Hypoxia and the Receptor for Advanced Glycation End Products (RAGE) Signaling in Cancer
Abstract
:1. Physiological and Pathological Hypoxia
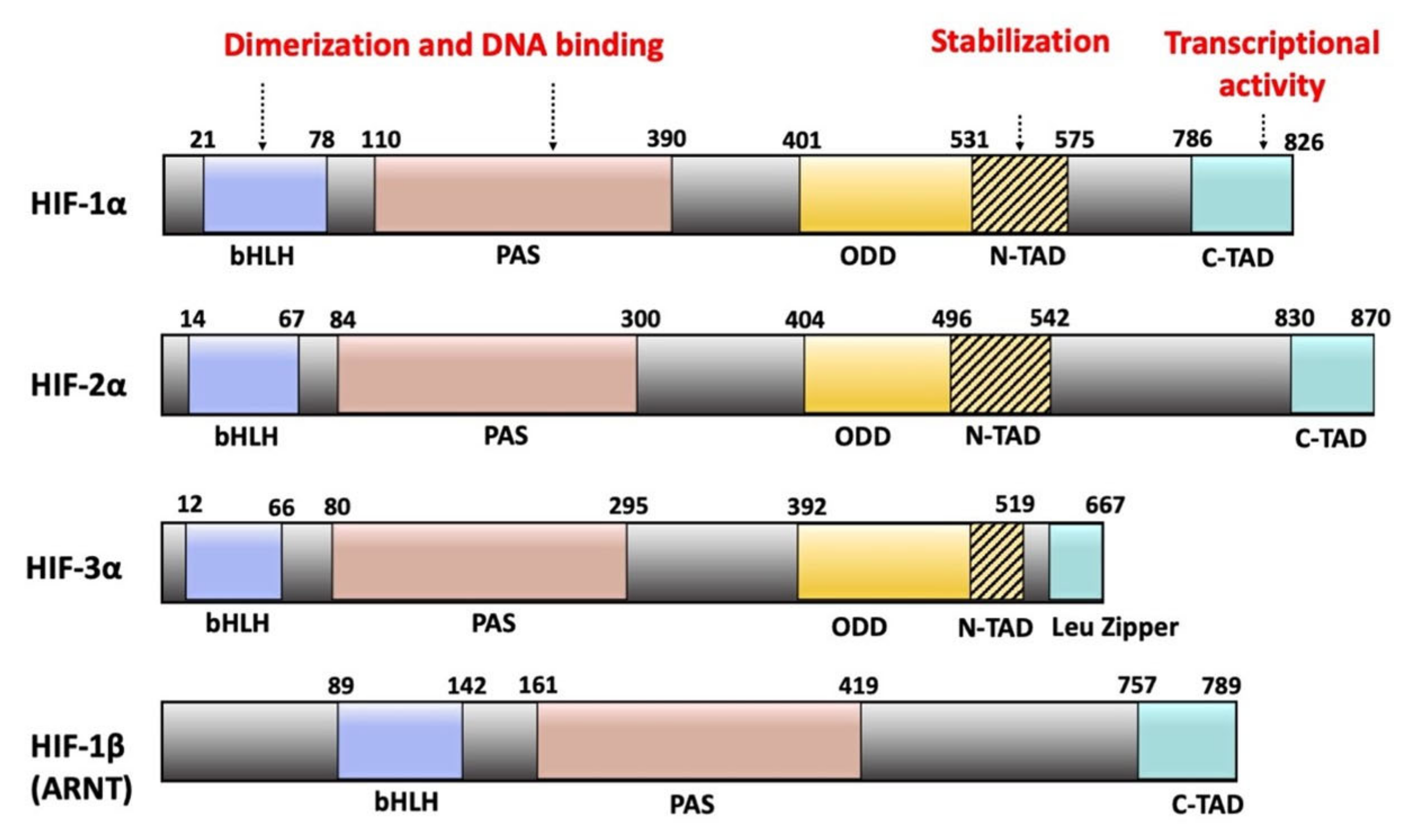
2. Hypoxia-Inducible Factors: Structure and Mechanisms
2.1. HIF Subunits
2.2. Domain Organization
2.3. Regulation Mechanisms
3. Effect of Hypoxia in Tumors
3.1. Metabolic Reprogramming
3.2. Angiogenesis
3.3. Apoptosis
3.4. Cell Invasion and Metastasis
3.5. Tumor Microenvironment and Inflammation
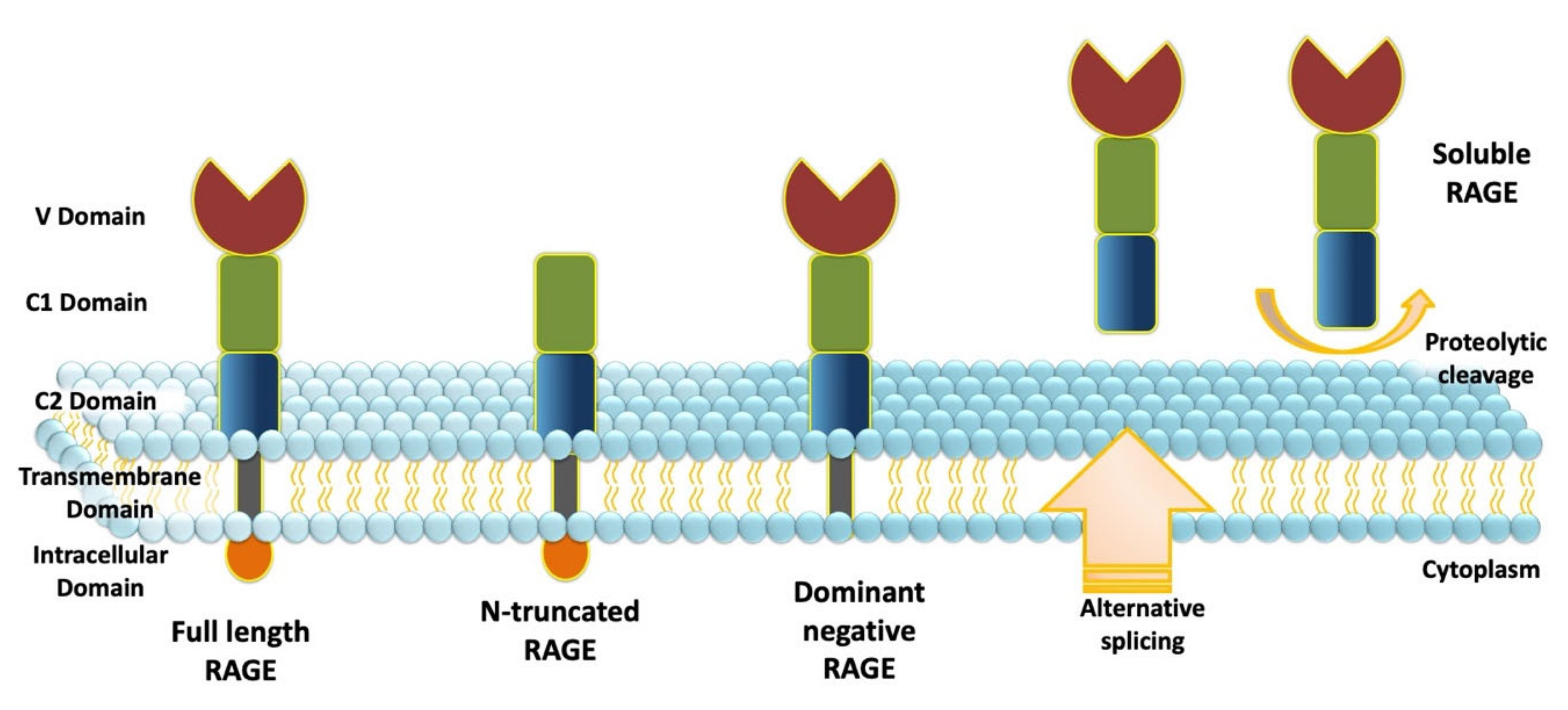
4. The Receptor for Advanced Glycation End Products
4.1. RAGE: Domain Structure and Isoforms
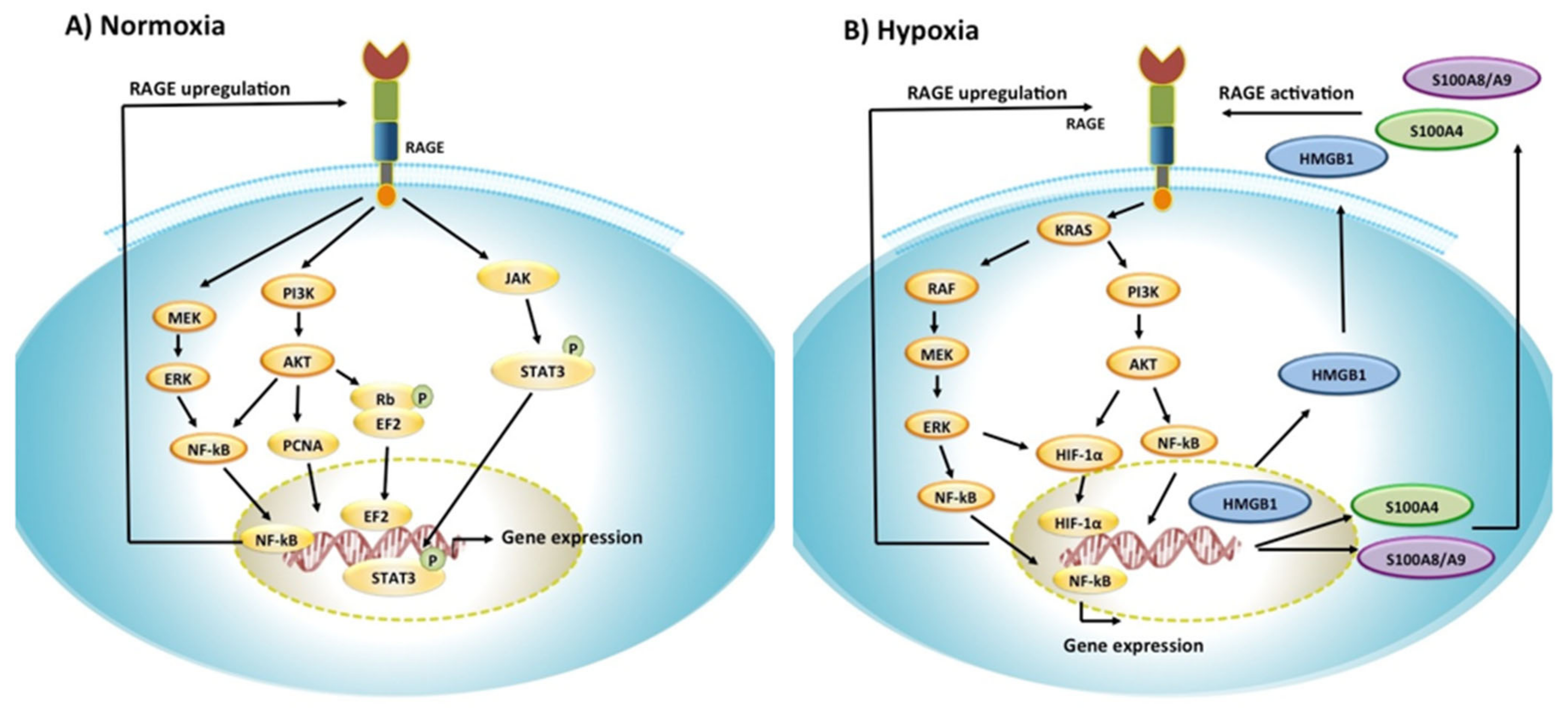
4.2. RAGE Signaling Pathways in Cancer
4.3. Hypoxia and RAGE Signaling in Cancer Tumors
4.4. RAGE Ligands in Hypoxic Tumors
4.4.1. HMGB1
4.4.2. S100 Proteins
4.4.3. S100B
4.4.4. S100P
4.4.5. S100A4
4.4.6. S100A7
4.4.7. S100A8/9
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AGEs | Advanced glycation endproducts |
| ALCAM | Activated leukocyte cell adhesion molecule |
| ANGPTL4 | Angiopoietin-like 4 |
| ARNT1 | Aryl hydrocarbon receptor nuclear translocator 1 |
| bHLH | Basic-helix-loop-helix |
| CBP/p300 | CREB binding protein/p300 coactivator |
| Cdc42 | Cell division control protein 42 |
| C-TAD | C-terminal transactivation domain |
| DAMP | Damage-associated molecular pattern |
| Dia-1 | Diaphanous 1 |
| EMMPRIN | Extracellular matrix metalloproteinase inducer |
| EMT | Epithelial mesenchymal transition |
| ERK | Extracellular-regulated kinase |
| HAF | Hypoxia-associated factor |
| HIF | Hypoxia-inducible factor |
| HIF-1α | Hypoxia-inducible Factor 1α |
| HMGB1 | High mobility group box 1 |
| HRE | Hypoxia-responsive elements |
| Ig | Immunoglobulin |
| Jak | Janus kinase |
| LZIP | Leucine zipper domain |
| MAP | Mitogen-activated protein |
| MCAM | Melanoma cells adhesion molecule |
| MDM2 | Mouse double minute 2 homolog |
| MHC | Major histocompatibility complex |
| MMP | Matrix metalloproteinases |
| mTOR | Mechanistic target of rapamycin kinase |
| MyD88 | Myeloid differentiation primary response 88 |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| N-TAD | N-terminal transactivation domain |
| NPTN | Neuroplastin β |
| OCT4 | Octamer binding transcription factor 4 |
| ODD | Oxygen-dependent degradation |
| PAS | Per-ARNT-Sim domains |
| PCNA | Proliferating cell nuclear antigen |
| PHD | Proline-hydroxylases |
| PI3K | Phosphoinositide 3-kinase |
| Rac 1 | Ras-related C3 botulinum toxin substrate 1 |
| RAGE | Receptor of advanced glycation end products |
| SRC-1 | Nuclear receptor coactivator 1 |
| SSSR | S100 soil sensor receptors |
| STAT | Signal transducer and activator of transcription |
| TIF2 | Transcriptional mediator/intermediary factor 2 |
| TIRAP | TIR domain-containing adaptor protein |
| TLR | Toll-like receptor |
| VHL | von Hippel-Lindau protein |
| VEGF | Vascular endothelial growth factor |
References
- McKeown, S.R. Defining normoxia, physoxia and hypoxia in tumours—implications for treatment response. Br. J. Radiol. 2014, 87, 20130676. [Google Scholar] [CrossRef] [Green Version]
- Michiels, C. Physiological and Pathological Responses to Hypoxia. Am. J. Pathol. 2004, 164, 1875–1882. [Google Scholar] [CrossRef] [Green Version]
- Hockel, M.; Vaupel, P. Tumor Hypoxia: Definitions and Current Clinical, Biologic, and Molecular Aspects. J. Natl. Cancer Inst. 2001, 93, 266–276. [Google Scholar] [CrossRef] [Green Version]
- Wilson, W.R.; Hay, M. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. [Google Scholar] [CrossRef]
- Ke, Q.; Costa, M. Hypoxia-Inducible Factor-1 (HIF-1). Mol. Pharmacol. 2006, 70, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Rankin, E.; Giaccia, A.J. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008, 15, 678–685. [Google Scholar] [CrossRef] [Green Version]
- Semenza, G.L.; Nejfelt, M.K.; Chi, S.M.; Antonarakis, S. Hypoxia-inducible nuclear factors bind to an enhancer element located 3’ to the human erythropoietin gene. Proc. Natl. Acad. Sci. USA 1991, 88, 5680–5684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Shen, S.-M.; Zhao, X.-Y.; Chen, G.-Q. Targeted genes and interacting proteins of hypoxia inducible factor. Int. J. Biochem. Mol. Boil. 2012, 3, 165–178. [Google Scholar]
- Rankin, E.; Giaccia, A.J. Hypoxic control of metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Downes, N.; Laham-Karam, N.; Kaikkonen, M.U.; Ylä-Herttuala, S. Differential but Complementary HIF1α and HIF2α Transcriptional Regulation. Mol. Ther. 2018, 26, 1735–1745. [Google Scholar] [CrossRef] [Green Version]
- Ratcliffe, P.J. HIF-1 and HIF-2: Working alone or together in hypoxia? J. Clin. Investig. 2007, 117, 862–865. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.L.; Jiang, B.H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen Sensing by Metazoans: The Central Role of the HIF Hydroxylase Pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Cummins, E.; Keogh, C.E.; Crean, D.; Taylor, C.T. The role of HIF in immunity and inflammation. Mol. Asp. Med. 2016, 47–48, 24–34. [Google Scholar] [CrossRef]
- Tian, H.; McKnight, S.L.; Russell, D.W. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively ex-pressed in endothelial cells. Genes Dev. 1997, 11, 72–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ema, M.; Taya, S.; Yokotani, N.; Sogawa, K.; Matsuda, Y.; Fujii-Kuriyama, Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. USA 1997, 94, 4273–4278. [Google Scholar] [CrossRef] [Green Version]
- Wiener, C.M.; Bootha, G.; Semenza, G.L. In Vivo Expression of mRNAs Encoding Hypoxia-Inducible Factor. Biochem. Biophys. Res. Commun. 1996, 225, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.-Z.; Moran, S.M.; HogenEsch, J.B.; Wartman, L.; Bradfield, C.A. Molecular Characterization and Chromosomal Localization of a Third α-Class Hypoxia Inducible Factor Subunit, HIF3α. Gene Expr. 2018, 7, 205–213. [Google Scholar]
- Hara, S.; Hamada, J.; Kobayashi, C.; Kondo, Y.; Imura, N. Expression and Characterization of Hypoxia-Inducible Factor (HIF)-3α in Human Kidney: Suppression of HIF-Mediated Gene Expression by HIF-3α. Biochem. Biophys. Res. Commun. 2001, 287, 808–813. [Google Scholar] [CrossRef]
- Li, Q.F.; Wang, X.R.; Yang, Y.W.; Lin, H. Hypoxia upregulates hypoxia inducible factor (HIF)-3α expression in lung epithelial cells: Characterization and comparison with HIF-1α. Cell Res. 2006, 16, 548–558. [Google Scholar] [CrossRef] [Green Version]
- Maynard, M.A.; Qi, H.; Chung, J.; Lee, E.H.L.; Kondo, Y.; Hara, S.; Conaway, R.C.; Conaway, J.; Ohh, M. Multiple Splice Variants of the Human HIF-3α Locus Are Targets of the von Hippel-Lindau E3 Ubiquitin Ligase Complex. J. Biol. Chem. 2003, 278, 11032–11040. [Google Scholar] [CrossRef] [Green Version]
- Pasanen, A.; Heikkila, M.; Rautavuoma, K.; Hirsilä, M.; Kivirikko, K.I.; Myllyharju, J. Hypoxia-inducible factor (HIF)-3α is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int. J. Biochem. Cell Biol. 2010, 42, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Maynard, M.A.; Evans, J.A.; Hosomi, T.; Hara, S.; Jewett, A.S.M.; Ohh, M. Human HIF-3alpha4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J. 2005, 19, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ko, H.P.; Whitlock, J.P. Induction of Phosphoglycerate Kinase 1 Gene Expression by Hypoxia. J. Biol. Chem. 1996, 271, 21262–21267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Potluri, N.; Lu, J.; Kim, Y.; Rastinejad, F. Structural integration in hypoxia-inducible factors. Nat. Cell Biol. 2015, 524, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Bruick, R.K.; McKnight, S.L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001, 294, 1337–1340. [Google Scholar] [CrossRef] [Green Version]
- Klinger, P.; Schietke, E.R.; Warnecke, C.; Swoboda, B.; Wiesener, M.; Hennig, F.F.; Gelse, K. Deletion of the oxygen-dependent degradation domain results in impaired transcriptional activity of hy-poxia-inducible factors. Transcription 2011, 2, 269–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, K.; Discher, D.J.; Hu, J.; Bishopric, N.H.; Webster, K.A. Molecular Regulation of the Endothelin-1 Gene by Hypoxia. J. Biol. Chem. 2001, 276, 12645–12653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Nock, S.; Nesper, M.; Sprinzl, A.M.; Sigler, P.B. Structure and Importance of the Dimerization Domain in Elongation Factor Ts from Thermus thermophilus. Biochemistry 1996, 35, 10269–10278. [Google Scholar] [CrossRef]
- Ravenna, L.; Salvatori, L.; Russo, M.A. HIF3alpha: The little we know. FEBS J. 2016, 283, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Webb, J.D.; Coleman, M.; Pugh, C.W. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell. Mol. Life Sci. 2009, 66, 3539–3554. [Google Scholar] [CrossRef]
- Koh, M.Y.; Spivak-Kroizman, T.R.; Powis, G. HIF-1 regulation: Not so easy come, easy go. Trends Biochem. Sci. 2008, 33, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Ohh, M.; Park, C.W.; Ivan, M.; Hoffman, M.A.; Kim, T.Y.; Huang, L.E.; Pavletich, N.; Chau, V.; Kaelin, W.G. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hip-pel-Lindau protein. Nat. Cell Biol. 2000, 2, 423–427. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J. Activation of the HIF pathway in cancer. Curr. Opin. Genet. Dev. 2001, 11, 293–299. [Google Scholar] [CrossRef]
- Jeong, J.-W.; Bae, M.-K.; Ahn, M.-Y.; Kim, S.-H.; Sohn, T.-K.; Bae, M.-H.; Yoo, M.-A.; Song, E.J.; Lee, K.-J.; Kim, K.-W. Regulation and Destabilization of HIF-1α by ARD1-Mediated Acetylation. Cell 2002, 111, 709–720. [Google Scholar] [CrossRef] [Green Version]
- Ravi, R.; Mookerjee, B.; Bhujwalla, Z.M.; Sutter, C.H.; Artemov, D.; Zeng, Q.; Dillehay, L.E.; Madan, A.; Semenza, G.L.; Bedi, A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1α. Genome Res. 2000, 14, 34–44. [Google Scholar]
- Mahon, P.C.; Hirota, K.; Semenza, G.L. FIH-1: A novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001, 15, 2675–2686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arany, Z.; Huang, E.; Eckner, R.; Bhattacharya, S.; Jiang, C.; Goldberg, M.A.; Bunn, H.F.; Livingston, D.M. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. USA 1996, 93, 12969–12973. [Google Scholar] [CrossRef] [Green Version]
- Ebert, B.L.; Bunn, H.F. Regulation of transcription by hypoxia requires a multiprotein complex that includes hypox-ia-inducible factor 1, an adjacent transcription factor, and p300/CREB binding protein. Mol. Cell Biol. 1998, 18, 4089–4096. [Google Scholar] [CrossRef] [Green Version]
- Kallio, P.J.; Okamoto, K.; O’Brien, S.; Carrero, P.; Makino, Y.; Tanaka, H.; Poellinger, L. Signal transduction in hypoxic cells: Inducible nuclear translocation and recruitment of the CBP/p300 co-activator by the hypoxia-inducible factor-1alpha. EMBO J. 1998, 17, 6573–6586. [Google Scholar] [CrossRef]
- Ema, M.; Hirota, K.; Mimura, J.; Abe, H.; Yodoi, J.; Sogawa, K.; Poellinger, L.; Fujii-Kuriyama, Y. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: Their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999, 18, 1905–1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lando, D.; Peet, D.; Whelan, D.A.; Gorman, J.J.; Whitelaw, M.L. Asparagine Hydroxylation of the HIF Transactivation Domain: A Hypoxic Switch. Science 2002, 295, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Lando, D.; Peet, D.J.; Gorman, J.J.; Whelan, D.A.; Whitelaw, M.L.; Bruick, R.K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypox-ia-inducible factor. Genes Dev. 2002, 16, 1466–1471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, M.Y.; Darnay, B.G.; Powis, G. Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hy-poxia-inducible factor 1alpha, leading to its oxygen-independent degradation. Mol. Cell Biol. 2008, 28, 7081–7095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, M.Y.; Lemos, R.; Liu, X.; Powis, G. The hypoxia-associated factor switches cells from HIF-1alpha- to HIF-2alpha-dependent signaling pro-moting stem cell characteristics, aggressive tumor growth and invasion. Cancer Res. 2011, 71, 4015–4027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burrows, N.; Babur, M.; Resch, J.; Williams, K.J.; Brabant, G. Hypoxia-Inducible Factor in Thyroid Carcinoma. J. Thyroid. Res. 2011, 2011, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, D.E.; Berra, E.; Gothié, E.; Roux, D.; Pouysségur, J. p42/p44 Mitogen-activated Protein Kinases Phosphorylate Hypoxia-inducible Factor 1α (HIF-1α) and Enhance the Transcriptional Activity of HIF-1. J. Biol. Chem. 1999, 274, 32631–32637. [Google Scholar] [CrossRef] [Green Version]
- Gradin, K.; Takasaki, C.; Fujii-Kuriyama, Y.; Sogawa, K. The Transcriptional Activation Function of the HIF-like Factor Requires Phosphorylation at a Conserved Threonine. J. Biol. Chem. 2002, 277, 23508–23514. [Google Scholar] [CrossRef] [Green Version]
- Kietzmann, T.; Mennerich, D.; Dimova, E.Y. Hypoxia-Inducible Factors (HIFs) and Phosphorylation: Impact on Stability, Localization, and Transactivity. Front. Cell Dev. Biol. 2016, 4, 11. [Google Scholar] [CrossRef]
- Stiehl, D.P.; Jelkmann, W.; Wenger, R.H.; Hellwig-Bürgel, T. Normoxic induction of the hypoxia-inducible factor 1alpha by insulin and interleukin-1beta involves the phosphatidylinositol 3-kinase pathway. FEBS Lett. 2002, 512, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Davidson, G.; Huang, Y.; Jiang, B.-H.; Shi, X.; Costa, M.; Huang, C. Nickel compounds act through phosphatidylinositol-3-kinase/Akt-dependent, p70(S6k)-independent pathway to induce hypoxia inducible factor transactivation and Cap43 expression in mouse epidermal Cl41 cells. Cancer Res. 2004, 64, 94–101. [Google Scholar] [CrossRef] [Green Version]
- Beppu, K.; Nakamura, K.; Linehan, W.M.; Rapisarda, A.; Thiele, C.J. Topotecan blocks hypoxia-inducible factor-1alpha and vascular endothelial growth factor expression in-duced by insulin-like growth factor-I in neuroblastoma cells. Cancer Res. 2005, 65, 4775–4781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohlin, S.; Hamidian, A.; von Stedingk, K.; Bridges, E.; Wigerup, C.; Bexell, D.; Påhlman, S. PI3K-mTORC2 but not PI3K-mTORC1 regulates transcription of HIF2A/EPAS1 and vascularization in neuroblastoma. Cancer Res. 2015, 75, 4617–4628. [Google Scholar] [CrossRef] [Green Version]
- Van Uden, P.; Kenneth, N.S.; Webster, R.; Müller, H.A.; Mudie, S.; Rocha, S. Evolutionary conserved regulation of HIF-1beta by NF-kappaB. PLoS Genet. 2011, 7, e1001285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talks, K.L.; Turley, H.; Gatter, K.C.; Maxwell, P.; Pugh, C.; Ratcliffe, P.; Harris, A.L. The Expression and Distribution of the Hypoxia-Inducible Factors HIF-1α and HIF-2α in Normal Human Tissues, Cancers, and Tumor-Associated Macrophages. Am. J. Pathol. 2000, 157, 411–421. [Google Scholar] [CrossRef]
- Zhong, H.; De Marzo, A.M.; Laughner, E.; Lim, M.; Hilton, D.A.; Zagzag, D.; Buechler, P.; Isaacs, W.B.; Semenza, G.L.; Simons, J.W. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999, 59, 5830–5835. [Google Scholar]
- Semenza, G.L. HIF-1: Upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 2010, 20, 51–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nussenbaum, F.; Herman, I.M. Tumor Angiogenesis: Insights and Innovations. J. Oncol. 2010, 2010, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wigerup, C.; Påhlman, S.; Bexell, D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol. Ther. 2016, 164, 152–169. [Google Scholar] [CrossRef] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krock, B.L.; Skuli, N.; Simon, M.C. Hypoxia-Induced Angiogenesis: Good and Evil. Genes Cancer 2011, 2, 1117–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkman, J. Antiangiogenesis in cancer therapy—endostatin and its mechanisms of action. Exp. Cell Res. 2006, 312, 594–607. [Google Scholar] [CrossRef]
- Folkman, J.; Ingber, D. Inhibition of angiogenesis. Semin. Cancer Biol. 1992, 3, 89–96. [Google Scholar]
- Carmeliet, P.; Dor, Y.; Herbert, J.-M.; Fukumura, D.; Brusselmans, K.; Dewerchin, M.; Neeman, M.; Bono, F.; Abramovitch, R.; Maxwell, P.; et al. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nat. Cell Biol. 1998, 394, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Ryan, H.E.; Lo, J.; Johnson, R.S. HIF-1alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998, 17, 3005–3015. [Google Scholar] [CrossRef] [Green Version]
- Greijer, A.E.; van der Wall, E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J. Clin. Pathol. 2004, 57, 1009–1014. [Google Scholar] [CrossRef]
- Aubrey, B.; Kelly, G.L.; Janic, A.; Herold, M.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Graeber, T.G.; Peterson, J.F.; Tsai, M.; Monica, K.; Fornace, A.J.; Giaccia, A.J. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol. Cell. Biol. 1994, 14, 6264–6277. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Johnson, D.E.; Jin, J.R.; Antoku, K.; Dou, Q.P. Bcl-2- and CrmA-inhibitable dephosphorylation and cleavage of retinoblastoma protein during etopo-side-induced apoptosis. Int. J. Mol. Med. 1998, 1, 131–136. [Google Scholar]
- Chen, D.; Li, M.; Luo, J.; Gu, W. Direct Interactions between HIF-1α and Mdm2 Modulate p53 Function. J. Biol. Chem. 2003, 278, 13595–13598. [Google Scholar] [CrossRef] [Green Version]
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef]
- Graeber, T.; Osmanian, C.; Jacks, T.; Housman, D.E.; Koch, C.J.; Lowe, S.W.; Giaccia, A.J. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nat. Cell Biol. 1996, 379, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Koumenis, C.; Alarcon, R.; Hammond, E.; Sutphin, P.; Hoffman, W.; Murphy, M.; Derr, J.; Taya, Y.; Lowe, S.W.; Kastan, M.; et al. Regulation of p53 by Hypoxia: Dissociation of Transcriptional Repression and Apoptosis from p53-Dependent Transactivation. Mol. Cell. Biol. 2001, 21, 1297–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rankin, E.; Nam, J.-M.; Giaccia, A.J. Hypoxia: Signaling the Metastatic Cascade. Trends Cancer 2016, 2, 295–304. [Google Scholar] [CrossRef] [Green Version]
- De Bock, K.; Mazzone, M.; Carmeliet, P. Antiangiogenic therapy, hypoxia, and metastasis: Risky liaisons, or not? Nat. Rev. Clin. Oncol. 2011, 8, 393–404. [Google Scholar] [CrossRef] [Green Version]
- Semenza, G.L. The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochim. Biophys. Acta 2016, 1863, 382–391. [Google Scholar] [CrossRef]
- Keith, B.; Simon, M.C. Hypoxia-Inducible Factors, Stem Cells, and Cancer. Cell 2007, 129, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Botta, C.; Misso, G.; Martino, E.C.; Pirtoli, L.; Cusi, M.G.; Tassone, P.; Tagliaferri, P.; Caraglia, M.; Correale, P. The route to solve the interplay between inflammation, angiogenesis and anti-cancer immune response. Cell Death Dis. 2016, 7, e2299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Triner, D.; Shah, Y.M. Hypoxia-inducible factors: A central link between inflammation and cancer. J. Clin. Investig. 2016, 126, 3689–3698. [Google Scholar] [CrossRef]
- Rius, J.; Guma, M.; Schachtrup, C.; Akassoglou, K.; Zinkernagel, A.S.; Nizet, V.; Johnson, R.S.; Haddad, G.G.; Karin, M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature 2008, 453, 807–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Uden, P.; Kenneth, N.S.; Rocha, S. Regulation of hypoxia-inducible factor-1alpha by NF-kappaB. Biochem. J. 2008, 412, 477–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sitkovsky, M.; Lukashev, D. Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nat. Rev. Immunol. 2005, 5, 712–721. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grivennikov, S.I.; Greten, F.; Karin, M. Immunity, Inflammation, and Cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, V.; Delovitch, T.L. Different subsets of natural killer T cells may vary in their roles in health and disease. Immunity 2014, 142, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Palazon, A.; Goldrath, A.W.; Nizet, V.; Johnson, R.S. HIF Transcription Factors, Inflammation, and Immunity. Immunity 2014, 41, 518–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, Y.; Yang, D.; Oppenheim, J.J. Alarmins and Antitumor Immunity. Clin. Ther. 2016, 38, 1042–1053. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Xie, J.; Sun, X.; Zeh, H.J.; Kang, R.; Lotze, M.T.; Tang, D. DAMPs, ageing, and cancer: The ’DAMP Hypothesis’. Ageing Res. Rev. 2015, 24 Pt A, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Lotze, M.T.; Tracey, K.J. High-mobility group box 1 protein (HMGB1): Nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 2005, 5, 331–342. [Google Scholar] [CrossRef]
- Kang, R.; Tang, D.; Schapiro, N.E.; Lotze, M.T.; Zeh, H.J. Abstract B96: The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Tumor. Biol. 2012, 33, 567–577. [Google Scholar] [CrossRef]
- Papale, M.; Buccarelli, M.; Mollinari, C.; Russo, M.A.; Pallini, R.; Ricci-Vitiani, L.; Tafani, M. Hypoxia, Inflammation and Necrosis as Determinants of Glioblastoma Cancer Stem Cells Progression. Int. J. Mol. Sci. 2020, 21, 2660. [Google Scholar] [CrossRef] [Green Version]
- Neeper, M.; Schmidt, A.M.; Brett, J.; Yan, S.D.; Wang, F.; Pan, Y.C.; Elliston, K.; Stern, D.; Shaw, A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J. Biol. Chem. 1992, 267, 14998–15004. [Google Scholar] [CrossRef]
- Sugaya, K.; Fukagawa, T.; Matsumoto, K.-I.; Mita, K.; Takahashi, E.-I.; Ando, A.; Inoko, H.; Ikemura, T. Three Genes in the Human MHC Class III Region near the Junction with the Class II: Gene for receptor of advanced glycosylation end products, PBX2 homeobox gene and a notch homolog, human counterpart of mouse mammary tumor gene int-3. Genomics 1994, 23, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Verweij, C.L. How RAGE turns in rage. Genes Immun. 2002, 3, 117–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yonekura, H.; Yamamoto, Y.; Sakurai, S.; Petrova, R.G.; Abedin, J.; Li, H.; Yasui, K.; Takeuchi, M.; Makita, Z.; Takasawa, S.; et al. Novel splice variants of the receptor for advanced glycation end-products expressed in human vascular endothelial cells and pericytes, and their putative roles in diabetes-induced vascular injury. Biochem. J. 2003, 370 Pt 3, 1097–1109. [Google Scholar] [CrossRef]
- Hudson, B.I.; Carter, A.M.; Harja, E.; Kalea, A.Z.; Arriero, M.; Yang, H.; Grant, P.J.; Schmidt, A.M. Identification, classification, and expression of RAGE gene splice variants. FASEB J. 2007, 22, 1572–1580. [Google Scholar] [CrossRef]
- Raucci, A.; Cugusi, S.; Antonelli, A.; Barabino, S.M.; Monti, L.; Bierhaus, A.; Reiss, K.; Saftig, P.; Bianchi, M.E. A soluble form of the receptor for advanced glycation endproducts (RAGE) is produced by proteolytic cleavage of the membrane-bound form by the sheddase a disintegrin and metalloprotease 10 (ADAM10). FASEB J. 2008, 22, 3716–3727. [Google Scholar] [CrossRef]
- Sorci, G.; Riuzzi, F.; Giambanco, I.; Donato, R. RAGE in tissue homeostasis, repair and regeneration. Biochim. Biophys. Acta 2013, 1833, 101–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plotkin, L.I.; Essex, A.L.; Davis, H.M. RAGE Signaling in Skeletal Biology. Curr. Osteoporos. Rep. 2019, 17, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Liliensiek, B.; Weigand, M.A.; Bierhaus, A.; Nicklas, W.; Kasper, M.; Hofer, S.; Plachky, J.; Gröne, H.-J.; Kurschus, F.C.; Schmidt, A.M.; et al. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J. Clin. Investig. 2004, 113, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhu, L.; Zhang, Z.; Li, H.; Li, P.; Wang, Y.; Leng, M. HMGB1-RAGE signaling pathway in pPROM. Taiwan. J. Obstet. Gynecol. 2018, 57, 211–216. [Google Scholar] [CrossRef]
- Kwak, T.; Drews-Elger, K.; Ergonul, A.; Miller, P.C.; Braley, A.; Hwang, G.H.; Zhao, D.; Besser, A.; Yamamoto, Y.; El-Ashry, D.; et al. Targeting of RAGE-ligand signaling impairs breast cancer cell invasion and metastasis. Oncogene 2017, 36, 1559–1572. [Google Scholar] [CrossRef]
- Yan, S.F.; Ramasamy, R.; Schmidt, A.M. Tempering the wrath of RAGE: An emerging therapeutic strategy against diabetic complications, neurodegeneration, and inflammation. Ann. Med. 2009, 41, 408–422. [Google Scholar] [CrossRef] [Green Version]
- Srikanth, V.; Maczurek, A.; Phan, T.; Steele, M.; Westcott, B.; Juskiw, D.; Münch, G. Advanced glycation endproducts and their receptor RAGE in Alzheimer’s disease. Neurobiol. Aging 2011, 32, 763–777. [Google Scholar] [CrossRef]
- Bierhaus, A.; Humpert, P.M.; Morcos, M.; Wendt, T.; Chavakis, T.; Arnold, B.; Stern, D.M.; Nawroth, P.P. Understanding RAGE, the receptor for advanced glycation end products. J. Mol. Med. 2005, 83, 876–886. [Google Scholar] [CrossRef]
- Logsdon, C.D.; Fuentes, M.K.; Huang, E.H.; Arumugam, T. RAGE and RAGE ligands in cancer. Curr. Mol. Med. 2007, 7, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.; Morales, M.; Gonzalez, I.; Araya, P. Inhibition of RAGE Axis Signaling: A Pharmacological Challenge. Curr. Drug Targets 2019, 20, 340–346. [Google Scholar] [CrossRef]
- Huttunen, H.J.; Fages, C.; Rauvala, H. Receptor for advanced glycation end products (RAGE)-mediated neurite out-growth and activation of NF-kappaB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J. Biol. Chem. 1999, 274, 19919–19924. [Google Scholar] [CrossRef] [Green Version]
- Kislinger, T.; Tanji, N.; Wendt, T.; Qu, W.; Lu, Y.; Ferran, L.J., Jr.; Taguchi, A.; Olson, K.; Bucciarelli, L.; Goova, M.; et al. Receptor for Advanced Glycation End Products Mediates Inflammation and Enhanced Expression of Tissue Factor in Vasculature of Diabetic Apolipoprotein E–Null Mice. Arter. Thromb. Vasc. Biol. 2001, 21, 905–910. [Google Scholar] [CrossRef] [Green Version]
- Sakaguchi, T.; Yan, S.F.; Belov, D.; Rong, L.L.; Sousa, M.; Andrassy, M.; Marso, S.P.; Duda, S.; Arnold, B.; Liliensiek, B.; et al. Central role of RAGE-dependent neointimal expansion in arterial restenosis. J. Clin. Investig. 2003, 111, 959–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudson, B.I.; Kalea, A.Z.; del Mar Arriero, M.; Harja, E.; Boulanger, E.; D’Agati, V.; Schmidt, A.M. Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42. J. Biol. Chem. 2008, 283, 34457–34468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakaguchi, M.; Murata, H.; Yamamoto, K.-I.; Ono, T.; Sakaguchi, Y.; Motoyama, A.; Hibino, T.; Kataoka, K.; Huh, N.-H. TIRAP, an Adaptor Protein for TLR2/4, Transduces a Signal from RAGE Phosphorylated upon Ligand Binding. PLoS ONE 2011, 6, e23132. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, A.; Blood, D.C.; Del Toro, G.; Canet, A.; Lee, D.C.; Qu, W.; Tanji, N.; Lu, Y.; Lalla, E.; Fu, C.; et al. Blockade of RAGE–amphoterin signalling suppresses tumour growth and metastases. Nat. Cell Biol. 2000, 405, 354–360. [Google Scholar] [CrossRef]
- Hermani, A.; De Servi, B.; Medunjanin, S.; Tessier, P.A.; Mayer, D. S100A8 and S100A9 activate MAP kinase and NF-kappaB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp. Cell Res. 2006, 312, 184–197. [Google Scholar] [CrossRef]
- Ghavami, S.; Kerkhoff, C.; Chazin, W.J.; Kadkhoda, K.; Xiao, W.; Zuse, A.; Hashemi, M.; Eshraghi, M.; Schulze-Osthoff, K.; Klonisch, T.; et al. S100A8/9 induces cell death via a novel, RAGE-independent pathway that involves selective release of Smac/DIABLO and Omi/HtrA2. Biochim. Biophys. Acta. 2008, 1783, 297–311. [Google Scholar] [CrossRef] [Green Version]
- Rouhiainen, A.; Kuja-Panula, J.; Tumova, S.; Rauvala, H. RAGE-Mediated Cell Signaling. Methods Mol. Biol. 2013, 963, 239–263. [Google Scholar]
- Sharaf, H.; Matou-Nasri, S.; Wang, Q.; Rabhan, Z.; Al-Eidi, H.; Al Abdulrahman, A.; Ahmed, N. Advanced glycation endproducts increase proliferation, migration and invasion of the breast cancer cell line MDA-MB-231. Biochim. Biophys. Acta 2015, 1852, 429–441. [Google Scholar] [CrossRef] [Green Version]
- Azizan, N.; Suter, M.A.; Liu, Y.; Logsdon, C.D. RAGE maintains high levels of NFkappaB and oncogenic Kras activity in pancreatic cancer. Biochem. Biophys. Res. Commun. 2017, 493, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.-N.; Seo, J.-H.; Lee, M.-H.; Yoon, G.; Cho, S.-S.; Liu, K.; Choi, H.; Oh, K.B.; Cho, Y.-S.; Kim, H.; et al. Oridonin induces apoptosis in oral squamous cell carcinoma probably through the generation of reactive oxygen species and the p38/JNK MAPK pathway. Int. J. Oncol. 2018, 52, 1749–1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dukic-Stefanovic, S.; Gasic-Milenkovic, J.; Deuther-Conrad, W.; Münch, G. Signal transduction pathways in mouse microglia N-11 cells activated by advanced glycation endproducts (AGEs). J. Neurochem. 2003, 87, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.-M.; He, M.-Y.; Liu, Y.-W.; Lu, Y.-J.; Hong, Y.-Q.; Luo, H.-H.; Ren, Z.-L.; Zhao, S.-C.; Jiang, Y. AGE/RAGE/Akt pathway contributes to prostate cancer cell proliferation by promoting Rb phosphorylation and degradation. Am. J. Cancer Res. 2015, 5, 1741–1750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Song, Y.; Zhou, L.; Li, W.; Zhu, X. Downregulation of RAGE Inhibits Cell Proliferation and Induces Apoptosis via Regulation of PI3K/AKT Path-way in Cervical Squamous Cell Carcinoma. Onco. Targets Ther. 2020, 13, 2385–2397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesina, M.; Kurkowski, M.U.; Ludes, K.; Rose-John, S.; Treiber, M.; Klöppel, G.; Yoshimura, A.; Reindl, W.; Sipos, B.; Akira, S.; et al. Stat3/Socs3 Activation by IL-6 Transsignaling Promotes Progression of Pancreatic Intraepithelial Neoplasia and Development of Pancreatic Cancer. Cancer Cell 2011, 19, 456–469. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, A.; Wang, S.; Morris, J.P.; Folias, A.E.; Liou, A.; Kim, G.E.; Akira, S.; Boucher, K.; Firpo, M.A.; Mulvihill, S.J.; et al. Stat3 and MMP7 Contribute to Pancreatic Ductal Adenocarcinoma Initiation and Progression. Cancer Cell 2011, 19, 441–455. [Google Scholar] [CrossRef] [Green Version]
- Kang, R.; Loux, T.; Tang, D.; Schapiro, N.E.; Vernon, P.; Livesey, K.M.; Krasinskas, A.; Lotze, M.T.; Zeh 3rd, H.J. The expression of the receptor for advanced glycation endproducts (RAGE) is permissive for early pancreatic neoplasia. Proc. Natl. Acad. Sci. USA 2012, 109, 7031–7036. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.J.; Snowden, J.A.; Zeidler, M.; Danson, S. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 2015, 113, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Pichiule, P.; Chavez, J.C.; Schmidt, A.M.; Vannucci, S.J. Hypoxia-inducible Factor-1 Mediates Neuronal Expression of the Receptor for Advanced Glycation End Products following Hypoxia/Ischemia. J. Biol. Chem. 2007, 282, 36330–36340. [Google Scholar] [CrossRef] [Green Version]
- Kamide, T.; Kitao, Y.; Takeichi, T.; Okada, A.; Mohri, H.; Schmidt, A.M.; Kawano, T.; Munesue, S.; Yamamoto, Y.; Yamamoto, H.; et al. RAGE mediates vascular injury and inflammation after global cerebral ischemia. Neurochem. Int. 2012, 60, 220–228. [Google Scholar] [CrossRef]
- Hassid, B.G.; Nair, M.N.; Ducruet, A.F.; Otten, M.L.; Komotar, R.J.; Pinsky, D.J.; Schmidt, A.M.; Yan, S.F.; Connolly, E.S. Neuronal RAGE expression modulates severity of injury following transient focal cerebral ischemia. J. Clin. Neurosci. 2009, 16, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Zhai, D.-X.; Kong, Q.-F.; Xu, W.-S.; Bai, S.-S.; Peng, H.-S.; Zhao, K.; Li, G.-Z.; Wang, D.-D.; Sun, B.; Wang, J.-H.; et al. RAGE expression is up-regulated in human cerebral ischemia and pMCAO rats. Neurosci. Lett. 2008, 445, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Feirt, N.; Goldstein, M.; Guarrera, J.; Ippagunta, N.; Ekong, U.; Dun, H.; Lu, Y.; Qu, W.; Schmidt, A.M.; et al. Blockade of receptor for advanced glycation end product (RAGE) attenuates ischemia and reperfusion injury to the liver in mice. Hepatology 2004, 39, 422–432. [Google Scholar] [CrossRef]
- Zeng, S.; Dun, H.; Ippagunta, N.; Rosario, R.; Zhang, Q.Y.; Lefkowitch, J.; Yan, S.F.; Schmidt, A.M.; Emond, J.C. Receptor for advanced glycation end product (RAGE)-dependent modulation of early growth response-1 in hepatic ischemia/reperfusion injury. J. Hepatol. 2009, 50, 929–936. [Google Scholar] [CrossRef]
- Loboda, A.; Jozkowicz, A.; Dulak, J. HIF-1 and HIF-2 transcription factors—Similar but not identical. Mol. Cells 2010, 29, 435–442. [Google Scholar] [CrossRef]
- Kiewitz, R.; Acklin, C.; Minder, E.; Huber, P.R.; Schafer, B.; Heizmann, C.W. S100A1, a New Marker for Acute Myocardial Ischemia. Biochem. Biophys. Res. Commun. 2000, 274, 865–871. [Google Scholar] [CrossRef]
- Mori, T.; Tan, J.; Arendash, G.W.; Koyama, N.; Nojima, Y.; Town, T. Overexpression of human S100B exacerbates brain damage and periinfarct gliosis after permanent focal is chemia. Stroke 2008, 39, 2114–2121. [Google Scholar] [CrossRef] [Green Version]
- Pelinka, L.E.; Harada, N.; Szalay, L.; Jafarmadar, M.; Redl, H.; Bahrami, S. Release of S100B Differs During Ischemia and Reperfusion of the Liver, the Gut, and the Kidney in Rats. Shock 2004, 21, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Tafani, M.; Schito, L.; Pellegrini, L.; Villanova, L.; Marfe, G.; Anwar, T.; Rosa, R.; Indelicato, M.; Fini, M.; Pucci, B.; et al. Hypoxia-increased RAGE and P2X7R expression regulates tumor cell invasion through phosphorylation of Erk1/2 and Akt and nuclear translocation of NF-[14]B. Carcinogenesis 2011, 32, 1167–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, R.; Hou, W.; Zhang, Q.; Chen, R.; Lee, Y.J.; Bartlett, D.L.; Lotze, M.T.; Tang, D.; Zeh, H.J. RAGE is essential for oncogenic KRAS-mediated hypoxic signaling in pancreatic cancer. Cell Death Dis. 2014, 5, e1480. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Rai, V.; Hudson, B.I.; Song, F.; Schmidt, A.M.; Barile, G.R. RAGE binds C1q and enhances C1q-mediated phagocytosis. Cell. Immunol. 2012, 274, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Ruan, B.H.; Li, X.; Winkler, A.R.; Cunningham, K.M.; Kuai, J.; Greco, R.M.; Nocka, K.H.; Fitz, L.J.; Wright, J.F.; Pittman, D.D.; et al. Complement C3a, CpG Oligos, and DNA/C3a Complex Stimulate IFN-α Production in a Receptor for Advanced Glycation End Product-Dependent Manner. J. Immunol. 2010, 185, 4213–4222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, A.M.; Hofmann, M.; Taguchi, A.; Yan, S.D.; Stern, D.M. RAGE: A multiligand receptor contributing to the cellular response in diabetic vasculopathy and in-flammation. Semin. Thromb Hemost. 2000, 26, 485–493. [Google Scholar] [CrossRef]
- Sims, G.P.; Rowe, D.C.; Rietdijk, S.T.; Herbst, R.; Coyle, A.J. HMGB1 and RAGE in Inflammation and Cancer. Annu. Rev. Immunol. 2010, 28, 367–388. [Google Scholar] [CrossRef]
- Volp, K. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut 2006, 55, 234–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiguro, H.; Nakaigawa, N.; Miyoshi, Y.; Fujinami, K.; Kubota, Y.; Uemura, H. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are overexpressed and associated with prostate cancer development. Prostate 2005, 64, 92–100. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Chihara, Y.; Kondo, H. Differential effects between amphoterin and advanced glycation end products on colon cancer cells. Int. J. Cancer 2003, 104, 722–727. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Oue, N.; Wakikawa, A.; Shigeishi, H.; Matsutani, N.; Kuraoka, K.; Ito, R.; Yokozaki, H.; Yasui, W. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the in-vasive and metastatic activity of gastric cancer. J. Pathol. 2002, 196, 163–170. [Google Scholar] [CrossRef]
- Huber, R.; Meier-Schiesser, B.; Otsuka, A.; Fenini, G.; Satoh, T.; Gehrke, S.; Widmer, D.; Levesque, M.P.; Mangana, J.; Kerl, K.; et al. Tumour hypoxia promotes melanoma growth and metastasis via High Mobility Group Box-1 and M2-like macrophages. Sci. Rep. 2016, 6, 29914. [Google Scholar] [CrossRef] [Green Version]
- Pezzolo, A.; Marimpietri, D.; Raffaghello, L.; Cocco, C.; Pistorio, A.; Gambini, C.; Cilli, M.; Horenstein, A.; Malavasi, F.; Pistoia, V. Failure of antitumor-derived endothelial cell immunotherapy depends on augmentation of tumor hypoxia. Oncotarget 2014, 5, 10368–10381. [Google Scholar] [CrossRef] [Green Version]
- Vicentino, A.R.R.; Carneiro, V.; Allonso, D.; Guilherme, R.D.F.; Benjamim, C.; Dos Santos, H.A.M.; Xavier, F.; Pyrrho, A.; Gomes, J.; Fonseca, M.; et al. Emerging Role of HMGB1 in the Pathogenesis of Schistosomiasis Liver Fibrosis. Front. Immunol. 2018, 9, 1979. [Google Scholar] [CrossRef] [Green Version]
- Gerlitz, G.; Hock, R.; Ueda, T.; Bustin, M. The dynamics of HMG protein-chromatin interactions in living cells. Biochem. Cell Biol. 2009, 87, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Travers, A.A. Priming the nucleosome: A role for HMGB proteins? EMBO Rep. 2003, 4, 131–136. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, G.H.; Sanders, C.; Johns, E.W. A New Group of Chromatin-Associated Proteins with a High Content of Acidic and Basic Amino Acids. JBIC J. Biol. Inorg. Chem. 1973, 38, 14–19. [Google Scholar] [CrossRef]
- Andersson, U.; Erlandsson-Harris, H.; Yang, H.; Tracey, K.J. HMGB1 as a DNA-binding cytokine. J. Leukoc. Biol. 2002, 72, 1084–1091. [Google Scholar]
- Thomas, J.O.; Travers, A.A. HMG1 and 2, and related ’architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001, 26, 167–174. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Y. HMGB1 in inflammation and cancer. J. Hematol. Oncol. 2020, 13, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Iii, H.J.Z.; Lotze, M.T. High-mobility group box 1 and cancer. Biochim. Biophys. Acta 2010, 1799, 131–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, S.; Dragoi, A.M.; Wang, X.; Dallacosta, C.; Louten, J.; Musco, G.; Sitia, G.; Yap, G.S.; Wan, Y.; Biron, C.A.; et al. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood 2007, 110, 1970–1981. [Google Scholar] [CrossRef] [PubMed]
- Parkkinen, J.; Raulo, E.; Merenmies, J.; Nolo, R.; Kajander, E.O.; Baumann, M.; Rauvala, H. Amphoterin, the 30-kDa protein in a family of HMG1-type polypeptides. Enhanced expression in transformed cells, leading edge localization, and interactions with plasminogen activation. J. Biol. Chem. 1993, 268, 19726–19738. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Chavan, S.S.; Andersson, U. High Mobility Group Box Protein 1 (HMGB1): The Prototypical Endogenous Danger Molecule. Mol. Med. 2015, 21, S6–S12. [Google Scholar] [CrossRef]
- Kang, R.; Chen, R.; Zhang, Q.; Hou, W.; Wu, S.; Cao, L.; Huang, J.; Yu, Y.; Fan, X.-G.; Yan, Z.; et al. HMGB1 in health and disease. Mol. Asp. Med. 2014, 40, 1–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasahira, T.; Kirita, T.; Oue, N.; Bhawal, U.K.; Yamamoto, K.; Fujii, K.; Ohmori, H.; Luo, Y.; Yasui, W.; Bosserhoff, A.K.; et al. High mobility group box-1-inducible melanoma inhibitory activity is associated with nodal metastasis and lymphangiogenesis in oral squamous cell carcinoma. Cancer Sci. 2008, 99, 1806–1812. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, H.; Rauvala, H. Amphoterin as an extracellular regulator of cell motility: From discovery to disease. J. Intern. Med. 2004, 255, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Riuzzi, F.; Sorci, G.; Donato, R. The amphoterin (HMGB1)/receptor for advanced glycation end products (RAGE) pair modulates myoblast proliferation, apoptosis, adhesiveness, migration, and invasiveness. Functional inactivation of RAGE in L6 myoblasts results in tumor formation in vivo. J. Biol. Chem. 2006, 281, 8242–8253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todorova, J.; Pasheva, E. High mobility group B1 protein interacts with its receptor RAGE in tumor cells but not in normal tissues. Oncol. Lett. 2011, 3, 214–218. [Google Scholar] [CrossRef] [Green Version]
- Hori, O.; Brett, J.; Slattery, T.; Cao, R.; Zhang, J.; Chen, J.X.; Nagashima, M.; Lundh, E.R.; Vijay, S.; Nitecki, D.; et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Media-tion of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J. Biol. Chem. 1995, 270, 25752–25761. [Google Scholar] [CrossRef] [Green Version]
- Apetoh, L.; Ghiringhelli, F.; Tesniere, A.; Criollo, A.; Ortiz, C.; Lidereau, R.; Mariette, C.; Chaput, N.; Mira, J.P.; Delaloge, S.; et al. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radio-therapy. Immunol. Rev. 2007, 220, 47–59. [Google Scholar] [CrossRef]
- Li, W.; Xu, Q.; Deng, Y.; Yang, Z.; Xing, S.; Zhao, X.; Zhu, P.; Wang, X.; He, Z.; Gao, Y. High-mobility group box 1 accelerates lipopolysaccharide-induced lung fibroblast proliferation in vitro: Involvment of the NF-κB signaling pathway. Lab. Investig. 2015, 95, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Andersson, U.; Wang, H.; Palmblad, K.; Aveberger, A.-C.; Bloom, O.; Erlandsson-Harris, H.; Janson, A.; Kokkola, R.; Zhang, M.; Yang, H.; et al. High Mobility Group 1 Protein (Hmg-1) Stimulates Proinflammatory Cytokine Synthesis in Human Monocytes. J. Exp. Med. 2000, 192, 565–570. [Google Scholar] [CrossRef]
- El Gazzar, M. HMGB1 modulates inflammatory responses in LPS-activated macrophages. Inflamm. Res. 2007, 56, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Fages, C.; Nolo, R.; Huttunen, H.J.; Eskelinen, E.; Rauvala, H. Regulation of cell migration by amphoterin. J. Cell Sci. 2000, 113, 611–620. [Google Scholar] [CrossRef]
- Degryse, B.; Bonaldi, T.; Scaffidi, P.; Müller-Knapp, S.; Resnati, M.; Sanvito, F.; Arrigoni, G.; Bianchi, M.E. The High Mobility Group (Hmg) Boxes of the Nuclear Protein Hmg1 Induce Chemotaxis and Cytoskeleton Reorganization in Rat Smooth Muscle Cells. J. Cell Biol. 2001, 152, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Schioppa, T.; Mantovani, A.; Allavena, P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: Po-tential targets of anti-cancer therapy. Eur. J. Cancer 2006, 42, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Huang, R.-R.; Wen, D.-R.; Paul, E.; Wünsch, P.H.; Cochran, A.J. IL-10 expression by primary tumor cells correlates with melanoma progression from radial to vertical growth phase and development of metastatic competence. Mod. Pathol. 2011, 24, 801–809. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, W.; Tohme, S.; Chen, M.; Fu, Y.; Tian, D.; Lotze, M.; Tang, D.; Tsung, A. Hypoxia induced HMGB1 and mitochondrial DNA interactions mediate tumor growth in hepatocellular car-cinoma through Toll-like receptor. J. Hepatol. 2015, 63, 114–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Wang, G.Z.; Wang, Y.; Huang, H.-Z.; Li, W.-T.; Qu, X.-D. Hypoxia-induced HMGB1 expression of HCC promotes tumor invasiveness and metastasis via regulating macrophage-derived IL-6. Exp. Cell Res. 2018, 367, 81–88. [Google Scholar] [CrossRef]
- Yan, W.; Chang, Y.; Liang, X.; Cardinal, J.S.; Huang, H.; Thorne, S.H.; Monga, S.P.S.; Geller, D.A.; Lotze, M.T.; Tsung, A. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology 2012, 55, 1863–1875. [Google Scholar] [CrossRef]
- Cheng, P.; Ma, Y.; Gao, Z.; Duan, L. High Mobility Group Box 1 (HMGB1) Predicts Invasion and Poor Prognosis of Glioblastoma Multiforme via Activating AKT Signaling in an Autocrine Pathway. Med Sci. Monit. 2018, 24, 8916–8924. [Google Scholar] [CrossRef]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leclerc, E.; Fritz, G.; Vetter, S.W.; Heizmann, C.W. Binding of S100 proteins to RAGE: An update. Biochim. Biophys. Acta 2009, 1793, 993–1007. [Google Scholar] [CrossRef] [Green Version]
- Donato, R. Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech. 2003, 60, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Seta, K.A.; Yuan, Y.; Spicer, Z.; Lu, G.; Bedard, J.; Ferguson, T.K.; Pathrose, P.; Cole-Strauss, A.; Kaufhold, A.; Millhorn, D.E. The role of calcium in hypoxia-induced signal transduction and gene expression. Cell Calcium 2004, 36, 331–340. [Google Scholar] [CrossRef]
- Lee, H.J.; Jung, Y.H.; Choi, G.E.; Kim, J.S.; Chae, C.W.; Han, H.J. Role of HIF1α Regulatory Factors in Stem Cells. Int. J. Stem Cells 2019, 12, 8–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donato, R.; Sorci, G.; Riuzzi, F.; Arcuri, C.; Bianchi, R.; Brozzi, F.; Tubaro, C.; Giambanco, I. S100B’s double life: Intracellular regulator and extracellular signal. Biochim. Biophys. Acta 2009, 1793, 1008–1022. [Google Scholar] [CrossRef] [Green Version]
- Rothermundt, M.; Peters, M.; Prehn, J.; Arolt, V. S100B in brain damage and neurodegeneration. Microsc. Res. Tech. 2003, 60, 614–632. [Google Scholar] [CrossRef]
- Zaigham, M.; Lundberg, F.; Olofsson, P. Protein S100B in umbilical cord blood as a potential biomarker of hypox-ic-ischemic encephalopathy in asphyxiated newborns. Early Hum. Dev. 2017, 112, 48–53. [Google Scholar] [CrossRef]
- Seguella, L.; Capuano, R.; Pesce, M.; Annunziata, G.; Pesce, M.; De Conno, B.; Sarnelli, G.; Aurino, L.; Esposito, G. S100B Protein Stimulates Proliferation and Angiogenic Mediators Release through RAGE/pAkt/mTOR Pathway in Human Colon Adenocarcinoma Caco-2 Cells. Int. J. Mol. Sci. 2019, 20, 3240. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Hu, H.; Tong, X.; Jiang, Q.; Zhu, H.; Zhang, S. Calcium-binding protein S100P and cancer: Mechanisms and clinical relevance. J. Cancer Res. Clin. Oncol. 2011, 138, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, T.; Simeone, D.M.; Schmidt, A.M.; Logsdon, C.D. S100P Stimulates Cell Proliferation and Survival via Receptor for Activated Glycation End Products (RAGE). J. Biol. Chem. 2004, 279, 5059–5065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penumutchu, S.R.; Chou, R.H.; Yu, C. Structural insights into calcium-bound S100P and the V domain of the RAGE complex. PLoS ONE 2014, 9, e103947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, M.; Wang, G.; Barsukov, I.L.; Gross, S.; Smith, R.; Rudland, P.S. Direct interaction of metastasis-inducing S100P protein with tubulin causes enhanced cell migration without changes in cell adhesion. Biochem. J. 2020, 477, 1159–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Ito, T.; Nakahara, T.; Nagae, K.; Fuyuno, Y.; Nakao, M.; Akahoshi, M.; Nakagawa, R.; Tu, Y.; Uchi, H.; et al. Upregulation of S100P, receptor for advanced glycation end products and ezrin in malignant melanoma. J. Dermatol. 2013, 40, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Sze Wong, S.Y.; Ting Tse, E.Y.; Fat Ko, F.C.; Tey, S.K.; Yeung, Y.S.; Man, K.; Lam Lo, R.C.; Lin Ng, I.O.; Ping Yam, J.W. Mechanisms through Which Hypoxia-Induced Caveolin-1 Drives Tumorigenesis and Metastasis in Hepato-cellular Carcinoma. Cancer Res. 2016, 76, 7242–7253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahlmann, M.; Okhrimenko, A.; Marcinkowski, P.; Osterland, M.; Herrmann, M.; Smith, J.; Heizmann, C.W.; Schlag, P.M.; Stein, U. RAGE mediates S100A4-induced cell motility via MAPK/ERK and hypoxia signaling and is a prognos-tic biomarker for human colorectal cancer metastasis. Oncotarget 2014, 5, 3220–3233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herwig, N.; Belter, B.; Wolf, S.; Haase-Kohn, C.; Pietzsch, J. Interaction of extracellular S100A4 with RAGE prompts prometastatic activation of A375 melanoma cells. J. Cell. Mol. Med. 2016, 20, 825–835. [Google Scholar] [CrossRef]
- Fei, F.; Qu, J.; Zhang, M.; Li, Y.; Zhang, S. S100A4 in cancer progression and metastasis: A systematic review. Oncotarget 2017, 8, 73219–73239. [Google Scholar] [CrossRef] [Green Version]
- Boye, K.; Maelandsmo, G.M. S100A4 and metastasis: A small actor playing many roles. Am. J. Pathol. 2010, 176, 528–535. [Google Scholar] [CrossRef]
- Garrett, S.C.; Varney, K.M.; Weber, D.J.; Bresnick, A.R. S100A4, a Mediator of Metastasis. J. Biol. Chem. 2006, 281, 677–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Z.-X.; Wu, X.-Y.; Wang, X.-H.; Fu, H.C. Effect of hypoxia-inducible factor 1-α on Survivin in colorectal cancer. Mol. Med. Rep. 2010, 3, 409–415. [Google Scholar] [CrossRef] [Green Version]
- Mao, Q.; Zhang, Y.; Fu, X.; Xue, J.; Guo, W.; Meng, M.; Zhou, Z.; Mo, X.; Lu, Y. A tumor hypoxic niche protects human colon cancer stem cells from chemotherapy. J. Cancer Res. Clin. Oncol. 2012, 139, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Hongo, K.; Tsuno, N.H.; Kawai, K.; Sasaki, K.; Kaneko, M.; Hiyoshi, M.; Murono, K.; Tada, N.; Nirei, T.; Sunami, E.; et al. Hypoxia enhances colon cancer migration and invasion through promotion of epithelial-mesenchymal transition. J. Surg. Res. 2013, 182, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Fu, H.; Chen, D.; Hua, J.; Hu, Y.; Sun, K.; Sun, X. Subcellular distribution of S100A4 and its transcriptional regulation under hypoxic conditions in gastric cancer cell line BGC823. Cancer Sci. 2010, 101, 1141–1146. [Google Scholar] [CrossRef]
- Horiuchi, A.; Hayashi, T.; Kikuchi, N.; Hayashi, A.; Fuseya, C.; Shiozawa, T.; Konishi, I. Hypoxia upregulates ovarian cancer invasiveness via the binding of HIF-1α to a hypoxia-induced, methylation-free hypoxia response element of S100A4 gene. Int. J. Cancer 2012, 131, 1755–1767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Z.; Yang, G.; Shahzidi, S.; Tkacz-Stachowska, K.; Suo, Z.; Nesland, J.M.; Peng, Q. Induction of hypoxia-inducible factor-1α overexpression by cobalt chloride enhances cellular resistance to photodynamic therapy. Cancer Lett. 2006, 244, 182–189. [Google Scholar] [CrossRef]
- Xuan, X.; Li, Q.; Zhang, Z.; Du, Y.; Liu, P. Increased expression levels of S100A4 associated with hypoxia-induced invasion and metastasis in esopha-geal squamous cell cancer. Tumour. Biol. 2014, 35, 12535–12543. [Google Scholar] [CrossRef]
- Madsen, P.S.; Rasmussen, H.H.; Leffers, H.; Honoré, B.; Dejgaard, K.; Olsen, E.; Kiil, J.; Walbum, E.; Andersen, A.H.; Basse, B.; et al. Molecular Cloning, Occurrence, and Expression of a Novel Partially Secreted Protein “Psoriasin” That Is Highly Up-Regulated in Psoriatic Skin. J. Investig. Dermatol. 1991, 97, 701–712. [Google Scholar] [CrossRef] [Green Version]
- Ye, L.; Sun, P.-H.; Martin, T.A.; Sanders, A.J.; Mason, M.D.; Jiang, W. Psoriasin (S100A7) is a positive regulator of survival and invasion of prostate cancer cells. Urol. Oncol. Semin. Orig. Investig. 2013, 31, 1576–1583. [Google Scholar] [CrossRef]
- Yao, R.; Lopez-Beltran, A.; MacLennan, G.T.; Montironi, R.; Eble, J.N.; Cheng, L. Expression of S100 protein family members in the pathogenesis of bladder tumors. Anticancer. Res. 2007, 27, 3051–3058. [Google Scholar]
- Bhalla, S.; Kaur, H.; Dhall, A.; Raghava, G.P.S. Prediction and Analysis of Skin Cancer Progression using Genomics Profiles of Patients. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkie, T.; Verma, A.K.; Zhao, H.; Charan, M.; Ahirwar, D.K.; Kant, S.; Pancholi, V.; Mishra, S.; Ganju, R.K. Lipopolysaccharide from the commensal microbiota of the breast enhances cancer growth: Role of S100A7 and TLR4. Mol. Oncol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Emberley, E.D.; Murphy, L.C.; Watson, P. S100 proteins and their influence on pro-survival pathways in cancer. Biochem. Cell Biol. 2004, 82, 508–515. [Google Scholar] [CrossRef]
- Muoio, M.; Talia, M.; Lappano, R.; Sims, A.; Vella, V.; Cirillo, F.; Manzella, L.; Giuliano, M.; Maggiolini, M.; Belfiore, A.; et al. Activation of the S100A7/RAGE Pathway by IGF-1 Contributes to Angiogenesis in Breast Cancer. Cancers 2021, 13, 621. [Google Scholar] [CrossRef] [PubMed]
- Kerkhoff, C.; Klempt, M.; Sorg, C. Novel insights into structure and function of MRP8 (S100A8) and MRP14 (S100A9). Biochim. Biophys. Acta 1998, 1448, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Tomonobu, N.; Kinoshita, R.; Sakaguchi, M. S100 Soil Sensor Receptors and Molecular Targeting Therapy against Them in Cancer Metastasis. Transl. Oncol. 2020, 13, 100753. [Google Scholar] [CrossRef]
- Hibino, T.; Sakaguchi, M.; Miyamoto, S.; Yamamoto, M.; Motoyama, A.; Hosoi, J.; Shimokata, T.; Ito, T.; Tsuboi, R.; Huh, N.-H. S100A9 Is a Novel Ligand of EMMPRIN That Promotes Melanoma Metastasis. Cancer Res. 2013, 73, 172–183. [Google Scholar] [CrossRef] [Green Version]
- Ruma, I.M.; Putranto, E.W.; Kondo, E.; Murata, H.; Watanabe, M.; Huang, P.; Kinoshita, R.; Futami, J.; Inoue, Y.; Yamauchi, A.; et al. MCAM, as a novel receptor for S100A8/A9, mediates progression of malignant melanoma through prominent activation of NF-κB and ROS formation upon ligand binding. Clin. Exp. Metastasis 2016, 33, 609–627. [Google Scholar] [CrossRef]
- Ghavami, S.; Chitayat, S.; Hashemi, M.; Eshraghi, M.; Chazin, W.J.; Halayko, A.J.; Kerkhoff, C. S100A8/A9: A Janus-faced molecule in cancer therapy and tumorgenesis. Eur. J. Pharmacol. 2009, 625, 73–83. [Google Scholar] [CrossRef]
- Ghavami, S.; Rashedi, I.; Dattilo, B.M.; Eshraghi, M.; Chazin, W.J.; Hashemi, M.; Wesselborg, S.; Kerkhoff, C.; Los, M. S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J. Leukoc. Biol. 2008, 83, 1484–1492. [Google Scholar] [CrossRef] [PubMed]
- Turovskaya, O.; Foell, D.; Sinha, P.; Vogl, T.; Newlin, R.; Nayak, J.; Nguyen, M.; Olsson, A.; Nawroth, P.P.; Bierhaus, A.; et al. RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis. Carcinogenesis 2008, 29, 2035–2043. [Google Scholar] [CrossRef] [PubMed]
- Grebhardt, S.; Veltkamp, C.; Ströbel, P.; Mayer, D. Hypoxia and HIF-1 increase S100A8 and S100A9 expression in prostate cancer. Int. J. Cancer 2012, 131, 2785–2794. [Google Scholar] [CrossRef] [PubMed]
| Ligands | Cancer Type | Pathological Response | Reference |
|---|---|---|---|
| HMGB1 | Melanoma | Under hypoxia, melanoma cells release HMGB1, which promotes melanoma growth and metastasis | [150] |
| Hepatocellular carcinoma | HMGB1 released under hypoxia promotes cellular invasion and metastasis | [177,179] | |
| Glioblastoma | HMGB1 released under hypoxia promotes cell proliferation and invasion | [180] | |
| S100B | Colon adenocarcinoma | Stimulation of RAGE by S100B activates HIF1-α and the expression of HIF-1α dependent genes | [190] |
| S100P | Hepatocellular carcinoma | Hypoxia upregulates S100P and further promotes metastasis of HCC | [196] |
| S100A4 | Colorectal Cancer | Activation of RAGE by S100A4 results in increased levels of HIF-1α | [197] |
| Gastric, ovarian cancer | Hypoxia upregulates S100A4 | [205] | |
| S100A7 | Breast Cancer | Activation of RAGE by S100A7 results in increased angiogenesis | [13] |
| S100A8/A9 | Prostate Cancer | Hypoxia upregulates S100A8 and S100A9 transcript and protein levels | [223] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taneja, S.; Vetter, S.W.; Leclerc, E. Hypoxia and the Receptor for Advanced Glycation End Products (RAGE) Signaling in Cancer. Int. J. Mol. Sci. 2021, 22, 8153. https://doi.org/10.3390/ijms22158153
Taneja S, Vetter SW, Leclerc E. Hypoxia and the Receptor for Advanced Glycation End Products (RAGE) Signaling in Cancer. International Journal of Molecular Sciences. 2021; 22(15):8153. https://doi.org/10.3390/ijms22158153
Chicago/Turabian StyleTaneja, Sakshi, Stefan W. Vetter, and Estelle Leclerc. 2021. "Hypoxia and the Receptor for Advanced Glycation End Products (RAGE) Signaling in Cancer" International Journal of Molecular Sciences 22, no. 15: 8153. https://doi.org/10.3390/ijms22158153





