Personal Glucose Meter for α-Glucosidase Inhibitor Screening Based on the Hydrolysis of Maltose
Abstract
:1. Introduction
2. Results and Discussion
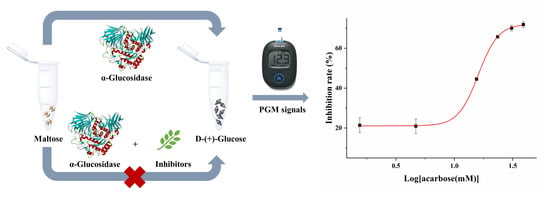
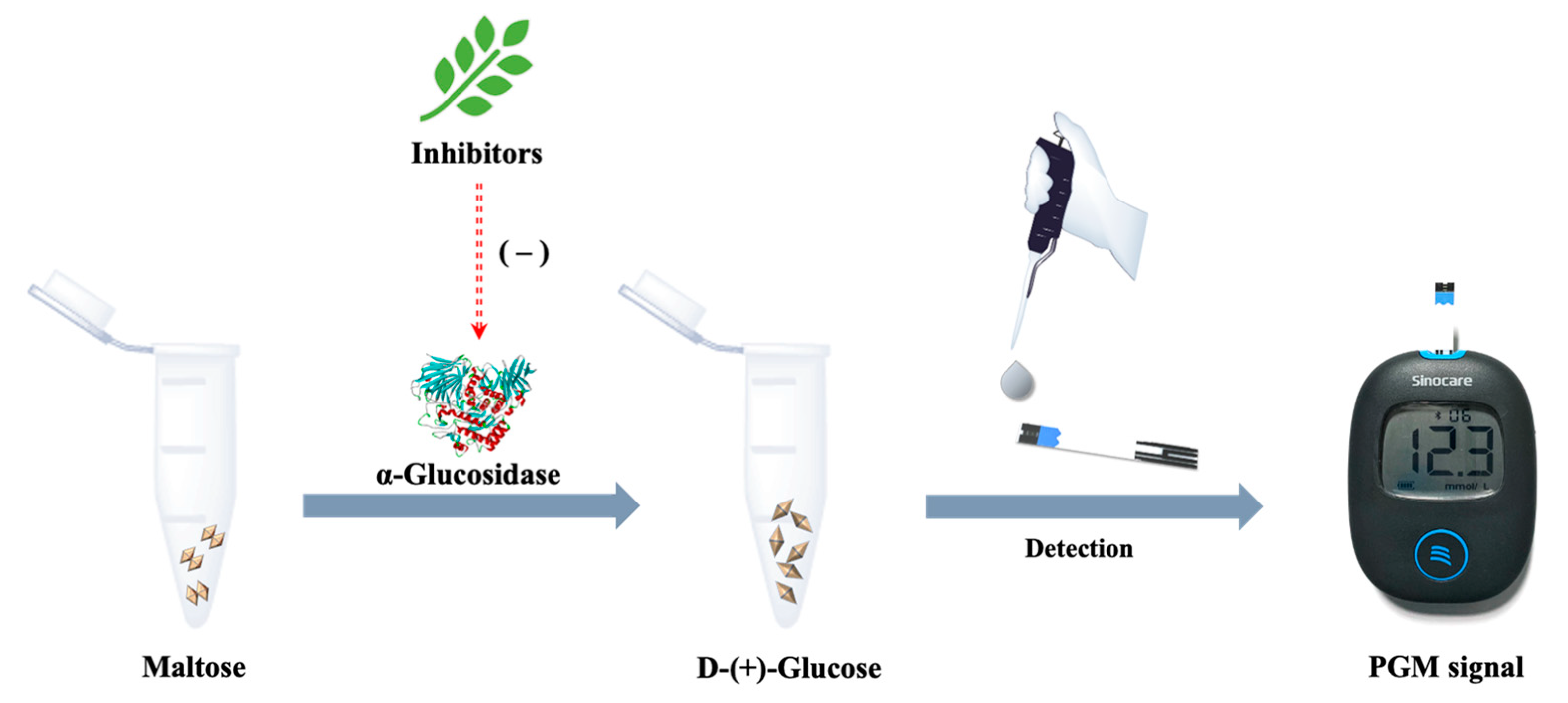
2.1. Principle of the PGM Method Based on α-Glucosidase-Mediated Cascade Reaction
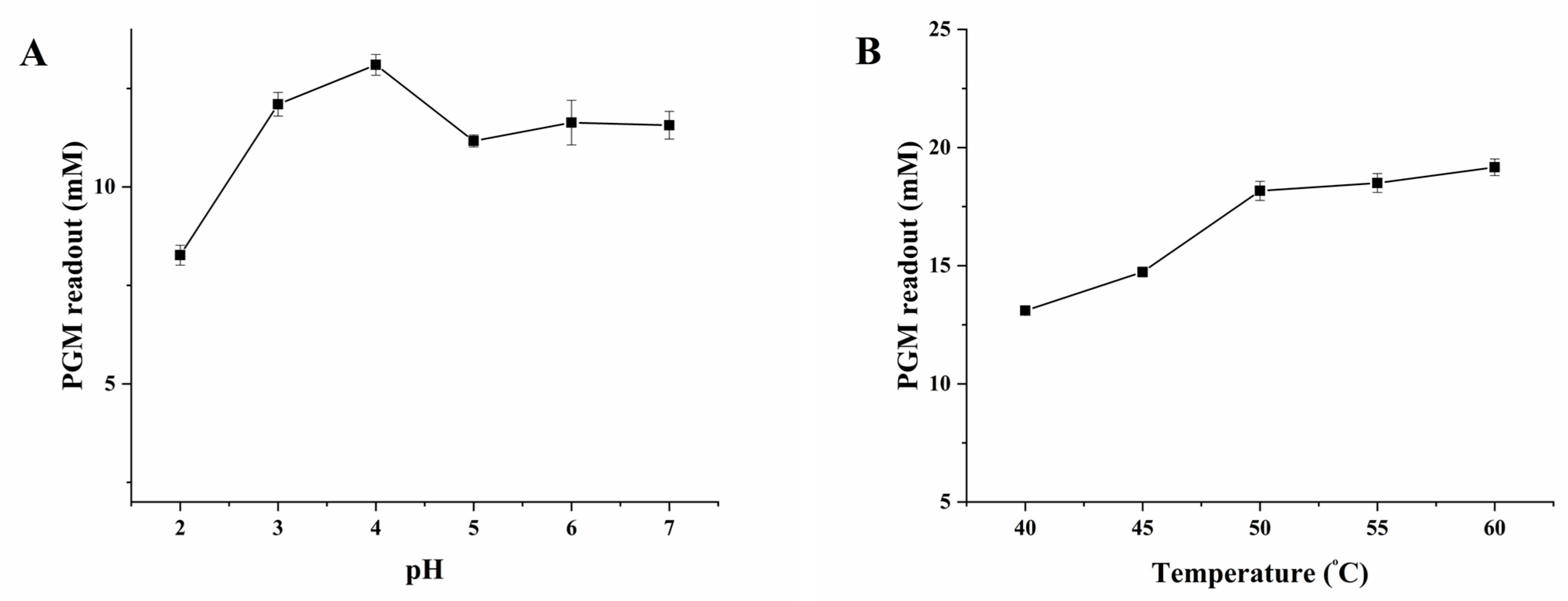
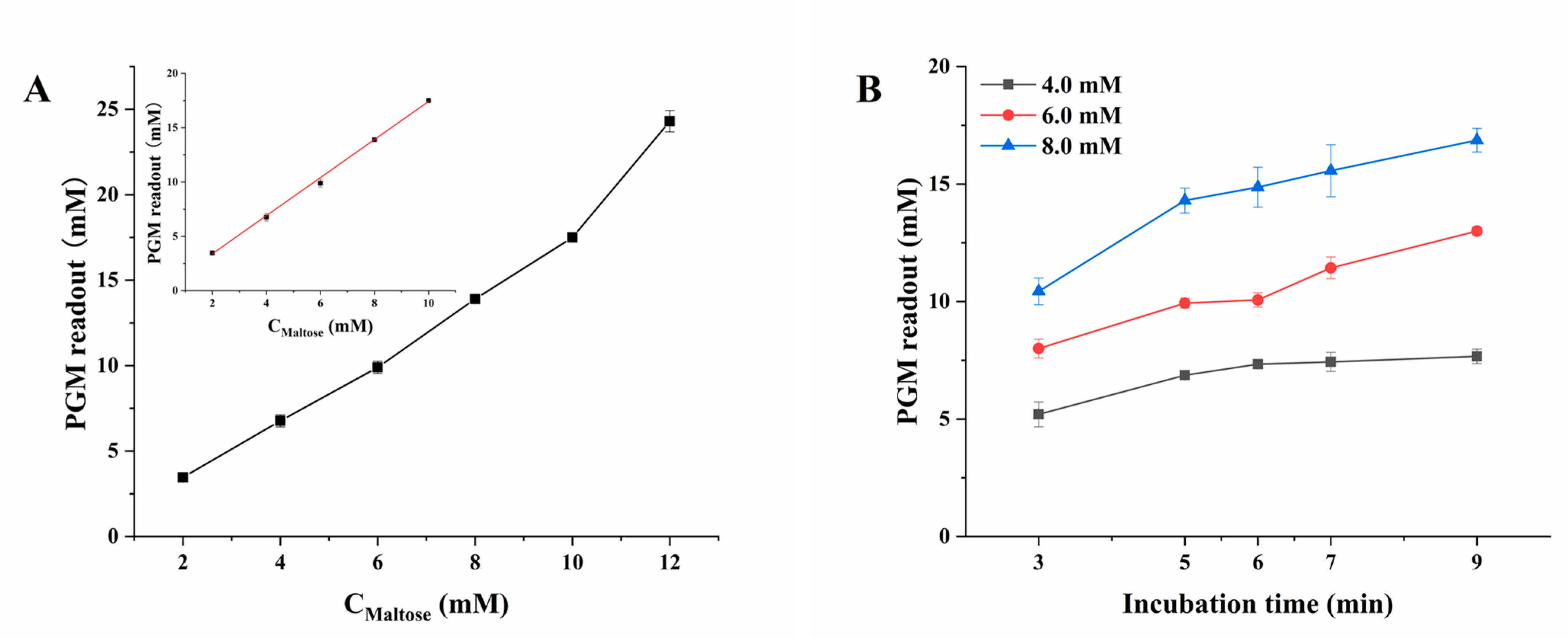
2.2. Optimization of the Experimental Conditions
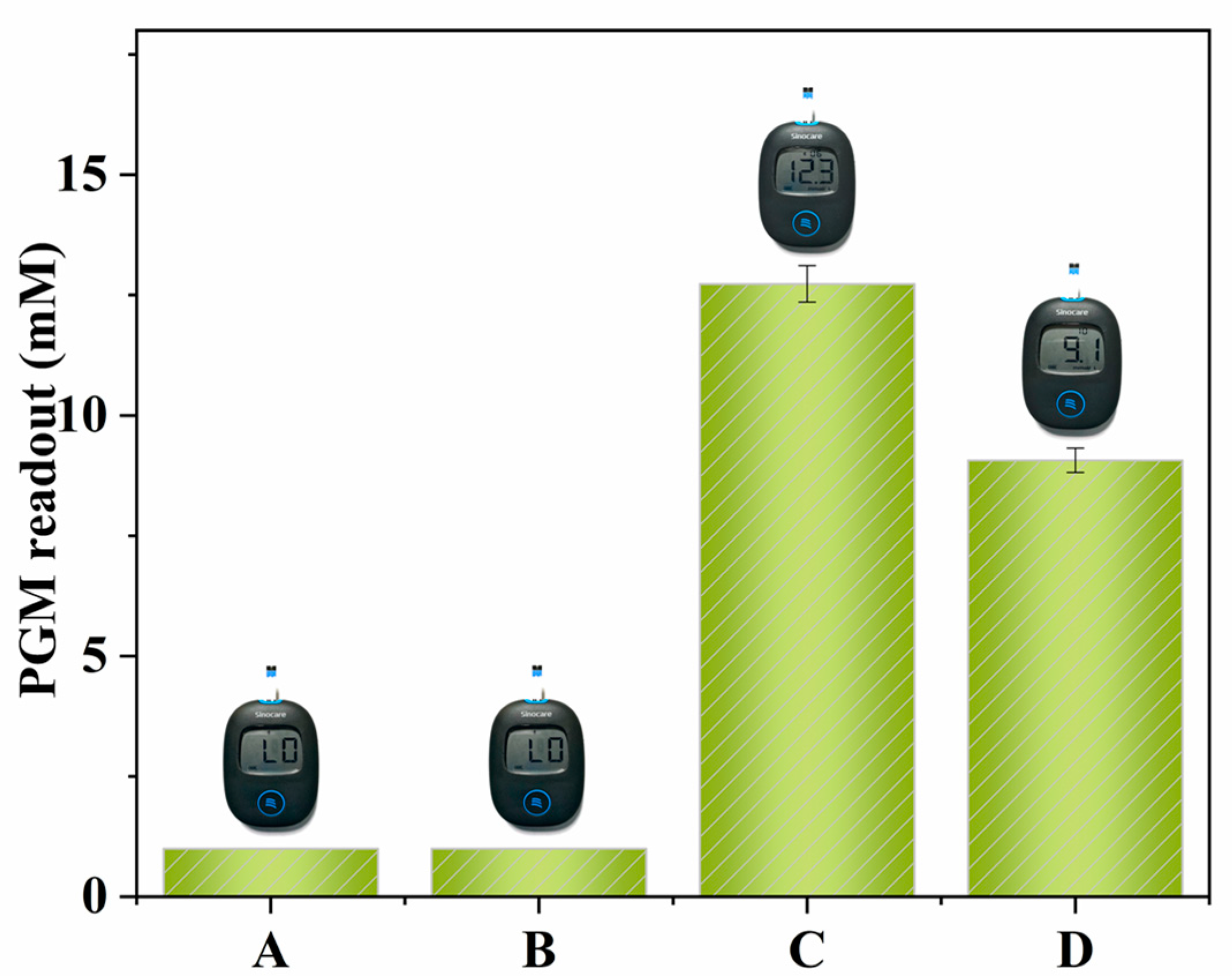
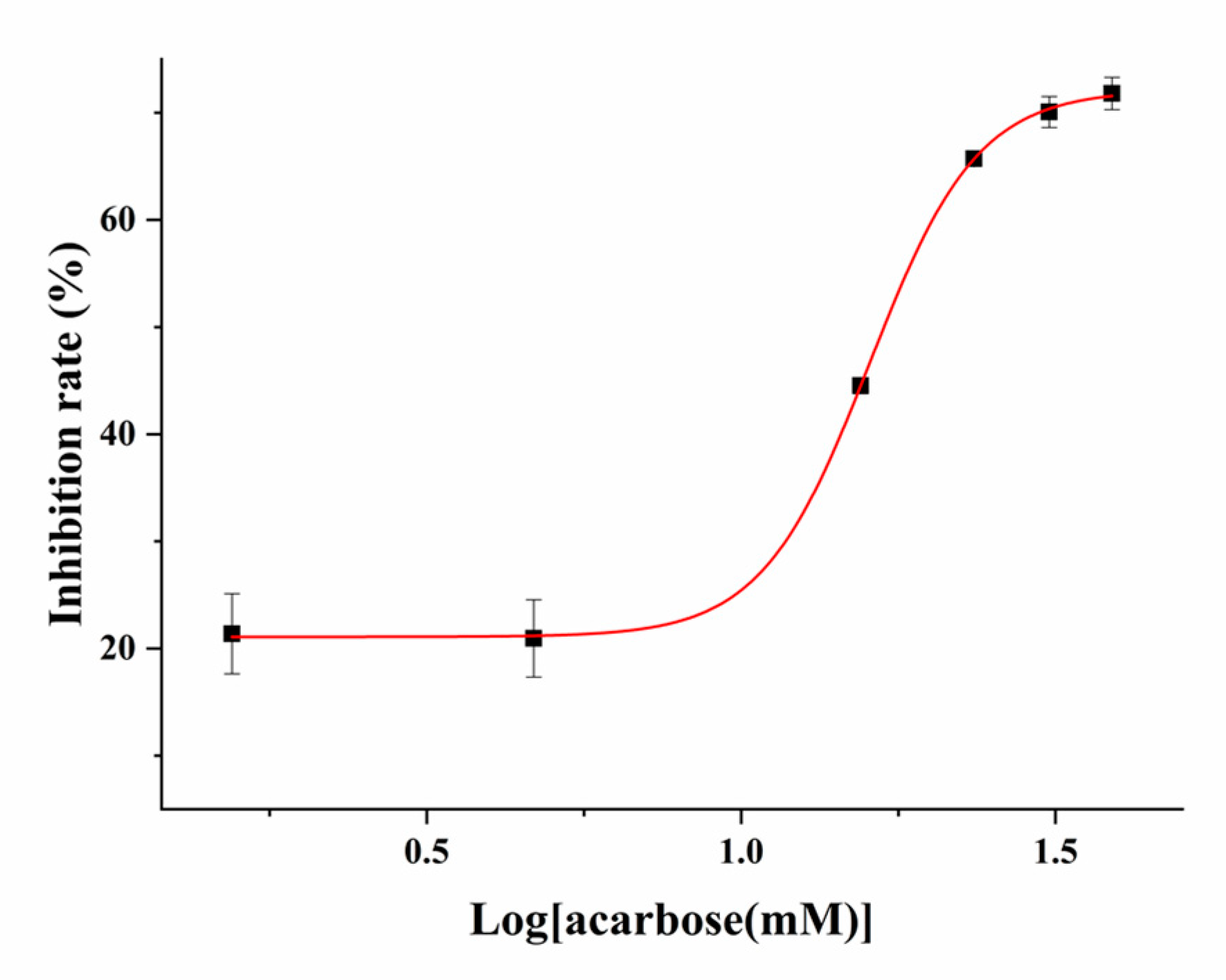
2.3. The Screening of Inhibitors of α-Glucosidase Using the PGM Method
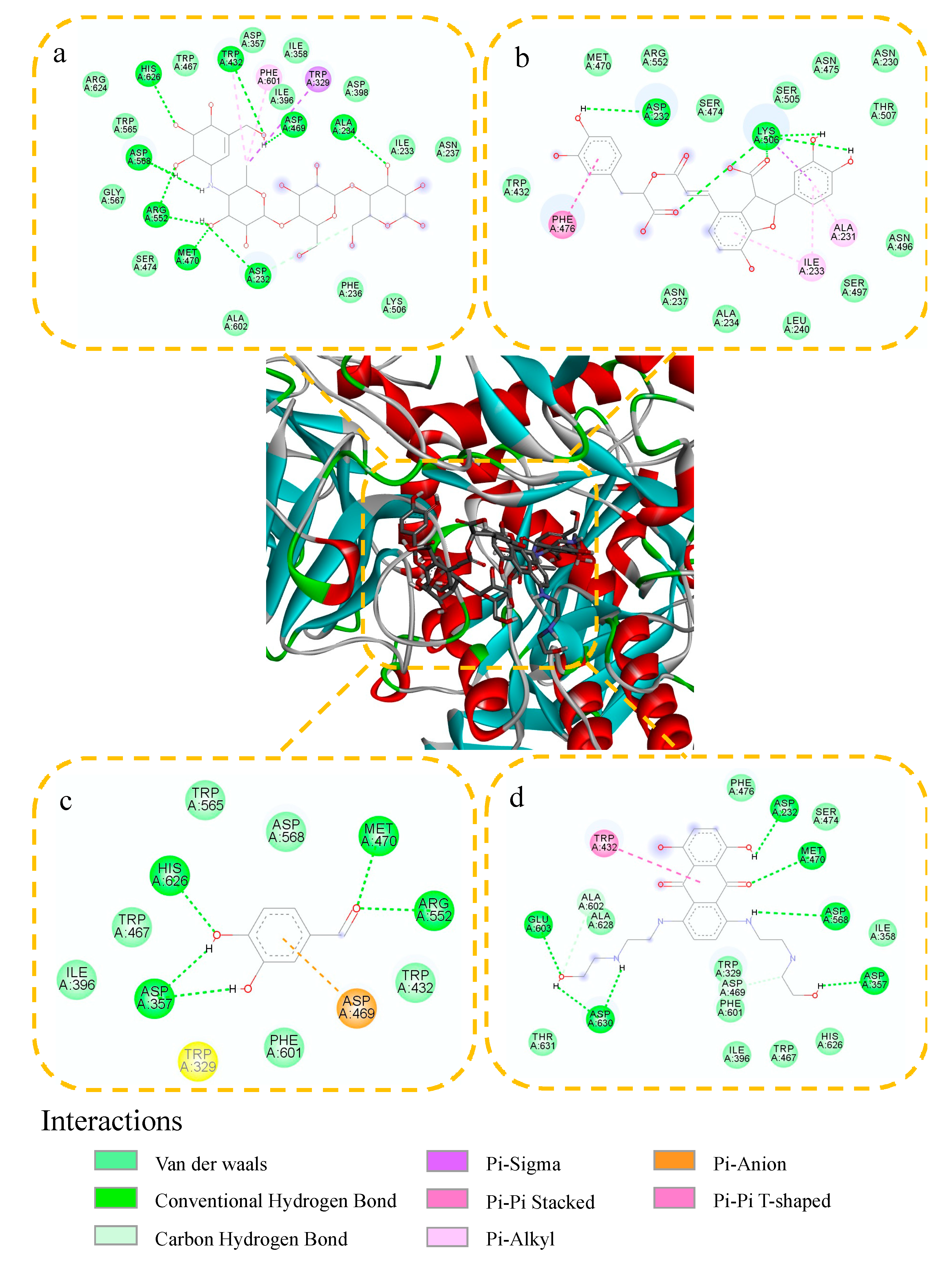
2.4. Molecular Docking Study
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Instruments
3.3. Preparation of Solutions and Samples
3.4. Analytical Procedure for α-Glucosidase Inhibitory Activity Evaluation
3.5. Inhibition Kinetics Study and Validation of the PGM-Based Method
3.6. Molecular Docking Study
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- World Health Organization. Global Report on Diabetes; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Chinsembu, K.C. Diabetes mellitus and nature’s pharmacy of putative antidiabetic plants. J. Herb. Med. 2019, 15, 100230. [Google Scholar] [CrossRef]
- World Health Organization. Definition, Diagnosis and Classification of Diabetes Mellitus and Its Complications: Report of a WHO Consultation. Part 1, Diagnosis and Classification of Diabetes Mellitus; World Health Organization: Geneva, Switzerland, 1999. [Google Scholar]
- American Diabetes Association. Expert Committee on the Diagnosis and Classification of Diabetes Mellitus Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2003, 26, S5–S20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, C.S.; Golden, S.H.; Anderson, C.; Bray, G.A.; Burke, L.E.; De Boer, I.H.; Vafiadis, D.K. Update on prevention of cardiovascular disease in adults with type 2 diabetes mellitus in light of recent evidence: A scientific statement from the American Heart Association and the American Diabetes Association. Circulation 2015, 132, 691–718. [Google Scholar] [CrossRef] [PubMed]
- Abbas, G.; Al Harrasi, A.; Hussain, H.; Hamaed, A.; Supuran, C.T. The management of diabetes mellitus-imperative role of natural products against dipeptidyl peptidase-4, α-glucosidase and sodium-dependent glucose co-transporter 2 (SGLT2). Bioorg. Chem. 2019, 86, 305–315. [Google Scholar] [CrossRef] [PubMed]
- de Melo, E.B.; da Silveira Gomes, A.; Carvalho, I. α-and β-Glucosidase inhibitors: Chemical structure and biological activity. Tetrahedron. 2006, 62, 10277–10302. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, G.; Pan, J.; Wang, Y. α-Glucosidase inhibition by luteolin: Kinetics, interaction and molecular docking. Int. J. Biol. Macromol. 2014, 64, 213–223. [Google Scholar] [CrossRef]
- Modak, M.; Dixit, P.; Londhe, J.; Ghaskadbi, S.; Devasagayam, T.P.A. Indian herbs and herbal drugs used for the treatment of diabetes. J. Clin. Biochem. Nutr. 2007, 40, 163–173. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.; Khan, A.; Halim, S.A.; Khan, M.; Zaib, S.; Al-Yahyaei, B.E.M.; Ibrar, A. Utilization of the common functional groups in bioactive molecules: Exploring dual inhibitory potential and computational analysis of keto esters against α-glucosidase and carbonic anhydrase-II enzymes. Int. J. Biol. Macromol. 2021, 167, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Tanaka, T.; Tamura, S.; Toshima, A.; Tamaya, K.; Miyata, Y.; Matsumoto, K. α-Glucosidase inhibitory profile of catechins and theaflavins. J. Agric. Food Chem. 2007, 55, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhang, Y.; Cheng, Y.; Wang, Y. Rapid screening and identification of α-glucosidase inhibitors from mulberry leaves using enzyme-immobilized magnetic beads coupled with HPLC/MS and NMR. Biomed. Chromatogr. 2013, 27, 148–155. [Google Scholar] [CrossRef]
- Li, G.; Kong, W.; Zhao, M.; Lu, S.; Gong, P.; Chen, G.; Wu, Y. A fluorescence resonance energy transfer (FRET) based “Turn-On” nanofluorescence sensor using a nitrogen-doped carbon dot-hexagonal cobalt oxyhydroxide nanosheet architecture and application to α-glucosidase inhibitor screening. Biosens. Bioelectron. 2016, 79, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Qian, Z.; Qian, Y.; Huang, Y.; Zhao, M.; Ao, H.; Feng, H. A fluorometric and real-time assay for α-glucosidase activity through supramolecular self-assembly and its application for inhibitor screening. Sens. Actuators B Chem. 2017, 245, 282–289. [Google Scholar] [CrossRef]
- Liu, D.M.; Dong, C.; Ma, R.T. A colorimetric method for screening α-glucosidase inhibitors from flavonoids using 3, 3’, 5, 5’-tetramethylbenzidine as a chromogenic probe. Colloids Surf. B Biointerfaces 2021, 197, 111400. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, J.; Wu, H.; Koh, K.; Yin, Y. Sensitive colorimetric assays for α-glucosidase activity and inhibitor screening based on unmodified gold nanoparticles. Anal. Chim. Acta 2015, 875, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Lisi, F.; Peterson, J.R.; Gooding, J.J. The application of personal glucose meters as universal point-of-care diagnostic tools. Biosens. Bioelectron. 2020, 148, 111835. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Li, X.; Sun, X.; Zhang, J.; Zhao, Y.; Liu, X.; Li, F. DNA tetrahedra-cross-linked hydrogel functionalized paper for onsite analysis of dna methyltransferase activity using a personal glucose meter. Anal. Chem. 2020, 92, 4592–4599. [Google Scholar] [CrossRef]
- Tang, J.; Huang, Y.; Liu, H.; Zhang, C.; Tang, D. Novel glucometer-based immunosensing strategy suitable for complex systems with signal amplification using surfactant-responsive cargo release from glucose-encapsulated liposome nanocarriers. Biosens. Bioelectron. 2016, 79, 508–514. [Google Scholar] [CrossRef]
- Fu, L.; Zhuang, J.; Lai, W.; Que, X.; Lu, M.; Tang, D. Portable and quantitative monitoring of heavy metal ions using DNAzyme-capped mesoporous silica nanoparticles with a glucometer readout. J. Mater. Chem. B 2013, 1, 6123–6128. [Google Scholar] [CrossRef]
- Xiang, Y.; Lu, Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011, 3, 697–703. [Google Scholar] [CrossRef]
- Zhao, Y.; Du, D.; Lin, Y. Glucose encapsulating liposome for signal amplification for quantitative detection of biomarkers with glucometer readout. Biosens. Bioelectron. 2015, 72, 348–354. [Google Scholar] [CrossRef]
- Chen, G.Y.; Zhang, H.; Yang, F.Q. A simple and portable method for beta-Glucosidase activity assay and its inhibitor screening based on a personal glucose meter. Anal. Chim. Acta 2021, 1142, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, G.Y.; Qian, Z.M.; Li, W.J.; Li, C.H.; Hu, Y.J.; Yang, F.Q. A portable personal glucose meter method for enzyme activity detection and inhibitory activity evaluation based on alkaline phosphatase-mediated reaction. Anal. Bioanal. Chem. 2021, 413, 2457–2466. [Google Scholar] [CrossRef] [PubMed]
- Giuberti, G.; Rocchetti, G.; Lucini, L. Interactions between phenolic compounds, amylolytic enzymes and starch: An updated overview. Curr. Opin. Food Sci. 2020, 31, 102–113. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, H.; Wang, X.; Jiang, H.; Hu, G.; Yang, F.Q. Preparation of phytic acid modified α-Glucosidase/Cu3(PO4)2·3H2O hybrid nanoflower and its application. Enzyme Microb. Technol. 2021, 146, 0141–0229. [Google Scholar] [CrossRef]
| Compounds | Inhibition (%) | Compounds | Inhibition (%) | Compounds | Inhibition (%) |
|---|---|---|---|---|---|
| Acarbose | 31.5 ± 2.5 | Protocatechuic acid | 11.9 ± 2.6 | Rosmarinic acid | 0 |
| Lithospermic acid | 52.5 ± 3.0 | Catechin | 10.3 ± 4.2 | (-)-Epigallocatechin gallate | 0 |
| Protocatechualdehyde | 36.8 ± 2.8 | Puerarin 6’’-O-xyloside | 7.5 ± 6.8 | Gallic acid monohydrate | 0 |
| 2,3,5,4′-Tetrahydroxy stilbene-2-O-β-d-glucoside | 27.0 ± 2.0 | Cryptochlorogenic acid | 4.6 ± 3.7 | Genipin | 0 |
| Anhydrous citric acid | 20.7 ± 3.4 | Theophylline | 4.5 ± 1.5 | Gastrodin | 0 |
| Salvianolic acid B | 19.4 ± 1.8 | Sorbic acid | 1.1 ± 1.4 | Brucine | 0 |
| Vanillic acid | 19.0 ± 1.8 | Chlorogenic acid | 0.9 ± 0 | Berbine | 0 |
| Vanillin | 17.6 ± 0.7 | Berberine hydrochloride | 0.9 ± 4.7 | 2,3,5,6-Tetramethylpyrazine | 0 |
| Danshensu | 17.2 ± 0.9 | α-Arbutin | 0 | trans-Cinnamaldehyde | 0 |
| (-)-Epigallocatechin | 14.1 ± 1.7 | Syringin | 0 | Tannic acid | - 1 |
| (S)-(+)-Mandelic acid | 13.6 ± 3.7 | Mogroside V | 0 | - | - |
| Paeoniflorin | 12.8 ± 1.6 | Salvianolic acid A | 0 | - | - |
| Extracts | Inhibition (%) | Extracts | Inhibition (%) | Extracts | Inhibition (%) |
|---|---|---|---|---|---|
| Lemon | 43.3 ± 3.5 | Common Anemarrhena | 5.1 ± 2.5 | Radix Polygoni multiflori preparata | 0 |
| Ligusticum chuanxiong hort | 14.1 ± 2.5 | Gengen | 4.1 ± 3.2 | Paeoniae Radix Alba | 0 |
| Mulberry Leaf | 8.6 ± 0.6 | Ural licorice | 3.8 ± 0.4 | Rhubarb | 0 |
| Salvia miltiorrhiza | 8.0 ± 1.9 | Polygonatum sibiricum | 2.8 ± 2.3 | Radix angelicae sinensis | 0 |
| Membranous milkvetch root | 6.9 ± 1.9 | Radix lithospermi | 0.6 ± 1.7 | Honeysuckle flower | - 1 |
| Cinnamon | 6.5 ± 3.5 | Polygonum cuspidatum | 1.2 ± 3.1 | Chinese nut-gall | - 1 |
| Compounds | Binding Energy (Kcal/mol) | Hydrogen Bonds | Other Amino Acid Residues |
|---|---|---|---|
| Acarbose | −5.03 | ASP568, ASP232, ARG552, ASP469, ALA234, MET470, HIS626, TRP432 | TRP467, TRP565, ARG624, ASP357, ILE358, ILE396, ASP398, ILE233, ASN237, LYS506, PHE236, LA602, SER474, GLY567, PHE601, TRP329 |
| Lithospermic acid | −4.68 | ASP232, LYS506 | TRP432, MET470, ARG552, SER474, SER505, ASN475, ASN230, THR507, ASN496, SER497, LEU240, ALA234, ASN237, PHE476, ILE233, ALA231 |
| Protocatechualdehyde | −4.96 | ASP357, ARG552, HIS626, MET470 | ILE396, TRP467, TRP565, ASP568, TRP432, PHE601, ASP469, TRP329 |
| 2,3,5,4′-Tetrahydroxy stilbene-2-O-β-D-glucoside | −8.74 | GLU603, ASP630, ASP357, ASP232, ASP568, MET470 | THR631, ALA602, ALA628, PHE476, SER474, ILE358, TRP329, ASP469, PHE601, ILE396, TRP467, HIS626, TRP432 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, T.; Chen, G.-Y.; Zhang, H.; Yang, F.-Q. Personal Glucose Meter for α-Glucosidase Inhibitor Screening Based on the Hydrolysis of Maltose. Molecules 2021, 26, 4638. https://doi.org/10.3390/molecules26154638
Tian T, Chen G-Y, Zhang H, Yang F-Q. Personal Glucose Meter for α-Glucosidase Inhibitor Screening Based on the Hydrolysis of Maltose. Molecules. 2021; 26(15):4638. https://doi.org/10.3390/molecules26154638
Chicago/Turabian StyleTian, Tao, Guo-Ying Chen, Hao Zhang, and Feng-Qing Yang. 2021. "Personal Glucose Meter for α-Glucosidase Inhibitor Screening Based on the Hydrolysis of Maltose" Molecules 26, no. 15: 4638. https://doi.org/10.3390/molecules26154638
APA StyleTian, T., Chen, G.-Y., Zhang, H., & Yang, F.-Q. (2021). Personal Glucose Meter for α-Glucosidase Inhibitor Screening Based on the Hydrolysis of Maltose. Molecules, 26(15), 4638. https://doi.org/10.3390/molecules26154638