MTHFR Knockdown Assists Cell Defense against Folate Depletion Induced Chromosome Segregation and Uracil Misincorporation in DNA
Abstract
1. Introduction
1.1. Folate Depletion Causes Chromosome Instability
1.2. Folate Metabolism, MTHFR and Hepatoma
1.3. Folate Co-Factors vs. the Balance between Methyl Group and Nucleotide Supplies
1.4. MTHFR and DNA Stability
2. Results
2.1. Lower MTHFR Gene Expression Increased Survival in HCC Patients
2.2. shRNA Lentivirus Production and Transfection in HEK 293T Cells
2.3. Efficiency of Different Target shRNA Sequences on MTHFR mRNA Reduction
2.4. Lower MTHFR Is Associated with Decreased Proportional Cell Populations in the G1 Phase and an Increase in Those in the G2/M Phase
2.5. Reduced MTHFR Is Associated with Decreased Intracellular SAM and SAH Contents
2.6. shMTHFR Did Not Intensify Low Folate Induced SAM Reduction in HepG2 Cells
2.7. shMTHFR Promoted Nuclei SHMT1/DHFR/TYMS Protein Expression under Folate Deficiency
2.8. shMTHFR Assisted Purine Synthesis in HepG2 Cells under Folate Deficiency; Such Impacts Were Amplified after Folinate Supplementation
2.9. shMTHFR Reduced Micronucleated Binucleated Cells and Uracil Misincorporation into DNA under Folate Deficiency
2.10. shMTHFR Promoted Nuclear MLH1/p53 Expression under Folate Deficiency
3. Discussion
4. Materials and Methods
4.1. mRNA Expression of MTHFR and Survival Rate in HCC Patients
4.2. Establishment of MTHFR Knockdown Cell-Line by RNA Interference
4.3. Cell Culture Conditions
4.4. MTHFR Gene Expression in HepG2 Cells with shMTHFR
4.5. Effects of siRNA on MTHFR Protein Expression
4.6. Effects of siRNA on MTHFR Enzyme Activity
4.7. Cell Cycle Analyses
4.8. Determination of SAM and SAH Contents
4.9. Western Blot Analyses
4.10. Stable Isotope Labeling Experiments
4.11. Determination of Micronuclei
4.12. Determination of Uracil Content in the DNA
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- MacFarlane, A.J.; Behan, N.A.; Field, M.S.; Williams, A.; Stover, P.J.; Yauk, C.L. Dietary folic acid protects against genotoxicity in the red blood cells of mice. Mutat. Res. 2015, 779, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Wu, M.T.; Lin, Y.J.; Tang, F.Y.; Ko, H.A.; Chiang, E.P. Regulation of Folate-Mediated One-Carbon Metabolism by Glycine N-Methyltransferase (GNMT) and Methylenetetrahydrofolate Reductase (MTHFR). J. Nutr. Sci. Vitaminol. 2015, 61, S148–S150. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.Y.; Chiang, E.P.; Shih, Y.T.; Lane, H.Y.; Lin, J.T.; Wu, C.Y. Lower serum folate is associated with development and invasiveness of gastric cancer. World J. Gastroenterol. 2014, 20, 11313–11320. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.H.; Quan, Z.Y.; Piao, J.M.; Zhang, T.T.; Jiang, M.H.; Shin, M.H.; Choi, J.S. Plasma Folate and Vitamin B12 Levels in Patients with Hepatocellular Carcinoma. Int. J. Mol. Sci. 2016, 17, 1032. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Wu, M.T.; Tang, F.Y.; Chen, D.Y.; Ko, H.A.; Shane, B.; Huang, W.N.; Chiang, E.P. MTHFR C677T polymorphism increases MTX sensitivity via the inhibition of S-adenosylmethionine and de novo purine synthesis. Clin. Sci. 2019, 133, 253–267. [Google Scholar] [CrossRef]
- Novikoff, A.B.; Mori, M.; Quintana, N.; Yam, A. Studies of the secretory process in the mammalian exocrine pancreas. I. The condensing vacuoles. J. Cell Biol. 1977, 75, 148–165. [Google Scholar] [CrossRef]
- Blount, B.C.; Mack, M.M.; Wehr, C.M.; MacGregor, J.T.; Hiatt, R.A.; Wang, G.; Wickramasinghe, S.N.; Everson, R.B.; Ames, B.N. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: Implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. USA 1997, 94, 3290–3295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Lin, W.L.; Lin, Y.J.; Tang, F.Y.; Chen, Y.M.; Chiang, E.P. A novel role of the tumor suppressor GNMT in cellular defense against DNA damage. Int. J. Cancer 2014, 134, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Duthie, S.J.; Hawdon, A. DNA instability (strand breakage, uracil misincorporation, and defective repair) is increased by folic acid depletion in human lymphocytes in vitro. FASEB J. 1998, 12, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Ni, J.; Zhou, T.; Xu, W.; Fenech, M.; Wang, X. Choline and/or folic acid deficiency is associated with genomic damage and cell death in human lymphocytes in vitro. Nutr. Cancer 2012, 64, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Crott, J.W. Micronuclei, nucleoplasmic bridges and nuclear buds induced in folic acid deficient human lymphocytes-evidence for breakage-fusion-bridge cycles in the cytokinesis-block micronucleus assay. Mutat. Res. 2002, 504, 131–136. [Google Scholar] [CrossRef]
- Leopardi, P.; Marcon, F.; Caiola, S.; Cafolla, A.; Siniscalchi, E.; Zijno, A.; Crebelli, R. Effects of folic acid deficiency and MTHFR C677T polymorphism on spontaneous and radiation-induced micronuclei in human lymphocytes. Mutagenesis 2006, 21, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Baghurst, P.; Luderer, W.; Turner, J.; Record, S.; Ceppi, M.; Bonassi, S. Low intake of calcium, folate, nicotinic acid, vitamin E, retinol, beta-carotene and high intake of pantothenic acid, biotin and riboflavin are significantly associated with increased genome instability—Results from a dietary intake and micronucleus index survey in South Australia. Carcinogenesis 2005, 26, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.Y.; Chih, H.M.; Lan, J.L.; Chang, H.Y.; Chen, W.W.; Chiang, E.P. Blood lipid profiles and peripheral blood mononuclear cell cholesterol metabolism gene expression in patients with and without methotrexate treatment. BMC Med. 2011, 9, 4. [Google Scholar] [CrossRef]
- Chang, H.Y.; Tzen, J.T.; Lin, S.J.; Wu, Y.T.; Chiang, E.P. Long-term prednisolone treatments increase bioactive vitamin B6 synthesis in vivo. J. Pharmacol. Exp. Ther. 2011, 337, 102–109. [Google Scholar] [CrossRef]
- Selhub, J.; Miller, J.W. The pathogenesis of homocysteinemia: Interruption of the coordinate regulation by S-adenosylmethionine of the remethylation and transsulfuration of homocysteine. Am. J. Clin. Nutr. 1992, 55, 131–138. [Google Scholar] [CrossRef]
- Frosst, P.; Blom, H.J.; Milos, R.; Goyette, P.; Sheppard, C.A.; Matthews, R.G.; Boers, G.J.; den Heijer, M.; Kluijtmans, L.A.; van den Heuvel, L.P.; et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995, 10, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Bagley, P.J.; Selhub, J. A common mutation in the methylenetetrahydrofolate reductase gene is associated with an accumulation of formylated tetrahydrofolates in red blood cells. Proc. Natl. Acad. Sci. USA 1998, 95, 13217–13220. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Peng, Q.; Chen, Z.; Deng, Y.; Huang, S.; Xu, J.; Li, H.; Li, S.; Zhao, J. The association between MTHFR gene polymorphisms and hepatocellular carcinoma risk: A meta-analysis. PLoS ONE 2013, 8, e56070. [Google Scholar] [CrossRef]
- Qi, Y.H.; Yao, L.P.; Cui, G.B.; Liang, J.; Shao, Q.J.; Yan, L.F.; Du, P. Meta-analysis of MTHFR C677T and A1298C gene polymorphisms: Association with the risk of hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2014, 38, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Han, B.; Zhai, H.; Cheng, X.; Ma, K. Significant association between MTHFR C677T polymorphism and hepatocellular carcinoma risk: A meta-analysis. Tumour Biol. 2014, 35, 189–193. [Google Scholar] [CrossRef]
- Chang, W.; Meng, Q.; Liu, J.H.; Wu, L.X.; Chen, Y.; Chen, S.D. Significant association between the MTHFR A1298C polymorphism and hepatocellular carcinoma risk: A meta-analysis. Genet. Mol. Res. 2015, 14, 15972–15980. [Google Scholar] [CrossRef]
- Herbig, K.; Chiang, E.P.; Lee, L.R.; Hills, J.; Shane, B.; Stover, P.J. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J. Biol. Chem. 2002, 277, 38381–38389. [Google Scholar] [CrossRef]
- Wang, Y.C.; Chiang, E.P. Low-dose methotrexate inhibits methionine S-adenosyltransferase in vitro and in vivo. Mol. Med. 2012, 18, 423–432. [Google Scholar] [CrossRef]
- Chang, H.Y.; Tang, F.Y.; Chen, D.Y.; Chih, H.M.; Huang, S.T.; Cheng, H.D.; Lan, J.L.; Chiang, E.P. Clinical use of cyclooxygenase inhibitors impairs vitamin B-6 metabolism. Am. J. Clin. Nutr. 2013, 98, 1440–1449. [Google Scholar] [CrossRef][Green Version]
- Chiang, E.P.; Wang, Y.C.; Tang, F.Y. Folate restriction and methylenetetrahydrofolate reductase 677T polymorphism decreases adoMet synthesis via folate-dependent remethylation in human-transformed lymphoblasts. Leukemia 2007, 21, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Sohn, K.J.; Jang, H.; Campan, M.; Weisenberger, D.J.; Dickhout, J.; Wang, Y.C.; Cho, R.C.; Yates, Z.; Lucock, M.; Chiang, E.P.; et al. The methylenetetrahydrofolate reductase C677T mutation induces cell-specific changes in genomic DNA methylation and uracil misincorporation: A possible molecular basis for the site-specific cancer risk modification. Int. J. Cancer 2009, 124, 1999–2005. [Google Scholar] [CrossRef] [PubMed]
- Stempak, J.M.; Sohn, K.J.; Chiang, E.P.; Shane, B.; Kim, Y.I. Cell and stage of transformation-specific effects of folate deficiency on methionine cycle intermediates and DNA methylation in an in vitro model. Carcinogenesis 2005, 26, 981–990. [Google Scholar] [CrossRef]
- Davis, S.R.; Quinlivan, E.P.; Shelnutt, K.P.; Ghandour, H.; Capdevila, A.; Coats, B.S.; Wagner, C.; Shane, B.; Selhub, J.; Bailey, L.B.; et al. Homocysteine synthesis is elevated but total remethylation is unchanged by the methylenetetrahydrofolate reductase 677C->T polymorphism and by dietary folate restriction in young women. J. Nutr. 2005, 135, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Quinlivan, E.P.; Davis, S.R.; Shelnutt, K.P.; Henderson, G.N.; Ghandour, H.; Shane, B.; Selhub, J.; Bailey, L.B.; Stacpoole, P.W.; Gregory, J.F., 3rd. Methylenetetrahydrofolate reductase 677C->T polymorphism and folate status affect one-carbon incorporation into human DNA deoxynucleosides. J. Nutr. 2005, 135, 389–396. [Google Scholar] [CrossRef][Green Version]
- James, S.J.; Miller, B.J.; Basnakian, A.G.; Pogribny, I.P.; Pogribna, M.; Muskhelishvili, L. Apoptosis and proliferation under conditions of deoxynucleotide pool imbalance in liver of folate/methyl deficient rats. Carcinogenesis 1997, 18, 287–293. [Google Scholar] [CrossRef]
- Kim, Y.I.; Shirwadkar, S.; Choi, S.W.; Puchyr, M.; Wang, Y.; Mason, J.B. Effects of dietary folate on DNA strand breaks within mutation-prone exons of the p53 gene in rat colon. Gastroenterology 2000, 119, 151–161. [Google Scholar] [CrossRef]
- Sekhon, J.; Pereira, P.; Sabbaghian, N.; Schievella, A.R.; Rozen, R. Antisense inhibition of methylenetetrahydrofolate reductase reduces survival of methionine-dependent tumour lines. Br. J. Cancer 2002, 87, 225–230. [Google Scholar] [CrossRef][Green Version]
- Stankova, J.; Shang, J.; Rozen, R. Antisense inhibition of methylenetetrahydrofolate reductase reduces cancer cell survival in vitro and tumor growth in vivo. Clin. Cancer Res. 2005, 11, 2047–2052. [Google Scholar] [CrossRef] [PubMed]
- Lawrance, A.K.; Deng, L.; Rozen, R. Methylenetetrahydrofolate reductase deficiency and low dietary folate reduce tumorigenesis in Apc min/+ mice. Gut 2009, 58, 805–811. [Google Scholar] [CrossRef]
- Chen, P.M.; Li, J.R.; Liu, C.C.; Tang, F.Y.; Chiang, E.I. Metabolic Pathways Enhancement Confers Poor Prognosis in p53 Exon Mutant Hepatocellular Carcinoma. Cancer Inform. 2020, 19, 1176935119899913. [Google Scholar] [CrossRef] [PubMed]
- Li, J.R.; Sun, C.H.; Li, W.; Chao, R.F.; Huang, C.C.; Zhou, X.J.; Liu, C.C. Cancer RNA-Seq Nexus: A database of phenotype-specific transcriptome profiling in cancer cells. Nucleic Acids Res. 2016, 44, D944–D951. [Google Scholar] [CrossRef] [PubMed]
- Menyhart, O.; Nagy, A.; Gyorffy, B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci. 2018, 5, 181006. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.L.; Sou, N.L.; Tang, F.Y.; Ko, H.A.; Yeh, W.T.; Peng, J.H.; Chiang, E.I. Tracing Metabolic Fate of Mitochondrial Glycine Cleavage System Derived Formate In Vitro and In Vivo. Int. J. Mol. Sci. 2020, 21, 8808. [Google Scholar] [CrossRef] [PubMed]
- Chiang, E.P.; Wang, Y.C.; Chen, W.W.; Tang, F.Y. Effects of insulin and glucose on cellular metabolic fluxes in homocysteine transsulfuration, remethylation, S-adenosylmethionine synthesis, and global deoxyribonucleic acid methylation. J. Clin. Endocrinol. Metab. 2009, 94, 1017–1025. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tibbetts, A.S.; Appling, D.R. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010, 30, 57–81. [Google Scholar] [CrossRef]
- Huang, R.F.; Ho, Y.H.; Lin, H.L.; Wei, J.S.; Liu, T.Z. Folate deficiency induces a cell cycle-specific apoptosis in HepG2 cells. J. Nutr. 1999, 129, 25–31. [Google Scholar] [CrossRef]
- Anderson, D.D.; Woeller, C.F.; Chiang, E.P.; Shane, B.; Stover, P.J. Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J. Biol. Chem. 2012, 287, 7051–7062. [Google Scholar] [CrossRef] [PubMed]
- Field, M.S.; Kamynina, E.; Stover, P.J. MTHFD1 regulates nuclear de novo thymidylate biosynthesis and genome stability. Biochimie 2016, 126, 27–30. [Google Scholar] [CrossRef]
- Sou, N.L.; Huang, Y.H.; Chen, D.Y.; Chen, Y.M.; Tang, F.Y.; Ko, H.A.; Fan, Y.H.; Lin, Y.Y.; Wang, Y.C.; Chih, H.M.; et al. Folinate Supplementation Ameliorates Methotrexate Induced Mitochondrial Formate Depletion In Vitro and In Vivo. Int. J. Mol. Sci. 2021, 22, 1350. [Google Scholar] [CrossRef] [PubMed]
- Anguera, M.C.; Field, M.S.; Perry, C.; Ghandour, H.; Chiang, E.P.; Selhub, J.; Shane, B.; Stover, P.J. Regulation of folate-mediated one-carbon metabolism by 10-formyltetrahydrofolate dehydrogenase. J. Biol. Chem. 2006, 281, 18335–18342. [Google Scholar] [CrossRef]
- Wang, Y.C.; Tang, F.Y.; Chen, S.Y.; Chen, Y.M.; Chiang, E.P. Glycine-N methyltransferase expression in HepG2 cells is involved in methyl group homeostasis by regulating transmethylation kinetics and DNA methylation. J. Nutr. 2011, 141, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Chen, Y.M.; Lin, Y.J.; Liu, S.P.; Chiang, E.P. GNMT expression increases hepatic folate contents and folate-dependent methionine synthase-mediated homocysteine remethylation. Mol. Med. 2011, 17, 486–494. [Google Scholar] [CrossRef]
- Krishnan, A.; Gopinath, V.R.; Johnson, A.; Nair, S.A.; Pillai, M.R. Cell-cycle analysis and micronuclei frequency reveals G0/G1 blockers as weak micronuclei inducers. Drug Chem. Toxicol. 2013, 36, 249–254. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, A.J.; Anderson, D.D.; Flodby, P.; Perry, C.A.; Allen, R.H.; Stabler, S.P.; Stover, P.J. Nuclear localization of de novo thymidylate biosynthesis pathway is required to prevent uracil accumulation in DNA. J. Biol. Chem. 2011, 286, 44015–44022. [Google Scholar] [CrossRef]
- Hawn, M.T.; Umar, A.; Carethers, J.M.; Marra, G.; Kunkel, T.A.; Boland, C.R.; Koi, M. Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res. 1995, 55, 3721–3725. [Google Scholar]
- Janic, A.; Valente, L.J.; Wakefield, M.J.; Di Stefano, L.; Milla, L.; Wilcox, S.; Yang, H.; Tai, L.; Vandenberg, C.J.; Kueh, A.J.; et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat. Med. 2018, 24, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Wani, Y.; Notohara, K.; Tsukayama, C.; Okada, S. Reduced expression of hMLH1 and hMSH2 gene products in high-grade hepatocellular carcinoma. Acta Med. Okayama 2001, 55, 65–71. [Google Scholar] [CrossRef]
- Gu, L.; Wu, J.; Qiu, L.; Jennings, C.D.; Li, G.M. Involvement of DNA mismatch repair in folate deficiency-induced apoptosis small star, filled. J. Nutr. Biochem. 2002, 13, 355–363. [Google Scholar] [CrossRef]
- Lloyd, J.C.; Raines, C.A.; John, U.P.; Dyer, T.A. The chloroplast FBPase gene of wheat: Structure and expression of the promoter in photosynthetic and meristematic cells of transgenic tobacco plants. Mol. Gen. Genet. 1991, 225, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.D.; Woeller, C.F.; Stover, P.J. Small ubiquitin-like modifier-1 (SUMO-1) modification of thymidylate synthase and dihydrofolate reductase. Clin. Chem. Lab. Med. 2007, 45, 1760–1763. [Google Scholar] [CrossRef]
- Woeller, C.F.; Anderson, D.D.; Szebenyi, D.M.; Stover, P.J. Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J. Biol. Chem. 2007, 282, 17623–17631. [Google Scholar] [CrossRef]
- Fridman, A.; Saha, A.; Chan, A.; Casteel, D.E.; Pilz, R.B.; Boss, G.R. Cell cycle regulation of purine synthesis by phosphoribosyl pyrophosphate and inorganic phosphate. Biochem. J. 2013, 454, 91–99. [Google Scholar] [CrossRef]
- Kondo, M.; Yamaoka, T.; Honda, S.; Miwa, Y.; Katashima, R.; Moritani, M.; Yoshimoto, K.; Hayashi, Y.; Itakura, M. The rate of cell growth is regulated by purine biosynthesis via ATP production and G(1) to S phase transition. J. Biochem. 2000, 128, 57–64. [Google Scholar] [CrossRef]
- Collins, A.R.; Black, D.T.; Waldren, C.A. Aberrant DNA repair and enhanced mutagenesis following mutagen treatment of Chinese hamster Ade-C cells in a state of purine deprivation. Mutat. Res. 1988, 193, 145–155. [Google Scholar] [CrossRef]
- Williams, A.B.; Schumacher, B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb. Perspect. Med. 2016, 6, a026070. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.M.; Sanchez, C.A.; Morgan, C.A.; Schimke, M.K.; Ramel, S.; Idzerda, R.L.; Raskind, W.H.; Reid, B.J. A p53-dependent mouse spindle checkpoint. Science 1995, 267, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- Linke, S.P.; Clarkin, K.C.; Di Leonardo, A.; Tsou, A.; Wahl, G.M. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996, 10, 934–947. [Google Scholar] [CrossRef]
- Guo, X.; Ni, J.; Zhu, Y.; Zhou, T.; Ma, X.; Xue, J.; Wang, X. Folate deficiency induces mitotic aberrations and chromosomal instability by compromising the spindle assembly checkpoint in cultured human colon cells. Mutagenesis 2017, 32, 547–560. [Google Scholar] [CrossRef]
- Wang, L.; Bani-Hani, A.; Montoya, D.P.; Roche, P.C.; Thibodeau, S.N.; Burgart, L.J.; Roberts, L.R. hMLH1 and hMSH2 expression in human hepatocellular carcinoma. Int. J. Oncol. 2001, 19, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Matsukura, S.; Miyazaki, K.; Yakushiji, H.; Ogawa, A.; Chen, Y.; Sekiguchi, M. Combined loss of expression of O6-methylguanine-DNA methyltransferase and hMLH1 accelerates progression of hepatocellular carcinoma. J. Surg. Oncol. 2003, 82, 194–200. [Google Scholar] [CrossRef]
- Chen, J.; Sadowski, I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. Proc. Natl. Acad. Sci. USA 2005, 102, 4813–4818. [Google Scholar] [CrossRef]
- Leung, W.K.; Kim, J.J.; Wu, L.; Sepulveda, J.L.; Sepulveda, A.R. Identification of a second MutL DNA mismatch repair complex (hPMS1 and hMLH1) in human epithelial cells. J. Biol. Chem. 2000, 275, 15728–15732. [Google Scholar] [CrossRef]
- Kondo, E.; Horii, A.; Fukushige, S. The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res. 2001, 29, 1695–1702. [Google Scholar] [CrossRef]
- Fedier, A.; Ruefenacht, U.B.; Schwarz, V.A.; Haller, U.; Fink, D. Increased sensitivity of p53-deficient cells to anticancer agents due to loss of Pms2. Br. J. Cancer 2002, 87, 1027–1033. [Google Scholar] [CrossRef][Green Version]
- Sun, D.F.; Weng, Y.R.; Chen, Y.X.; Lu, R.; Wang, X.; Fang, J.Y. Knock-down of methylenetetrahydrofolate reductase reduces gastric cancer cell survival: An in vitro study. Cell Biol. Int. 2008, 32, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.Y.; Wu, H.J.; Wang, S.M.; Chen, P.M.; Tang, F.Y.; Chiang, E.I. MAT2A Localization and Its Independently Prognostic Relevance in Breast Cancer Patients. Int. J. Mol. Sci. 2021, 22, 5382. [Google Scholar] [CrossRef]
- Lathrop Stern, L.; Shane, B.; Bagley, P.J.; Nadeau, M.; Shih, V.; Selhub, J. Combined marginal folate and riboflavin status affect homocysteine methylation in cultured immortalized lymphocytes from persons homozygous for the MTHFR C677T mutation. J. Nutr. 2003, 133, 2716–2720. [Google Scholar] [CrossRef][Green Version]
- Chung, Y.C.; Tang, F.Y.; Liao, J.W.; Chung, C.H.; Jong, T.T.; Chen, S.S.; Tsai, C.H.; Chiang, E.P. Isatis indigotica induces hepatocellular cancer cell death via caspase-independent apoptosis-inducing factor translocation apoptotic pathway in vitro and in vivo. Integr. Cancer Ther. 2011, 10, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Fell, D.; Benjamin, L.E.; Steele, R.D. Determination of adenosine and S-adenosyl derivatives of sulfur amino acids in rat liver by high-performance liquid chromatography. J. Chromatogr. 1985, 345, 150–156. [Google Scholar] [CrossRef]
- Chiang, E.P.; Chiu, S.C.; Pai, M.H.; Wang, Y.C.; Wang, F.Y.; Kuo, Y.H.; Tang, F.Y. Organosulfur garlic compounds induce neovasculogenesis in human endothelial progenitor cells through a modulation of MicroRNA 221 and the PI3-K/Akt signaling pathways. J. Agric. Food Chem. 2013, 61, 4839–4849. [Google Scholar] [CrossRef]
- Tang, F.Y.; Chiang, E.P.; Pai, M.H. Consumption of S-allylcysteine inhibits the growth of human non-small-cell lung carcinoma in a mouse xenograft model. J. Agric. Food Chem. 2010, 58, 11156–11164. [Google Scholar] [CrossRef]
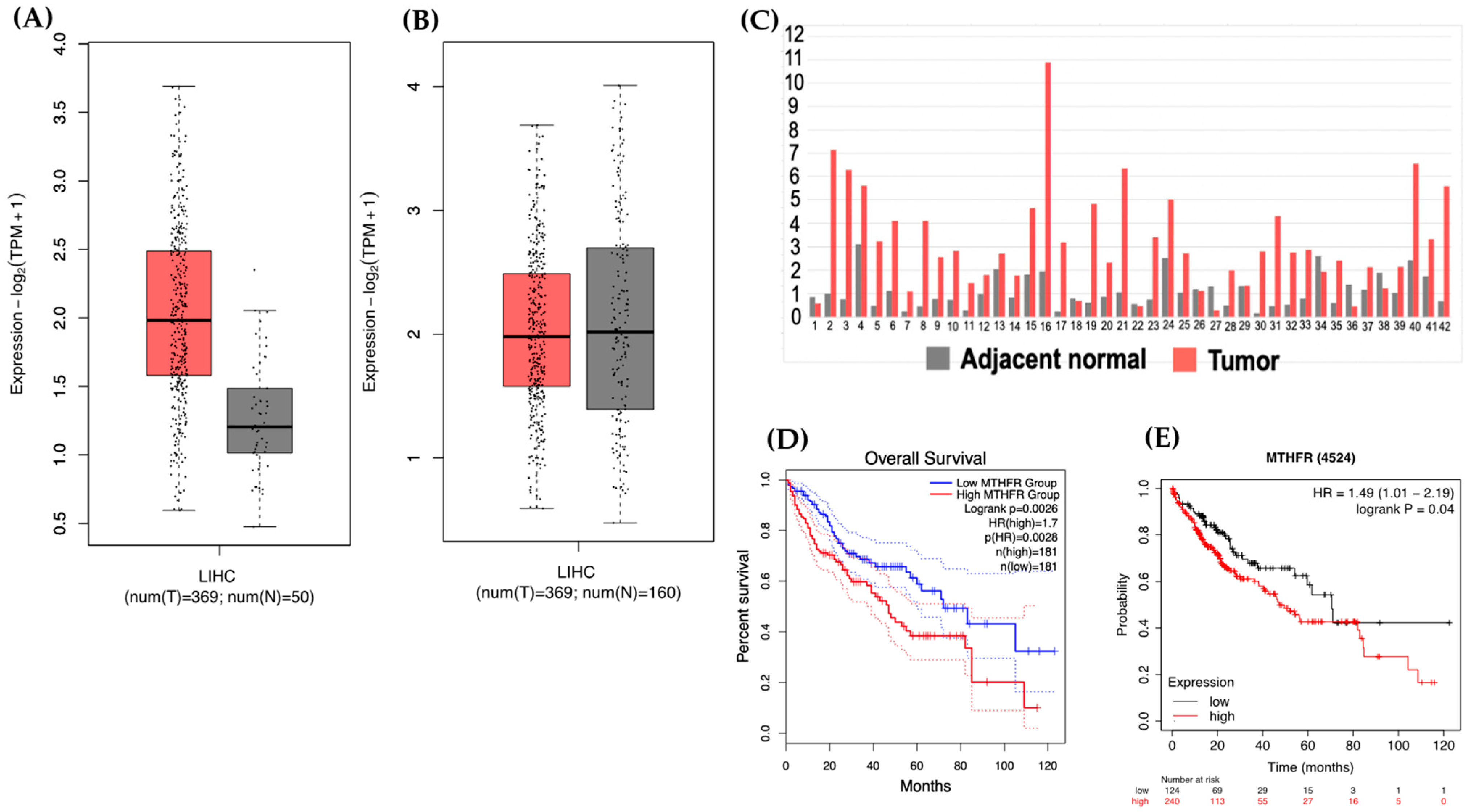

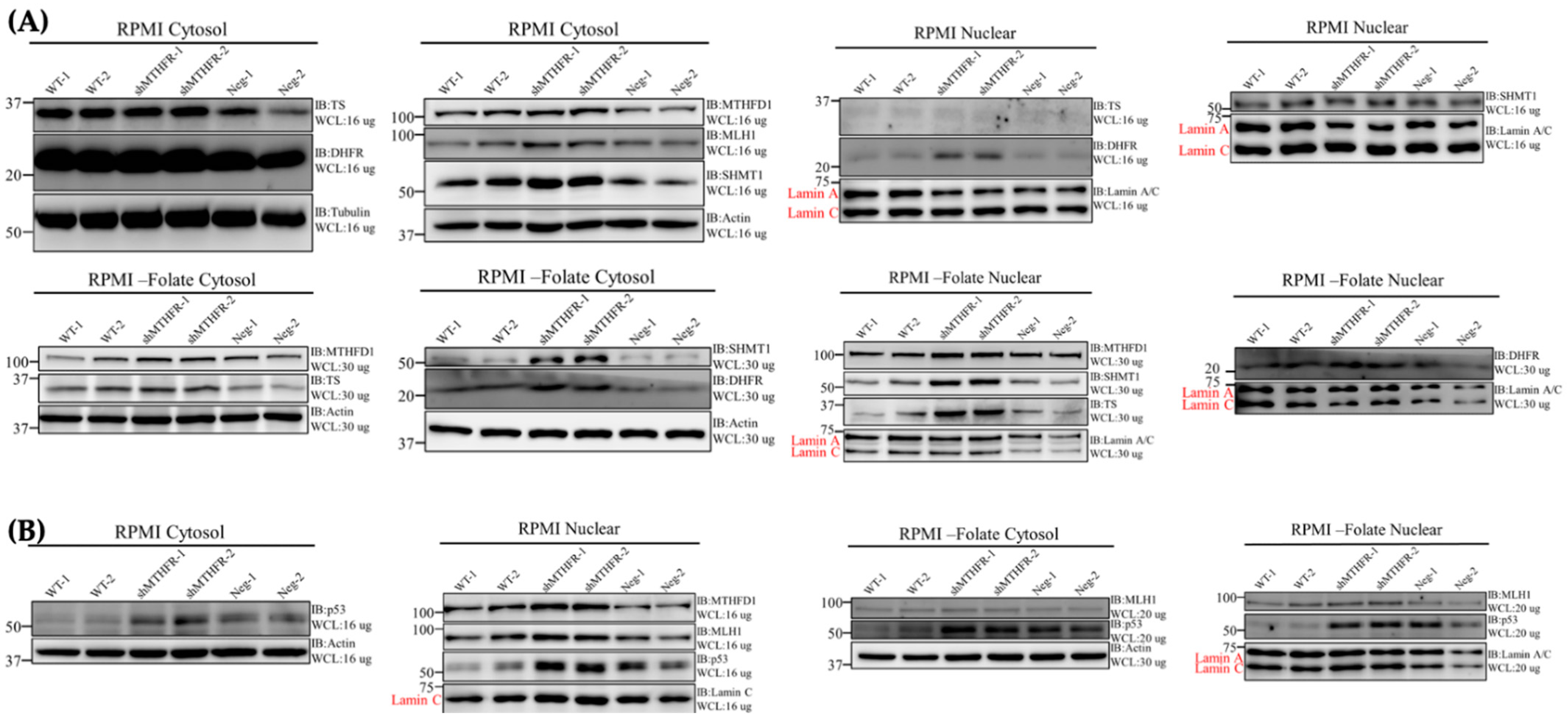
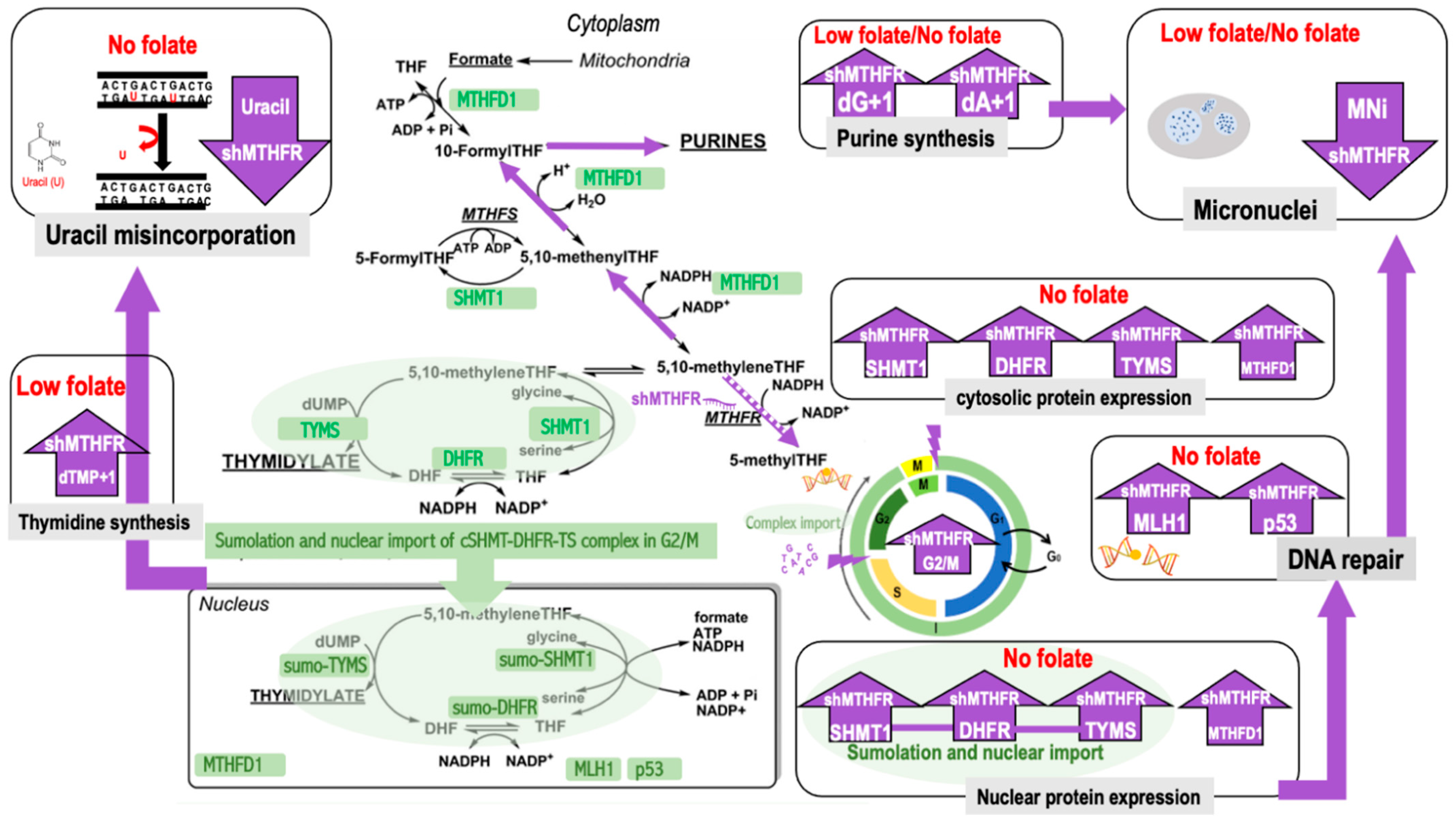
| (A) | |||||||
| shRNA (Clone ID) | Target sequence | Source | |||||
| TRCN0000046468/sh3′UTR | CCTCAGTTTCTCCATCAGCTT | National RNAi Core Facility, Academia Sinica, Taiwan | |||||
| TRCN0000046469/sh697 | GCTGACACATTCTTCCGCTTT | ||||||
| TRCN0000046470/sh77 | CCAAAGATAGTTCGAGATGTT | ||||||
| TRCN0000046471/sh546 | CCGAAGTGAGTTTGGTGACTA | ||||||
| TRCN0000046472/sh1618 | CTTGTCAATGTGAAGGGTGAA | ||||||
| TRCN0000072178/shGFP/Neg | CAACAGCCACAACGTCTATAT | ||||||
| (B) Gene expression and cell cycle distribution | |||||||
| Cell-line | MTHFR expression | G1 (%) 1 | S (%) 1 | G2/M (%) 1 | |||
| Wild type | 100 % | 71.2 ± 0.6 ab | 19.7 ± 1.0 f | 9.1 ± 0.6 m | |||
| RNAi shGFP 1 | 110 ± 1% | 73.8 ± 2.3 a | 17.3± 1.9 fgh | 8.9 ± 0.5 m | |||
| RNAi sh3′UTR 1 | 96 ± 10% | 70.1 ± 1.1 bc | 14.0 ± 0.2 i | 15.9 ± 1.0 k | |||
| RNAi sh1618 1 | 61 ± 2% | 68.5 ± 1.0 bc | 17.8 ± 0.6 gh | 13.5 ± 1.1 l | |||
| RNAi sh546 1 | 44 ± 3% | 64.8 ± 1.0 de | 18.0 ± 1.7 fg | 17.1 ± 0.3 k | |||
| RNAi sh697 1 | 42 ± 1% | 67.3 ± 0.7 cd | 16.1 ± 0.2 ghi | 16.6 ± 0.8 k | |||
| RNAi sh77 1 | 37 ± 6% | 63.5 ± 3.6 e | 14.9 ± 3.2 hi | 21.6 ± 0.4 j | |||
| (C) The associations between MTHFR gene expression and cell cycle distributions 2 | |||||||
| G1 (%) | S (%) | G2/M (%) | |||||
| n | r2 | p-value 2 | r2 | p-value 2 | r2 | p-value 2 | |
| MTHFR mRNA | 7 | 0.86 | <0.0001 | 0.034 | 0.907 | −0.808 | <0.0001 |
| (D) The associations between MTHFR gene expression and the cell cycle distribution 2 | |||||||
| MTHFR | n | G1 (%) | S (%) | G2/M (%) | |||
| r2 | p-value 2 | r2 | p-value 2 | r2 | p-value 2 | ||
| Gene expression | 7 | 0.86 | <0.0001 | 0.03 | 0.91 | −0.810 | <0.0001 |
| Protein expression | 3 | 0.56 | 0.12 | 0.760 | 0.020 | −0.800 | 0.010 |
| Enzyme Activity | 3 | 0.592 | 0.094 | 0.812 | 0.008 | −0.842 | 0.005 |
| (A) 2 | SAM 1 | % | SAH 1 | % | SAM: SAH ratio 1 % | ||
| Wt 1 | 438.5 ± 18.0 a | −30.8% 3 | 16.39 ± 0.46 | NS | 26.75 d | −20.0% 3,* | |
| Neg 1 | 523.1 ± 59.4 a | −42.0% 4 | 15.98 ± 0.36 | NS | 32.72 e | −34.6% 4 | |
| shMTHFR 1 | 303.5 ± 75.4 b | 14.18 ± 2.86 | 21.41 d | ||||
| (B) | SAM | SAH | SAM: SAH ratio 1 | ||||
| MTHFR 1 | n | r 7 | p-value 7 | R 7 | p-value 7 | r 7 | p-value 7 |
| mRNA 5 | 3 | 0.693 | 0.127 | 0.133 | 0.802 | 0.719 | 0.107 |
| Protein 6 | 3 | 0.353 | 0.351 | −0.153 | 0.694 | 0.362 | 0.339 |
| Activity 6 | 3 | 0.868 | 0.003 | 0.062 | 0.874 | 0.687 | 0.041 |
| (A) Cell cycle distributions in folate deficiency in Wt, Neg, and shMTHFR cells | ||||
| Genotype | G1 (%) | S (%) | G2/M (%) | |
| Folate replete 2 | 71.2 ± 1.1 a | 20.4 ± 1.1 ef | 9.1 ± 1.2 k | |
| Wt 1 | No folate 2 | 50.3 ± 4.2 c | 37.2 ± 4.2 g | 14.2 ± 1.3 j |
| % change | −21 ± 4 | +17 ± 4 | +4 ± 1 | |
| Folate replete 2 | 74. ± 2 a | 18.2 ± 2.1 ef | 9.1 ± 1.1 k | |
| Neg 1 | No folate 2 | 67.6 ± 2 ab | 20.6 ± 1.4 e | 13.6 ± 1 j |
| % change | −6 ± 2 | 3 ± 11 | +4 ± 1 | |
| Folate replete 2 | 64.3 ± 4 b | 15.7 ± 3.1 f | 22.2 ± 0.4 h | |
| shMTHFR 1 | No folate 2 | 49.4 ± 3 c | 33.2 ± 4.2 g | 19.5 ± 1.3 i |
| % change | −15 ± 3 | +18.1 ± 4 | −3 ± 1 | |
| (B) SAM and SAH contents in folate deficiency in Wt, Neg, and shMTHFR cells | ||||
| Genotype | SAM 4 | SAH 4 | SAM/SAH 5 | |
| Folate replete | 2457.3 ± 237.2 a | 92.3 ± 14.1 a | 26.8 ± 1.6 e | |
| Wt 1 | Low Folate 3 | 1753.6 ± 289.0 b | 78.1 ± 1.7 a | 22.4 ± 3.6 ef |
| No folate | 19,44.1 ± 501.8 ab | 406.2 ± 101.4 b | 4.78 ± 0.04 g | |
| Folate replete | 2577.9 ± 67.7 a | 79.3 ± 7.4 a | 32.7 ± 3.03 h | |
| Neg 1 | Low Folate 3 | 1444.0 ± 257.0 b | 72.1 ± 16.0 a | 20.2 ± 0.9 ef |
| No folate | 2076.8 ± 623.6 ab | 109.1 ± 2.6 a | 19.1 ± 6.1 ef | |
| Folate replete | 2021.9 ± 546.6 ab | 97.3 ± 34.6 a | 21.5 ± 3.55 f | |
| shMTHFR 1 | Low Folate 3 | 1618.5 ± 267.1 b | 120.8 ± 46.5 a | 14.1 ± 2.9 i |
| No folate | 1696.2 ± 277.0 b | 108.9 ± 5.4 a | 15.6 ± 2.7 i | |
| (A) Cytosol | ||||
| Cytosol | ||||
| Folate replete 4 | SHMT1 | DHFR | TYMS | MTHFD |
| WT 3 | 0.329 ± 0.038 a | 0.237 ± 0.032 ae | 0.293 ± 0.000 a | 0.340 ± 0.011 a |
| Neg 3 | 0.197 ± 0.197 c | 0.306 ± 0.016 a | 0.204 ± 0.054 abce | 0.227 ± 0.011 c |
| shMTHFR 3 | 0.411 ± 0.411 abf* | 0.289 ± 0.000 a | 0.320 ± 0.006 bc* | 0.370 ± 0.011 ab* |
| Cytosol | ||||
| Folate deplete 4 | SHMT1 | DHFR | TYMS | MTHFD |
| WT 3 | 0.081 ± 0.029 d | 0.170 ± 0.029 de | 0.186 ± 0.013 d | 0.135 ± 0.022 d |
| Neg 3 | 0.064 ± 0.001 df | 0.089 ± 0.016 df | 0.106 ± 0.015 f | 0.162 ± 0.023 def |
| shMTHFR 3 | 0.355 ± 0.029 ae | 0.241 ± 0.022 e* | 0.208 ± 0.001 de* | 0.203 ± 0.000 e |
| (B) Nucleus | ||||
| Nucleus | ||||
| Folate replete 4 | SHMT1 | DHFR | TYMS | MTHFD1 |
| WT 3 | 0.137 ± 0.020 a | 0.024 ± 0.000 a | 0.086 ± 0.020 a | 0.321 ± 0.001 a |
| Neg 3 | 0.105 ± 0.004 a | 0.019 ± 0.001 ac | 0.074 ± 0.028 a | 0.216 ± 0.001 c |
| shMTHFR 3 | 0.123 ± 0.006 a | 0.064 ± 0.001 b | 0.100 ± 0.007 a | 0.400 ± 0.006 b |
| Nucleus | ||||
| Folate deplete 4 | SHMT1 | DHFR | TYMS | MTHFD1 |
| WT 3 | 0.162 ± 0.033 de | 0.159 ± 0.019 de | 0.115 ± 0.071 de | 0.141 ± 0.003 df |
| Neg 3 | 0.093 ± 0.019 df | 0.096 ± 0.024 df | 0.096 ± 0.040 df | 0.129 ± 0.040 ef |
| shMTHFR 3 | 0.245 ± 0.044 e* | 0.206 ± 0.000 e* | 0.289 ± 0.022 e* | 0.231 ± 0.013 e |
| dA + 1 4 | dA + 2 4 | dA (MIA) 4 | dG + 1 4 | dG + 2 4 | dG (MIA) 4 | dT + 1 5 | |
|---|---|---|---|---|---|---|---|
| Wt 2 | |||||||
| Control 3 | 0.275 ± 0.000 a | 0.056 ± 0.000 ab | 0.413 ± 0.005 ab | 0.287 ± 0.000 a | 0.052 ± 0.000 a | 0.362 ± 0.000 a | 0.244 ± 0.001 a |
| −FA 3 | 0.173 ± 0.007 d | 0.025 ± 0.000 d | 0.294 ± 0.003 df | 0.188 ± 0.007 d | 0.022 ± 0.00 d | 0.240 ± 0.005 d | 0.217 ± 0.005 d |
| −FA + folinate 3 | 0.348 ± 0.001 gh | 0.086 ± 0.000 g | 0.499 ± 0.000 g | 0.340 ± 0.005 ghi | 0.074 ± 0.004 g | 0.439 ± 0.018 g | 0.278 ± 0.002 gi |
| Neg 2 | |||||||
| Control 3 | 0.291 ± 0.001 bc | 0.066 ± 0.001 c | 0.455 ± 0.005 c | 0.290 ± 0.006 a | 0.056 ± 0.003 a | 0.389 ± 0.035 ac | 0.270 ± 0.002 c |
| −FA 3 | 0.135 ± 0.003 f | 0.020 ± 0.001 f | 0.302 ± 0.011 f | 0.154 ± 0.004 f | 0.018 ± 0.002 f | 0.239 ± 0.026 df | 0.177 ± 0.001 f |
| −FA + folinate 3 | 0.343 ± 0.002 gi | 0.089 ± 0.000 i | 0.522 ± 0.001 i | 0.328 ± 0.002 i | 0.077 ± 0.003 g | 0.470 ± 0.015 gi | 0.284 ± 0.001 i |
| shMTHFR 2 | |||||||
| Control 3 | 0.269 ± 0.007 ab | 0.059 ± 0.003 bc | 0.442 ± 0.016 ac | 0.284 ± 0.006 a | 0.059 ± 0.002 a | 0.419 ± 0.011 b | 0.255 ± 0.001 b |
| −FA 3 | 0.179 ± 0.001 de | 0.032 ± 0.000 e | 0.358 ± 0.008 e | 0.197 ± 0.003 de | 0.029 ± 0.000 e | 0.298 ± 0.000 e | 0.216 ± 0.001 de |
| −FA + folinate 3 | 0.355 ± 0.002 h | 0.097 ± 0.000 h | 0.550 ± 0.003 h | 0.347 ± 0.005 h | 0.087 ± 0.000 g | 0.501 ± 0.008 h | 0.292 ± 0.000 h |
| (A) Micronuclei determined by cytokinesis-blocked micronucleus (CBMN) assay | ||||
| Number of micronuclei (MNi)/500 counts 1 | ||||
| Cell line 2 | Folate replete 3 | Low folate 3 | No folate 3 | |
| WT | 16.3 ± 3.1 a | 56.7 ± 2.5 d | 106.7 ± 9.0 g | |
| Neg | 25.0 ± 4.0 c | 71.0 ± 1.0 f | 68.0 ± 1.0 h | |
| shMTHFR | 6.0 ± 3.6 b | 27.3 ± 2.3 e | 28.3 ± 10.0 i | |
| (B) Uracil contents in the DNA 4 | ||||
| Folate replete 3 | No folate 3 | p-value 5 | % change 5 | |
| WT | 1.34 ± 0.02 | 2.14 ± 0.02 | <0.001 | +60.1 ± 1.69 |
| Neg | 1.33 ± 0.02 | 2.16 ± 0.01 | <0.001 | +62.4 ± 1.12 |
| shMTHFR | 1.24 ± 0.02 | 1.79 ± 0.08 | <0.001 | +45.1 ± 6.56 |
| Neg vs. WT | ||||
| p-value | 0.642 | 0.304 | ||
| % change | −0.50 ± 1.35 | 0.87 ± 0.69 | ||
| shMTHFR vs. WT | ||||
| p-value | 0.002 | 0.002 | ||
| % change | −7.44 ± 1.20 | −16.2 ± 3.79 | ||
| shMTHFR vs. Neg | ||||
| p-value | 0.002 | 0.001 | ||
| % change | −6.97 ± 1.21 | −16.9 ± 3.76 | ||
| 2-Way ANOVA 6 | ||||
| MTHFR effect | <0.001 | |||
| Folate effect | <0.001 | |||
| MTHFR x Folate | <0.001 | |||
| (C) shMTHFR induced p53, MLH1 proteins expression in cytosol and nuclear 7 | ||||
| Cytosol | Nuclear | |||
| Folate replete 8 | p53 | MLH1 | p53 | MLH1 |
| WT | 0.108 ± 0.015 a | 0.179 ± 0.035 ab | 0.126 ± 0.030 a | 0.278 ± 0.029 a |
| Neg | 0.239 ± 0.001 c | 0.173 ± 0.022 a | 0.218 ± 0.030 a | 0.248 ± 0.051 a |
| shMTHFR | 0.278 ± 0.032 bc# | 0.273 ± 0.007 b* | 0.281 ± 0.022 b | 0.411 ± 0.038 a*# |
| Cytosol | Nuclear | |||
| Folate deplete 8 | p53 | MLH1 | p53 | MLH1 |
| WT | 0.079 ± 0.031 de | 0.161 ± 0.038 d | 0.079 ± 0.002 df | 0.174 ± 0.036 d |
| Neg | 0.179 ± 0.008 f | 0.142 ± 0.016 d | 0.179 ± 0.068 ef | 0.096 ± 0.026 d |
| shMTHFR | 0.242 ± 0.053 ef* | 0.197 ± 0.011 d*# | 0.242 ± 0.016 e* | 0.229 ± 0.001 e |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.-T.; Ye, W.-T.; Wang, Y.-C.; Chen, P.-M.; Liu, J.-Y.; Tai, C.-K.; Tang, F.-Y.; Li, J.-R.; Liu, C.-C.; Chiang, E.-P.I. MTHFR Knockdown Assists Cell Defense against Folate Depletion Induced Chromosome Segregation and Uracil Misincorporation in DNA. Int. J. Mol. Sci. 2021, 22, 9392. https://doi.org/10.3390/ijms22179392
Wu M-T, Ye W-T, Wang Y-C, Chen P-M, Liu J-Y, Tai C-K, Tang F-Y, Li J-R, Liu C-C, Chiang E-PI. MTHFR Knockdown Assists Cell Defense against Folate Depletion Induced Chromosome Segregation and Uracil Misincorporation in DNA. International Journal of Molecular Sciences. 2021; 22(17):9392. https://doi.org/10.3390/ijms22179392
Chicago/Turabian StyleWu, Ming-Tsung, Wei-Ting Ye, Yi-Cheng Wang, Po-Ming Chen, Jun-You Liu, Chien-Kuo Tai, Feng-Yao Tang, Jian-Rong Li, Chun-Chi Liu, and En-Pei Isabel Chiang. 2021. "MTHFR Knockdown Assists Cell Defense against Folate Depletion Induced Chromosome Segregation and Uracil Misincorporation in DNA" International Journal of Molecular Sciences 22, no. 17: 9392. https://doi.org/10.3390/ijms22179392
APA StyleWu, M.-T., Ye, W.-T., Wang, Y.-C., Chen, P.-M., Liu, J.-Y., Tai, C.-K., Tang, F.-Y., Li, J.-R., Liu, C.-C., & Chiang, E.-P. I. (2021). MTHFR Knockdown Assists Cell Defense against Folate Depletion Induced Chromosome Segregation and Uracil Misincorporation in DNA. International Journal of Molecular Sciences, 22(17), 9392. https://doi.org/10.3390/ijms22179392






