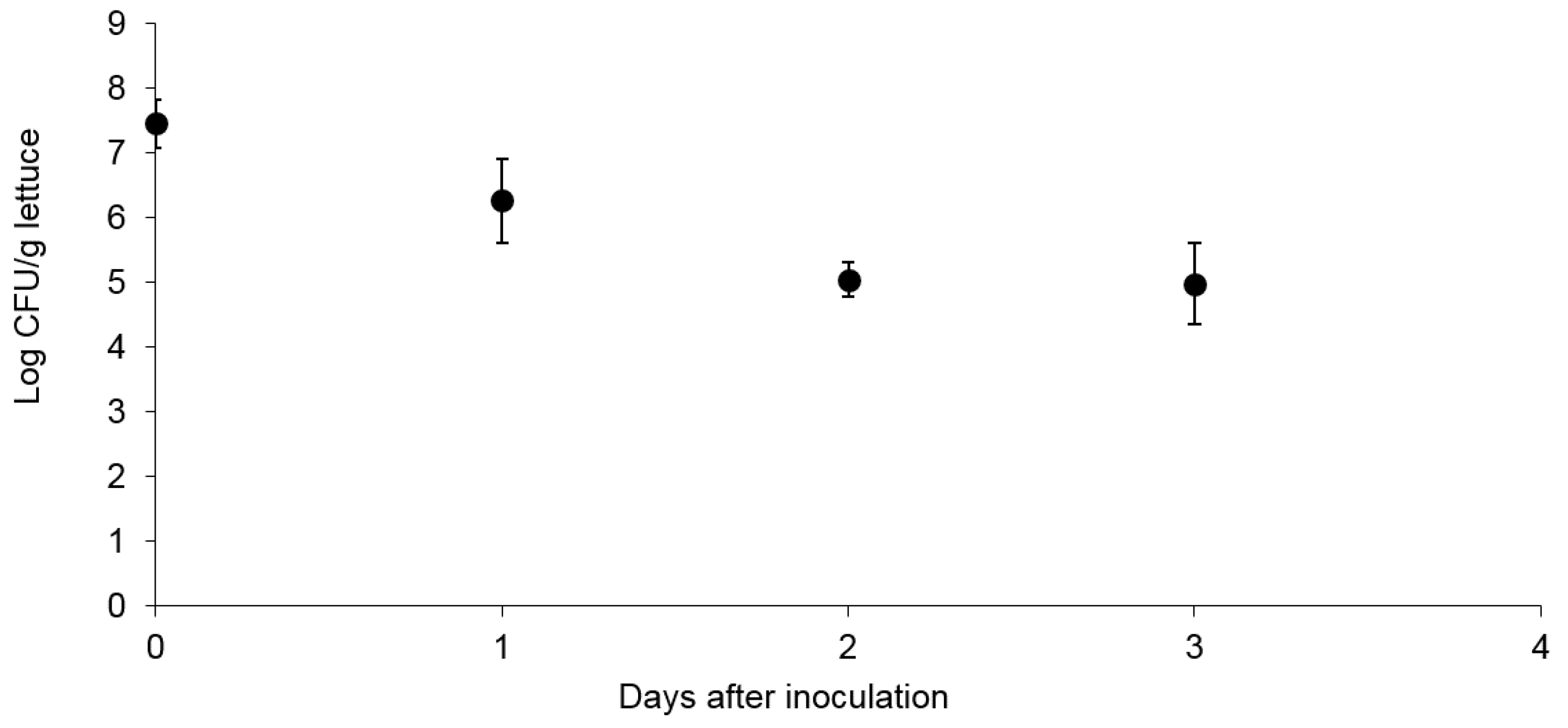
3.1. Survival and Association of E. coli Sakai on/with Growing Butterhead Lettuce
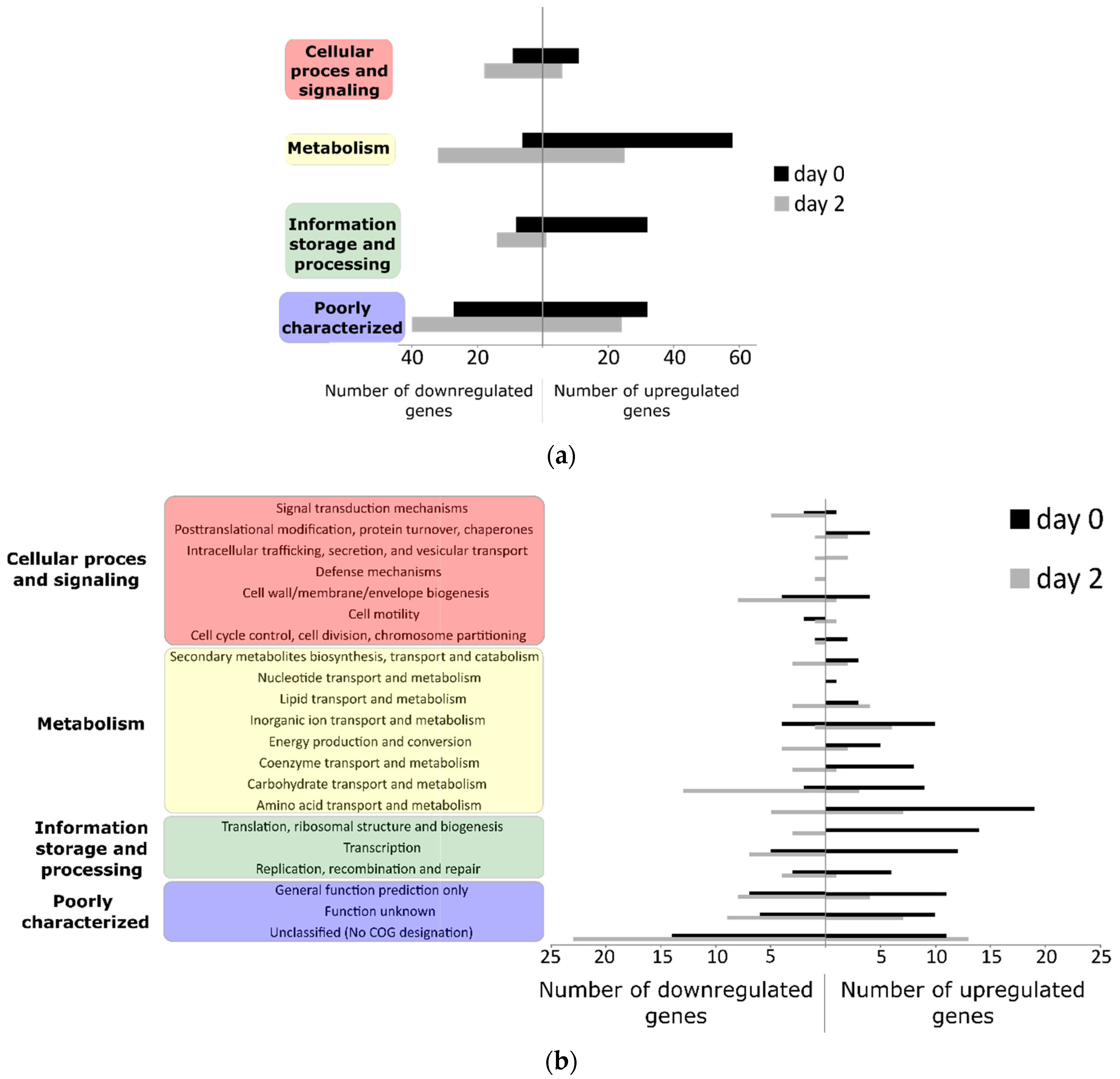
3.2. Differentially Regulated Transcriptome of E. coli O157:H7 Sakai on Growing Butterhead Lettuce
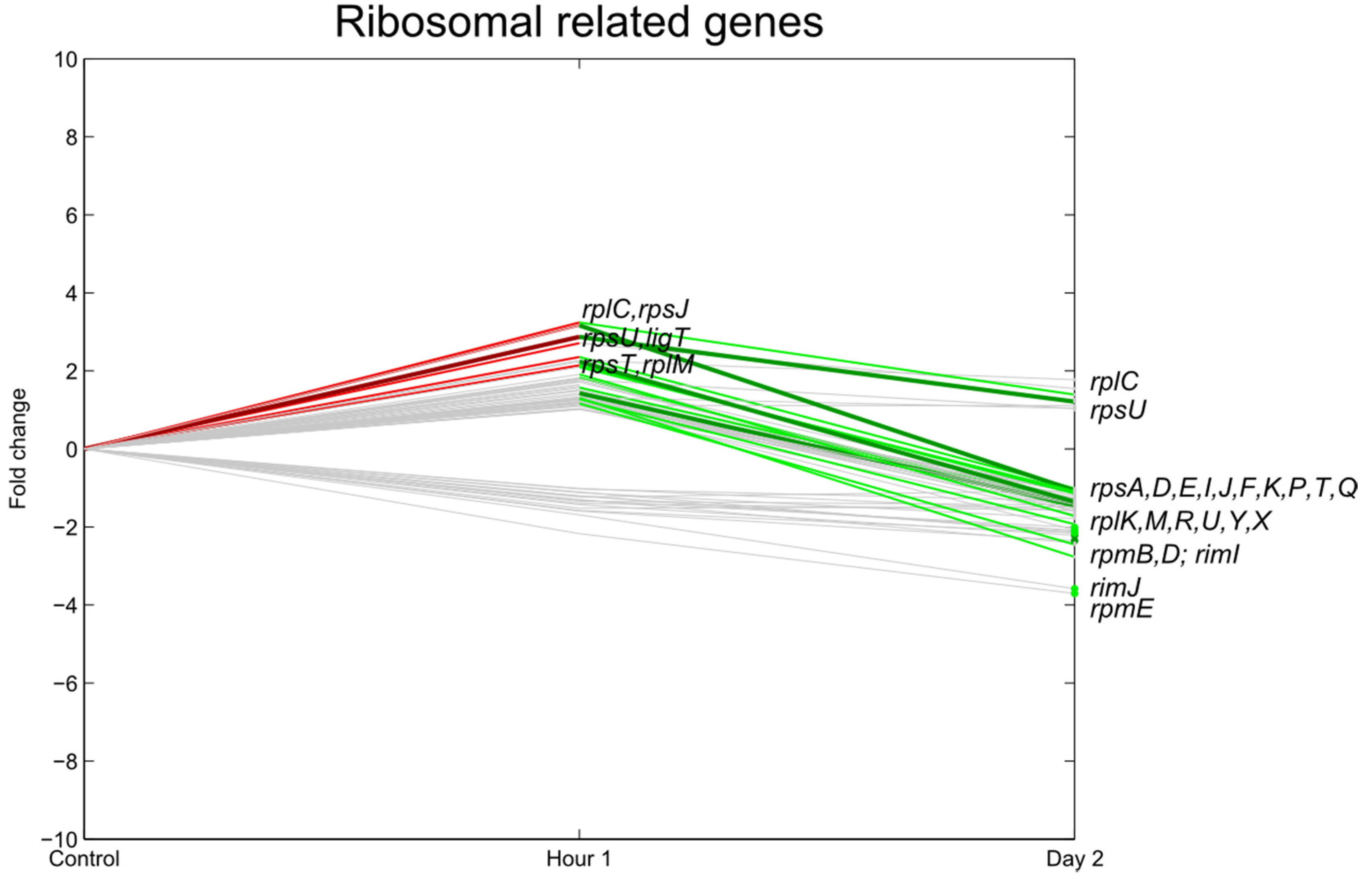
3.3. Transcription, Translation
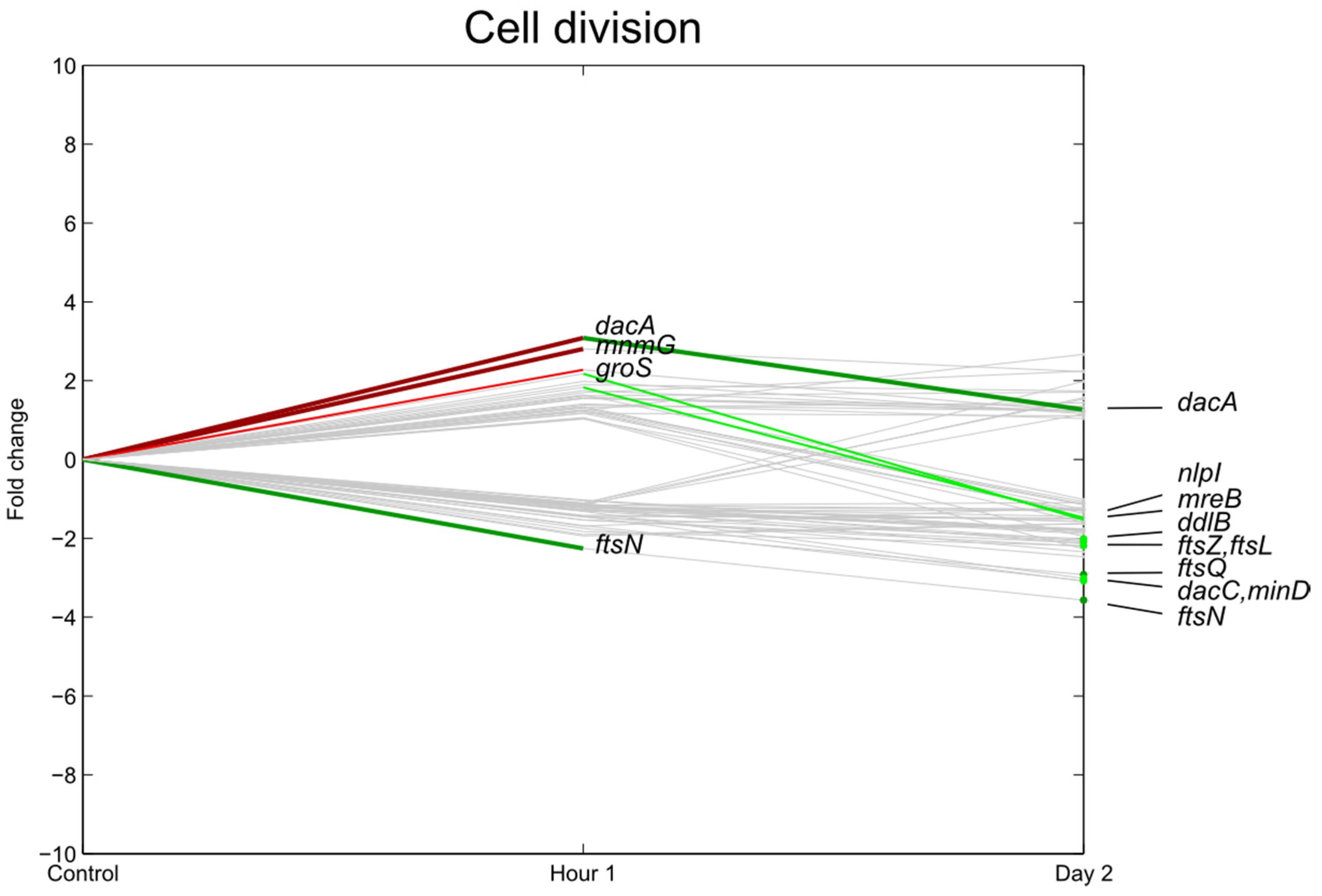
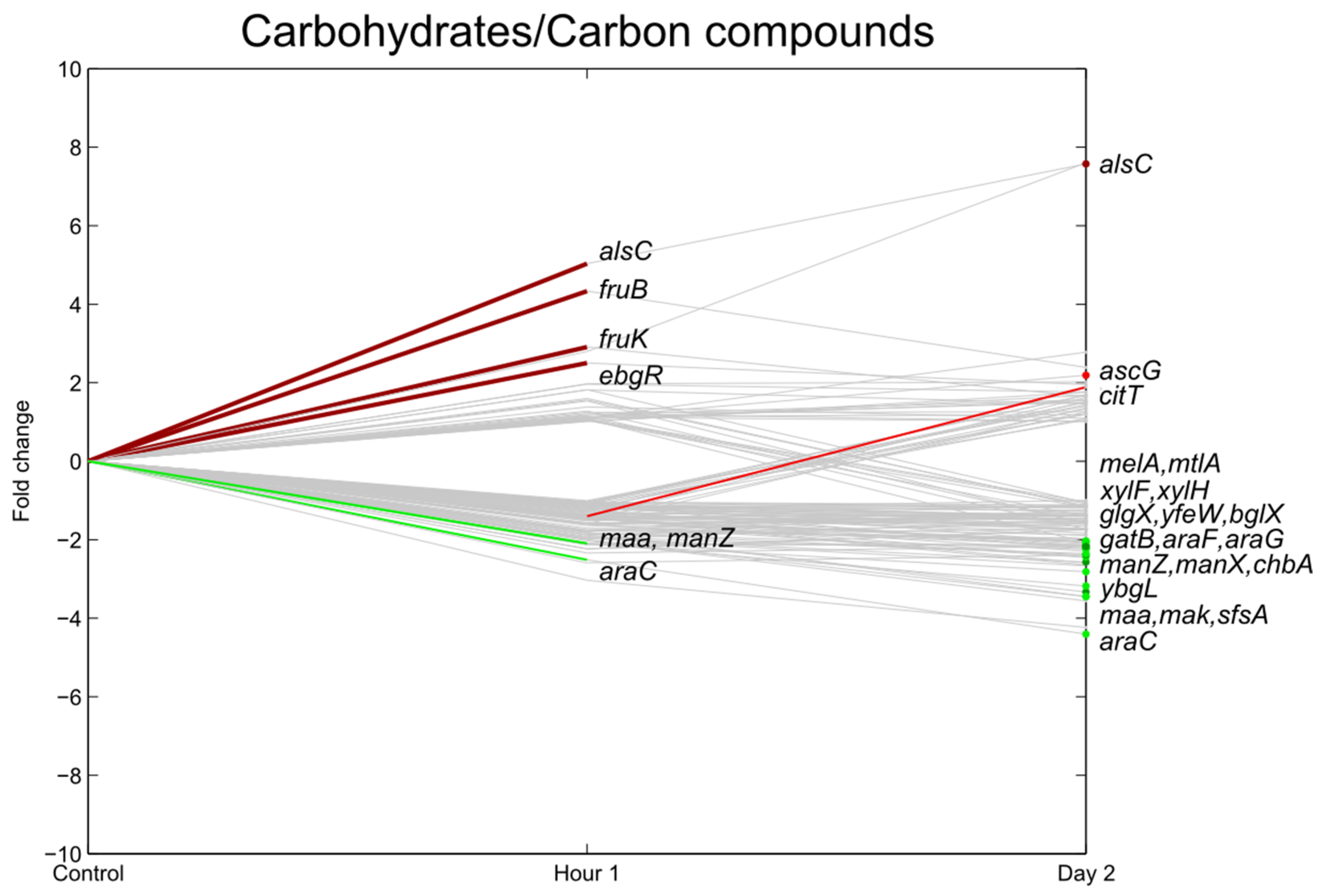
3.4. Carbohydrate Transport
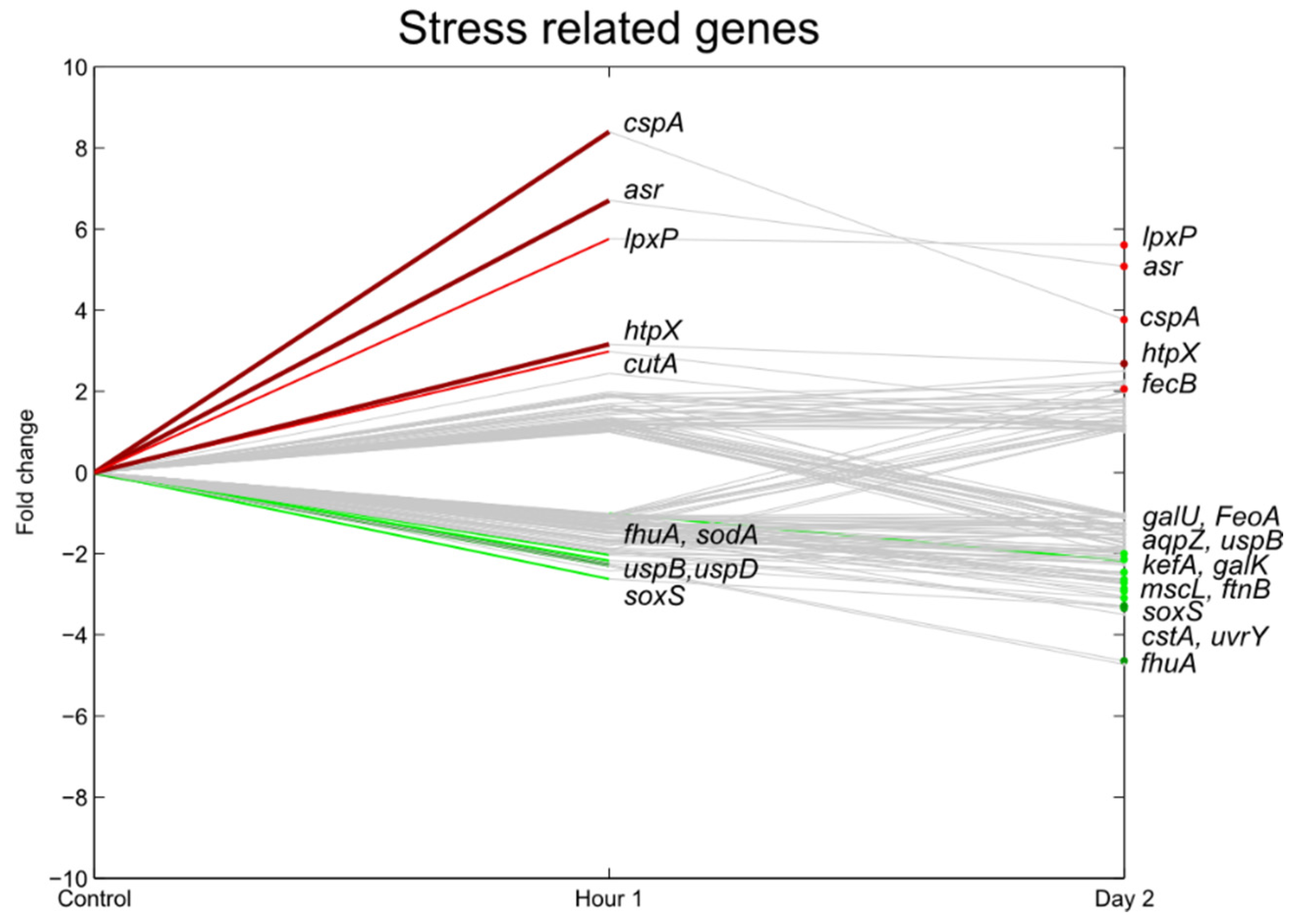
3.5. Stress Responsive Genes
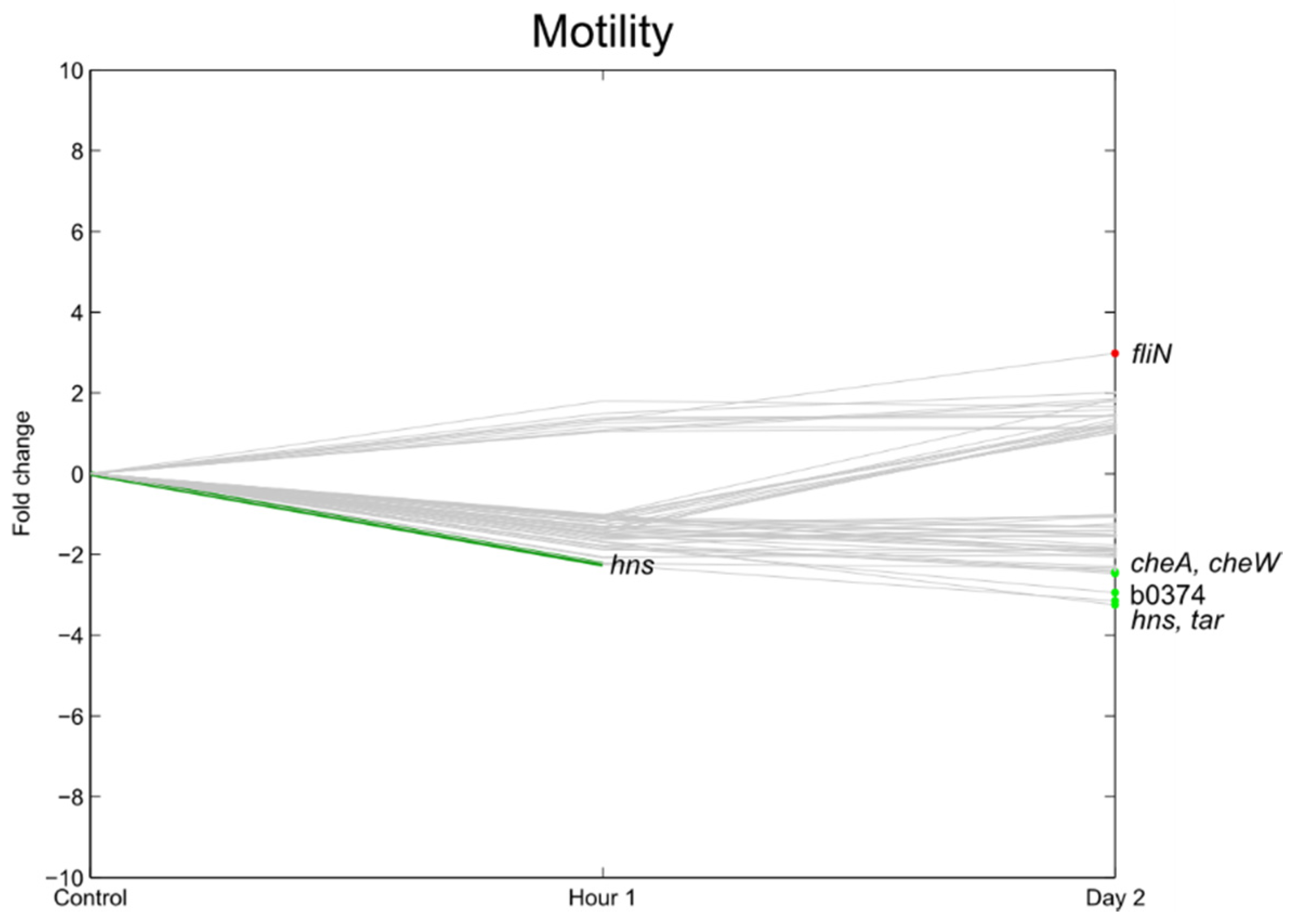
3.6. Flagella, Motility, Fimbrial Expression, Biofilm
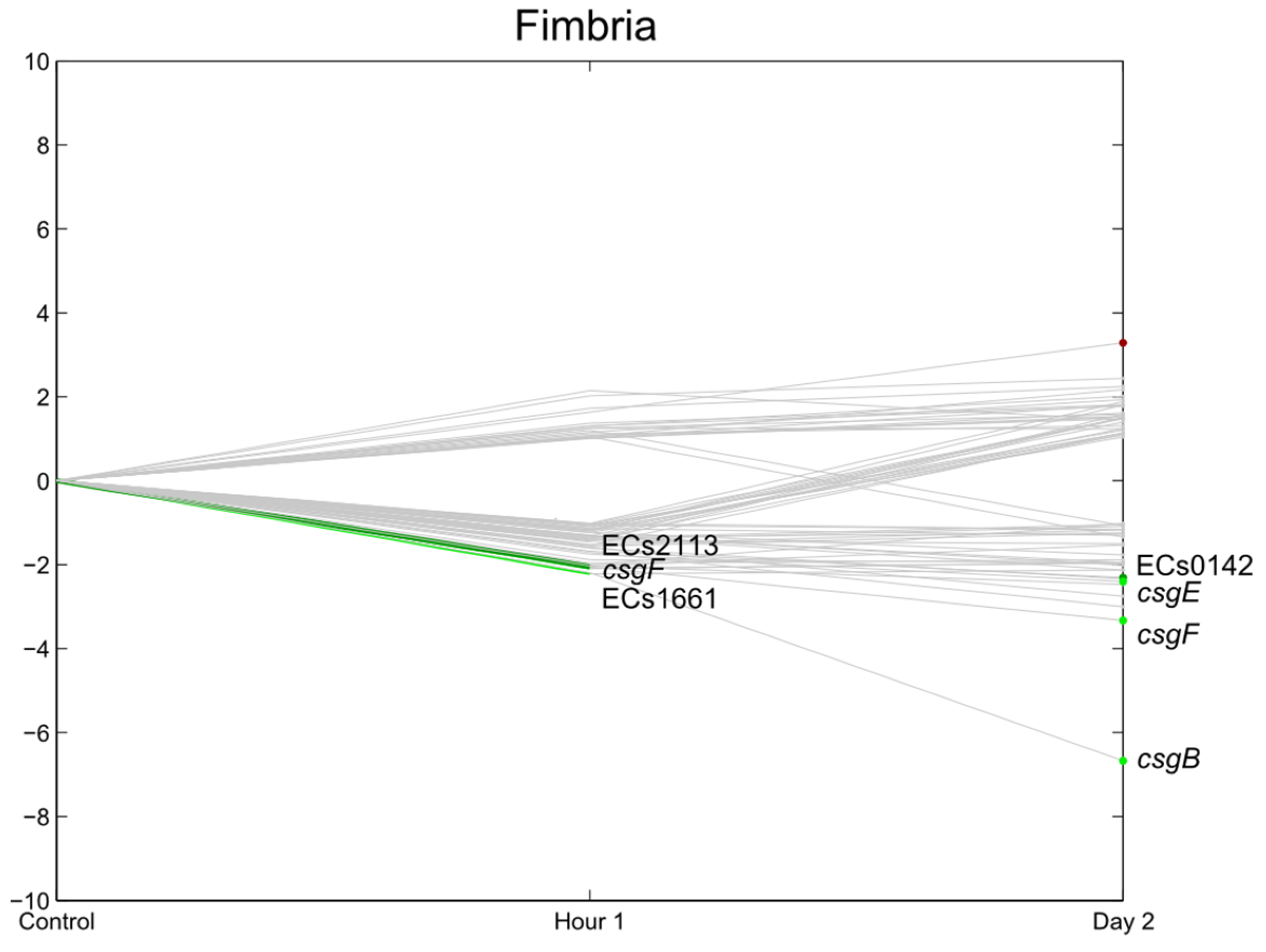
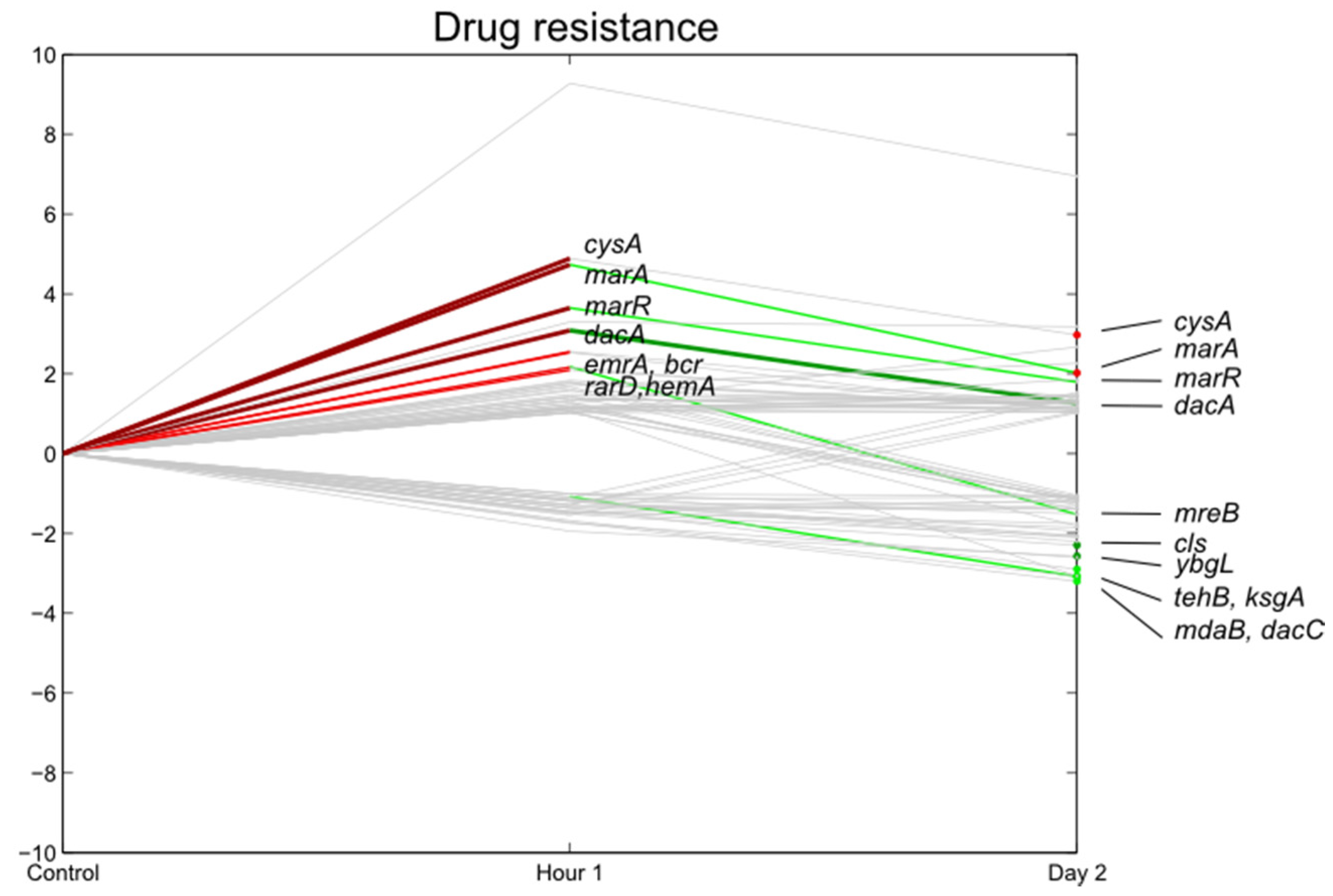
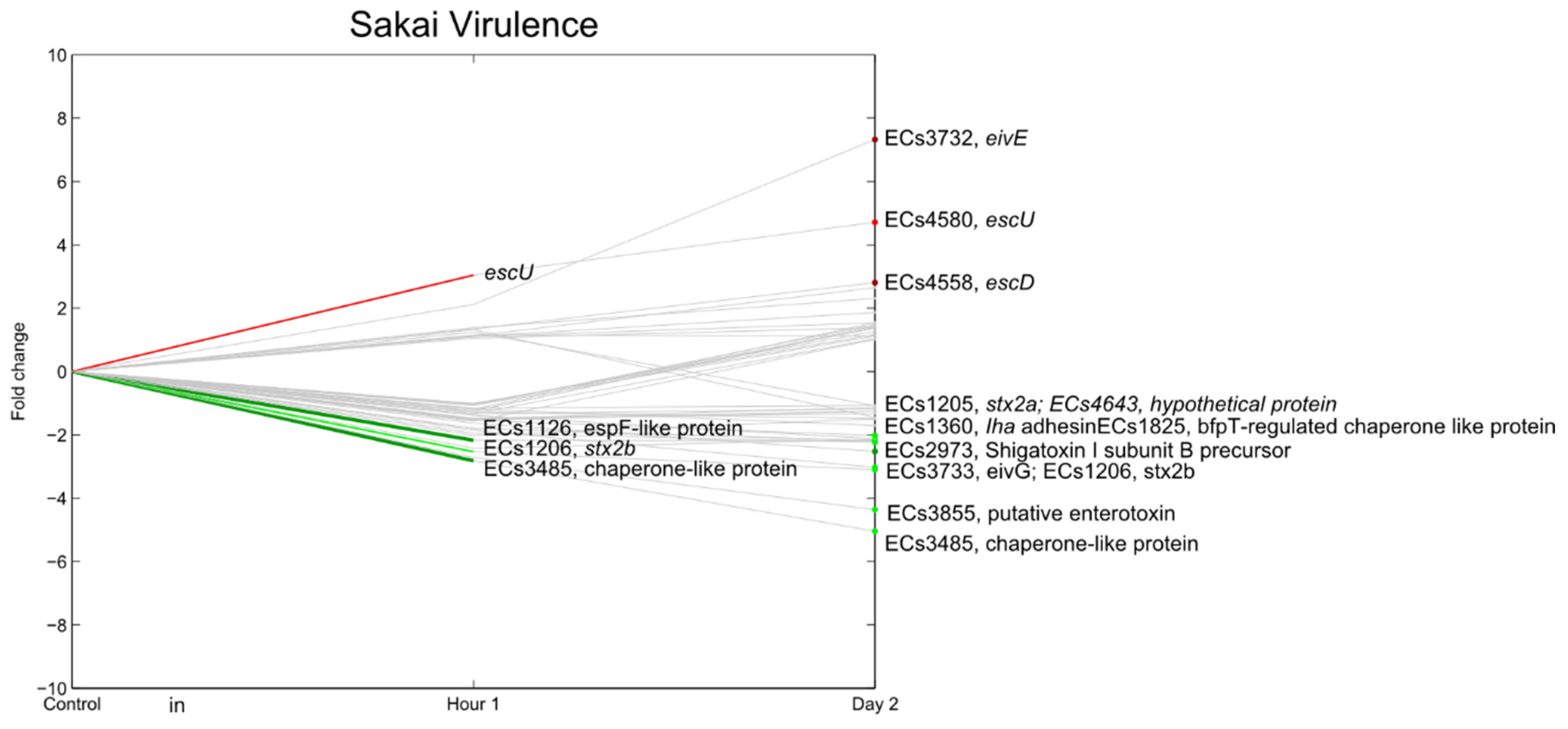
3.7. E. coli O157:H7 Sakai Virulence Genes
3.8. Unknown Genes
| GeneName | Foldchange Day 0 | Foldchange Day 2 | Similarity with Known Gene | Strain with Known Gene with Similar Sequence |
|---|---|---|---|---|
| ECs0073 | −2.3 | −2.5 | non-LEE-encoded type III secreted effector | Escherichia coli O157:H7 str. TW14359 |
| ECs0230 | 2.0 | 3.7 | VCA0109-like protein (RNA methyltransferase type VI secretion protein) | Escherichia coli O157:H7 str. EDL933 |
| ECs0238 | −2.4 | −4.4 | / | / |
| ECs0281 | −1.9 | −2.4 | Putative tail fiber assembly protein | Escherichia coli O157:H7 str. EDL933 |
| ECs0541 | −2.1 | −2.7 | PKD domain protein | Escherichia coli O157:H7 str. EC4115 |
| ECs1125 | −1.7 | −2.7 | EspF like protein or Tir-cytoskeleton coupling protein | Escherichia coli Xuzhou21/Escherichia coli O157:H7 str. TW14359 |
| ECs1204 | 1.7 | 2.2 | Putative DNA methylase | Escherichia coli O157:H7 str. EDL933, complete genome |
| ECs1238 | −2.0 | −3.1 | / | / |
| ECs1239 | −1.8 | −2.3 | / | / |
| ECs1246 | −2.2 | −3.7 | / | / |
| ECs1445 | −1.9 | −3.4 | / | / |
| ECs1567 | −2.6 | −3.3 | T3SS effector protein EspO | Escherichia coli O145:H28 str. RM12761 |
| ECs1586 | −2.9 | −4.0 | / | / |
| ECs2291 | −1.6 | −2.2 | Lipoprotein YnfC precursor | Escherichia coli O157:H7 str. SS17, complete genome |
| ECs2473 | −2.0 | −1.5 | Putative lipoprotein | |
| ECs2526 | 14.7 | 8.2 | Mg and Co efflux protein CorC | Escherichia coli O157:H7 str. EDL933, complete genome |
| ECs3238 | −3.8 | −6.2 | / | / |
| ECs3250 | −2.1 | −2.2 | 56 bp at 5′ side: hybrid sensory histidine kinase in regulatory system with EvgA 289 bp at 3’ side predicted transporter | a.o. Escherichia coli O157:H7 str. EDL933, complete genome |
| ECs3772 | −2.2 | −2.9 | / | / |
| ECs3966 | 2.3 | 3.5 | 23S rRNA (guanine−N−2−) −methyltransferase rlmG | a.o. Escherichia coli O157:H7 str. EDL933, complete genome |
| ECs4115 | 7.6 | 25.8 | similar to yhcR which is also known as aaeX | Escherichia coli O157:H7 str. EC4115 |
| ECs4465 | −2.6 | −4.4 | Fic family protein | Escherichia coli O157:H7 str. EC4115, complete genome |
| ECs4491 | 3.1 | 1.8 | Periplasmic septal ring factor with murein hydrolase activity EnvC | Escherichia coli O157:H7 str. EDL933, complete genome |
| ECs4537 | −2.2 | −2.4 | transposase ORF B, IS3 family protein | Escherichia coli O55:H7 str. RM12579, complete genome |
| ECs4816 | −2.4 | −2.4 | / | / |
| ECs5266 | 2.0 | 2.6 | transposase | Escherichia coli O157:H7 str. EDL933, complete genome |
| ECs5291 | −1.9 | −2.9 | adherence and invasion outermembrane protein (Inv, enhances Peyer's patches colonization) | Escherichia coli O157:H7 str. EDL933, complete genome |
3.9. Limitations of the Data
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Klein, S.; Tian, A.; Witmer, J.; DeWaal, C.S. The FDA Top Ten: The Riskiest Foods Regulated by the US Food and Drug Administration; The Center for Science in the Public Interest: Washington, DC, USA, 2009. [Google Scholar]
- Delaquis, P.; Bach, S.; Dinu, L.D. Behavior of Escherichia coli O157:H7 in leafy vegetables. J. Food Prot. 2007, 70, 1966–1974. [Google Scholar] [PubMed]
- Kyle, J.L.; Parker, C.T.; Goudeau, D.; Brandl, M.T. Transcriptome analysis of Escherichia coli O157:H7 exposed to lysates of lettuce leaves. Appl. Environ. Microbiol. 2010, 76, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Fink, R.C.; Black, E.P.; Hou, Z.; Sugawara, M.; Sadowsky, M.J.; Diez-Gonzalez, F. Transcriptional responses of Escherichia coli K-12 and O157:H7 associated with lettuce leaves. Appl. Environ. Microbiol. 2012, 78, 1752–1764. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Fink, R.C.; Black, E.P.; Sugawara, M.; Zhang, Z.; Diez-Gonzalez, F.; Sadowsky, M.J. Gene expression profiling of Escherichia coli in response to interactions with the lettuce rhizosphere. J. Appl. Microbiol. 2012, 113, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Fink, R.C.; Sugawara, M.; Diez-Gonzalez, F.; Sadowsky, M.J. Transcriptional and functional responses of Escherichia coli O157:H7 growing in the lettuce rhizoplane. Food Microbiol. 2013, 35, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Landstorfer, R.; Simon, S.; Schober, S.; Keim, D.; Scherer, S.; Neuhaus, K. Comparison of strand-specific transcriptomes of enterohemorrhagic Escherichia coli O157:H7 EDL933 (EHEC) under eleven different environmental conditions including radish sprouts and cattle feces. BMC Genomics 2014, 15, 353. [Google Scholar] [CrossRef] [PubMed]
- Dahan, S.; Knutton, S.; Shaw, R.K.; Crepin, V.F.; Dougan, G.; Frankel, G. Transcriptome of enterohemorrhagic Escherichia coli O157 adhering to eukaryotic plasma membranes. Infect. Immun. 2004, 72, 5452–5459. [Google Scholar] [CrossRef] [PubMed]
- Michino, H.; Araki, K.; Minami, S.; Takaya, S.; Sakai, N.; Miyazaki, M.; Ono, A.; Yanagawa, H. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 1999, 150, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Ozasa, K.; Mermin, J.H.; Griffin, P.M.; Masuda, K.; Imashuku, S.; Sawada, T. Factory outbreak of Escherichia coli O157:H7 infection in Japan. Emerg. Infect. Dis. 1999, 5, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Neidhardt, F.C.; Bloch, P.L.; Smith, D.F. Culture medium for enterobacteria. J. Bacteriol. 1974, 119, 736–747. [Google Scholar] [PubMed]
- Kisluk, G.; Hoover, D.G.; Kneil, K.E.; Yaron, S. Quantification of low and high levels of Salmonella enterica serovar Typhimurium on leaves. LWT Food Sci. Technol. 2012, 45, 36–42. [Google Scholar] [CrossRef]
- Fislage, R.; Berceanu, M.; Humboldt, Y.; Wendt, M.; Oberender, H. Primer design for a prokaryotic differential display RT-PCR. Nucleic Acids Res. 1997, 25, 1830–1835. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Demeke, T.; Clear, R.M.; Patrick, S.K. Simultaneous detection by PCR of Escherichia coli, Listeria monocytogenes and Salmonella typhimurium in artificially inoculated wheat grain. Int. J. Food Microbiol. 2006, 111, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, R.R.; Pallen, M.J. xBASE, a collection of online databases for bacterial comparative genomics. Nucleic Acids Res. 2006, 34, D335–D337. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Chen, I.-M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Pillay, M.; Ratner, A.; Huang, J.; Woyke, T.; Huntemann, M. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2013. [Google Scholar] [CrossRef] [PubMed]
- GenProTec. E. coli genome and proteome database. Available online: http://genprotec.mbl.edu/ (accessed on 31 December 2013).
- E. coli O157:H7 Sakai genome project. Available online: http://genome.bio.titech.ac.jp/bacteria/o157/search.html (accessed on 10 September 2014).
- Hayashi, T.; Makino, K.; Ohnishi, M.; Kurokawa, K.; Ishii, K.; Yokoyama, K.; Han, C.-G.; Ohtsubo, E.; Nakayama, K.; Murata, T. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001, 8, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Rocha, D.J.; Santos, C.S.; Pacheco, L.G. Bacterial reference genes for gene expression studies by RT-qPCR: Survey and analysis. Antonie Van Leeuwenhoek 2015, 108, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Zhou, L.; Lim, Q.E.; Zou, R.; Stephanopoulos, G.; Too, H.-P. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, R. Sucrose transporters in plants: Update on function and structure. Biochim. Biophys. Acta 2000, 1465, 246–262. [Google Scholar] [CrossRef]
- Kang, Y.; Outlaw, W.H.; Andersen, P.C.; Fiore, G.B. Guard-cell apoplastic sucrose concentration—A link between leaf photosynthesis and stomatal aperture size in the apoplastic phloem loader Vicia faba L. Plant Cell Environ. 2007, 30, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Kroupitski, Y.; Golberg, D.; Belausov, E.; Pinto, R.; Swartzberg, D.; Granot, D.; Sela, S. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 2009, 75, 6076–6086. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H.J.; Lindow, S.E. Appetite of an epiphyte: Quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 2001, 98, 3446–3453. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.J.; Djaman, O.; Imlay, J.A.; Kiley, P.J. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 9009–9014. [Google Scholar] [CrossRef] [PubMed]
- Hidese, R.; Mihara, H.; Esaki, N. Bacterial cysteine desulfurases: Versatile key players in biosynthetic pathways of sulfur-containing biofactors. Appl. Microbiol. Biotechnol. 2011, 91, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Brankatschk, K.; Kamber, T.; Pothier, J.F.; Duffy, B.; Smits, T.H. Transcriptional profile of Salmonella enterica subsp. enterica serovar Weltevreden during alfalfa sprout colonization. Microb. Biotechnol. 2013, 7, 528–544. [Google Scholar] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA. 2004, 101, 3160–3165. [Google Scholar] [CrossRef] [PubMed]
- Matilla, M.A.; Espinosa-Urgel, M.; Rodríguez-Herva, J.J.; Ramos, J.L.; Ramos-González, M.I. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol 2007. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhou, J.; Fu, Q.S.; Wang, J. The Escherichia coli azoreductase AzoR is involved in resistance to thiol-specific stress caused by electrophilic quinones. J. Bacteriol. 2009, 191, 6394–6400. [Google Scholar] [CrossRef] [PubMed]
- Goudeau, D.M.; Parker, C.T.; Zhou, Y.; Sela, S.; Kroupitski, Y.; Brandl, M.T. The Salmonella transcriptome in lettuce and cilantro soft rot reveals a niche overlap with the animal host intestine. Appl. Environ. Microbiol. 2013, 79, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Kortmann, J.; Narberhaus, F. Bacterial RNA thermometers: Molecular zippers and switches. Nat. Rev. Microbiol. 2012, 10, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Duffitt, A.D.; Reber, R.T.; Whipple, A.; Chauret, C. Gene expression during survival of Escherichia coli O157:H7 in soil and water. Int. J. Microbiol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Lesley, S.A.; Graziano, J.; Cho, C.Y.; Knuth, M.W.; Klock, H.E. Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng. 2002, 15, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Xicohtencatl-Cortes, J.; Chacon, E.S.; Saldana, Z.; Freer, E.; Giron, J.A. Interaction of Escherichia coli O157:H7 with leafy green produce. J. Food Prot. 2009, 72, 1531–1537. [Google Scholar] [PubMed]
- Berger, C.N.; Shaw, R.K.; Brown, D.J.; Mather, H.; Clare, S.; Dougan, G.; Pallen, M.J.; Frankel, G. Interaction of Salmonella enterica with basil and other salad leaves. ISME J. 2009, 3, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Lemuth, K.; Hardiman, T.; Winter, S.; Pfeiffer, D.; Keller, M.; Lange, S.; Reuss, M.; Schmid, R.; Siemann-Herzberg, M. Global transcription and metabolic flux analysis of Escherichia coli in glucose-limited fed-batch cultivations. Appl. Environ. Microbiol. 2008, 74, 7002–7015. [Google Scholar] [CrossRef] [PubMed]
- Jozefczuk, S.; Klie, S.; Catchpole, G.; Szymanski, J.; Cuadros-Inostroza, A.; Steinhauser, D.; Selbig, J.; Willmitzer, L. Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 2010, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Thilmony, R.; Underwood, W.; He, S.Y. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006, 46, 34–53. [Google Scholar] [CrossRef] [PubMed]
- Blomfield, I.C.V.D.M. Regulation of fimbrial expression. In EcoSal; Curtiss, R.I., Kaper, J.B., Squires, C.L., Karp, P.D., Neidhardt, F.C., Slauch, J.M., Eds.; ASM Press: Washington, DC, USA, 2007. [Google Scholar]
- Boyer, R.R.; Sumner, S.S.; Williams, R.C.; Pierson, M.D.; Popham, D.L.; Kniel, K.E. Influence of curli expression by Escherichia coli O157:H7 on the cell's overall hydrophobicity, charge, and ability to attach to lettuce. J. Food Prot. 2007, 70, 1339–1345. [Google Scholar] [PubMed]
- Römling, U.; Bian, Z.; Hammar, M.; Sierralta, W.D.; Normark, S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 1998, 180, 722–731. [Google Scholar] [PubMed]
- Keseler, I.M.; Collado-Vides, J.; Santos-Zavaleta, A.; Peralta-Gil, M.; Gama-Castro, S.; Muñiz-Rascado, L.; Bonavides-Martinez, C.; Paley, S.; Krummenacker, M.; Altman, T. EcoCyc: A comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011, 39, D583–D590. [Google Scholar] [CrossRef] [PubMed]
- Macarisin, D.; Patel, J.; Bauchan, G.; Giron, J.A.; Sharma, V.K. Role of curli and cellulose expression in adherence of Escherichia coli O157:H7 to spinach leaves. Foodborne Pathog. Dis. 2012, 9, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Vidal, O.; Longin, R.; Prigent-Combaret, C.; Dorel, C.; Hooreman, M.; Lejeune, P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998, 180, 2442–2449. [Google Scholar] [PubMed]
- Pomposiello, P.J.; Bennik, M.H.J.; Demple, B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 2001, 183, 3890–3902. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.S.; Garcia-Contreras, R.; Wood, T.K. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J. Bacteriol. 2007, 189, 3051–3062. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Feng, S.; Thate, T.E.; Kaper, J.B.; Boedeker, E.C. Towards a vaccine for attaching/effacing Escherichia coli: A LEE encoded regulator (ler) mutant of rabbit enteropathogenic Escherichia coli is attenuated, immunogenic, and protects rabbits from lethal challenge with the wild-type virulent strain. Vaccine 2006, 24, 3845–3855. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.M.; Kostrzynska, M.; Thompson, S. Escherichia coli O157:H7 stress and virulence gene expression on Romaine lettuce using comparative real-time PCR. J. Microbiol. Methods 2009, 77, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Lakshman, S.; Ferguson, S.; Ingram, D.T.; Luo, Y.G.; Patel, J. Effect of modified atmosphere packaging on the persistence and expression of virulence factors of Escherichia coli O157:H7 on shredded iceberg lettuce. J. Food Prot. 2011, 74, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Dinu, L.D.; Bach, S. Induction of viable but non-culturable Escherichia coli O157:H7 on the phyllosphere of lettuce: A food safety risk factor. Appl. Environ. Microbiol. 2011, 77, 8295–8302. [Google Scholar] [CrossRef] [PubMed]
- Wagner, P.L.; Livny, J.; Neely, M.N.; Acheson, D.W.; Friedman, D.I.; Waldor, M.K. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 2002, 44, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Grau-Leal, F.; Quiros, P.; Martinez-Castillo, A.; Muniesa, M. Free Shiga toxin 1-encoding bacteriophages are less prevalent than Shiga toxin 2 phages in extraintestinal environments. Environ. Microbiol. 2015, 17, 4790–4801. [Google Scholar] [CrossRef] [PubMed]
- Noel, J.T.; Arrach, N.; Alagely, A.; McClelland, M.; Teplitski, M. Specific responses of Salmonella enterica to tomato varieties and fruit ripeness identified by in vivo expression technology. PLoS ONE 2010, 5, e12406. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.-T.; Gratwick, K.S.; Kollman, J.; Park, D.; Nies, D.H.; Goffeau, A.; Saier, M.H., Jr. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1999, 1, 107–125. [Google Scholar] [PubMed]
- Van Dyk, T.K.; Templeton, L.J.; Cantera, K.A.; Sharpe, P.L.; Sariaslani, F.S. Characterization of the Escherichia coli AaeAB efflux pump: A metabolic relief valve? J. Bacteriol. 2004, 186, 7196–7204. [Google Scholar] [CrossRef] [PubMed]
- Kvist, M.; Hancock, V.; Klemm, P. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 2008, 74, 7376–7382. [Google Scholar] [CrossRef] [PubMed]
- Monnappa, A.K.; Lee, S.; Mitchell, R.J. Sensing of plant hydrolysate-related phenolics with an aaeXAB::luxCDABE bioreporter strain of Escherichia coli. Bioresour. Technol. 2013, 127, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Kapitein, N.; Mogk, A. Deadly syringes: Type VI secretion system activities in pathogenicity and interbacterial competition. Curr. Opin. Microbiol. 2013, 16, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Van der Linden, I.; Cottyn, B.; Uyttendaele, M.; Vlaemynck, G.; Heyndrickx, M.; Maes, M. Survival of enteric pathogens during butterhead lettuce growth: Crop stage, leaf age, and irrigation. Foodborne Pathog. Dis. 2013, 10, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H.J. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812–1822. [Google Scholar] [CrossRef] [PubMed]
- Brandl, M.T.; Amundson, R. Leaf age as a risk factor in contamination of lettuce with Escherichia coli O157:H7 and Salmonella enterica. Appl. Environ. Microbiol. 2008, 74, 2298–2306. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Frank, J.F. Penetration of Escherichia coli O157:H7 into lettuce tissues as affected by inoculum size and temperature and the effect of chlorine treatment on cell viability. J. Food Prot. 2000, 63, 434–440. [Google Scholar] [PubMed]
- Iturriaga, M.H.; Escartin, E.F.; Beuchat, L.R.; Martinez-Peniche, R. Effect of inoculum size, relative humidity, storage temperature, and ripening stage on the attachment of Salmonella Montevideo to tomatoes and tomatillos. J. Food Prot. 2003, 66, 1756–1761. [Google Scholar] [PubMed]
- Schikora, A.; Garcia, A.V.; Hirt, H. Plants as alternative hosts for Salmonella. Trends Plant Sci. 2012, 17, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Schikora, A.; Garcia, A.V.; Charrier, A.; Hirt, H. Infection of plants by the human pathogen Salmonella Typhimurium: Challenges and new insights. Biocommun. Plants 2012, 14, 349–360. [Google Scholar]
- Schikora, A.; Carreri, A.; Charpentier, E.; Hirt, H. The dark side of the salad: Salmonella typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS ONE 2008, 3, e2279. [Google Scholar] [CrossRef] [PubMed]
- Shirron, N.; Yaron, S. Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLoS ONE 2011, 6, e18855. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Matthews, K.R. Influence of the plant defense response to Escherichia coli O157:H7 cell surface structures on survival of that enteric pathogen on plant surfaces. Appl. Environ. Microbiol. 2012, 78, 5882–5889. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.; Usall, J.; Solsona, C.; Alegre, I.; Viñas, I.; Abadias, M. Effects of packaging type and storage temperature on the growth of foodborne pathogens on shredded ‘Romaine’ lettuce. Food Microbiol. 2010, 27, 375–380. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van der Linden, I.; Cottyn, B.; Uyttendaele, M.; Vlaemynck, G.; Heyndrickx, M.; Maes, M.; Holden, N. Microarray-Based Screening of Differentially Expressed Genes of E. coli O157:H7 Sakai during Preharvest Survival on Butterhead Lettuce. Agriculture 2016, 6, 6. https://doi.org/10.3390/agriculture6010006
Van der Linden I, Cottyn B, Uyttendaele M, Vlaemynck G, Heyndrickx M, Maes M, Holden N. Microarray-Based Screening of Differentially Expressed Genes of E. coli O157:H7 Sakai during Preharvest Survival on Butterhead Lettuce. Agriculture. 2016; 6(1):6. https://doi.org/10.3390/agriculture6010006
Chicago/Turabian StyleVan der Linden, Inge, Bart Cottyn, Mieke Uyttendaele, Geertrui Vlaemynck, Marc Heyndrickx, Martine Maes, and Nicola Holden. 2016. "Microarray-Based Screening of Differentially Expressed Genes of E. coli O157:H7 Sakai during Preharvest Survival on Butterhead Lettuce" Agriculture 6, no. 1: 6. https://doi.org/10.3390/agriculture6010006
APA StyleVan der Linden, I., Cottyn, B., Uyttendaele, M., Vlaemynck, G., Heyndrickx, M., Maes, M., & Holden, N. (2016). Microarray-Based Screening of Differentially Expressed Genes of E. coli O157:H7 Sakai during Preharvest Survival on Butterhead Lettuce. Agriculture, 6(1), 6. https://doi.org/10.3390/agriculture6010006







