Molecular Approaches to Agri-Food Traceability and Authentication: An Updated Review
Abstract
:1. Introduction
2. Analytical Methods for the Traceability and Authentication of Food Deriving from Plant Species
3. Molecular Approaches to Agri-Food Analysis
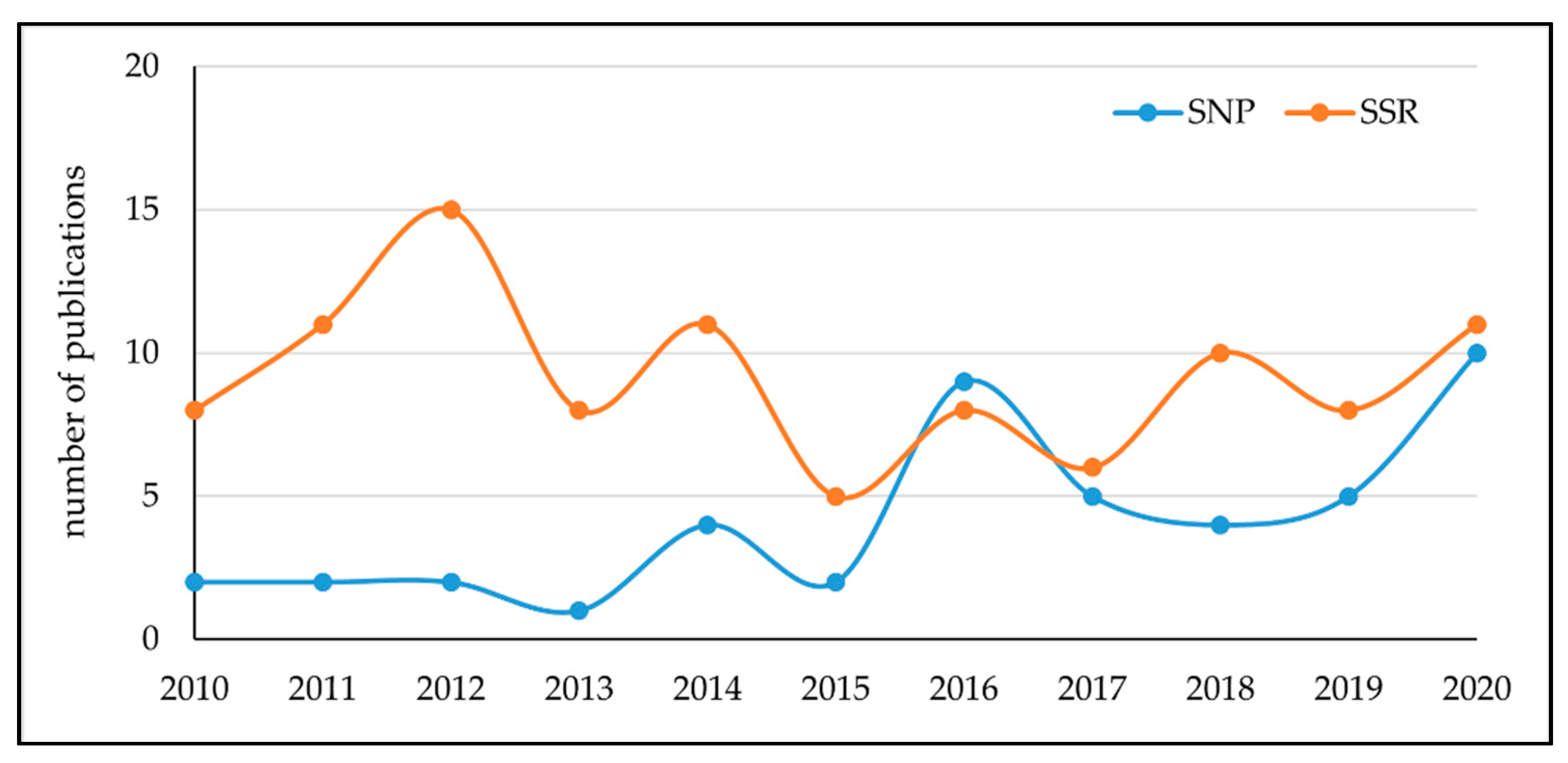
3.1. Molecular Marker-Based Methods
3.1.1. Simple Sequence Repeats (SSR)
3.1.2. Single Nucleotide Polymorphism (SNP)
3.2. Single Region Approaches
3.2.1. Species-Specific Primer PCR
3.2.2. DNA Barcoding
3.2.3. Isothermal Amplification-Based Methods
3.3. Next Generation Sequencing-Based Methods
3.3.1. Whole Metagenome Sequencing
3.3.2. DNA Metabarcoding
4. Advantages and Limits of Molecular Methods in Agri-Food Authentication and Traceability
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
List of Abbreviations
| HPLC | High-performance liquid chromatography |
| ELISA | Enzyme-linked immunosorbent assay |
| NMR | Nuclear magnetic resonance |
| SSR | Simple sequence repeat |
| SNP | Single nucleotide polymorphism |
| HRM | High resolution melting |
| RFLP | Restriction fragment length polymorphism |
| RPA | Recombinase polymerase amplification |
| LFD | Lateral flow device |
| LAMP | Loop-mediated isothermal amplification |
| GMO | Genetically modified organism |
| PDO | Protected designation of origin |
| PGI | Protected geographical indication |
| NGS | Next generation sequencing |
| WMS | Whole metagenome sequencing |
References
- Aung, M.M.; Chang, Y.S. Traceability in a food supply chain: Safety and quality perspectives. Food Control 2014, 39, 172–184. [Google Scholar] [CrossRef]
- Ballin, N.Z.; Laursen, K.H. To target or not to target? Definitions and nomenclature for targeted versus non-targeted analytical food authentication. Trends Food Sci. Technol. 2019, 86, 537–543. [Google Scholar] [CrossRef]
- Maitiniyazi, S.; Canavari, M. Exploring Chinese consumers’ attitudes toward traceable dairy products: A focus group study. J. Dairy Sci. 2020, 103, 11257–11267. [Google Scholar] [CrossRef] [PubMed]
- Pelegrino, B.O.; Silva, R.; Guimarães, J.T.; Coutinho, N.F.; Pimentel, T.C.; Castro, B.G.; Freitas, M.Q.; Esmerino, E.A.; Sant’Ana, A.S.; Silva, M.C.; et al. Traceability: Perception and attitudes of artisanal cheese producers in Brazil. J. Dairy Sci. 2020, 103, 4874–4879. [Google Scholar] [CrossRef]
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food authentication: Techniques, trends & emerging approaches. Trends Analyt. Chem. 2016, 85, 123–132. [Google Scholar]
- Lo, Y.T.; Shaw, P.C. DNA-based techniques for authentication of processed food and food supplements. Food Chem. 2018, 240, 767–774. [Google Scholar] [CrossRef]
- Galimberti, A.; Casiraghi, M.; Bruni, I.; Guzzetti, L.; Cortis, P.; Berterame, N.M.; Labra, M. From DNA barcoding to personalized nutrition: The evolution of food traceability. Curr. Opin. Food Sci. 2019, 28, 41–48. [Google Scholar] [CrossRef]
- Regulation (EU) No 1151/2012 of the European Parliament and of the Council of 21 November 2012 on Quality Schemes for Agricultural Products and Foodstuffs. Available online: https://eur-lex.europa.eu (accessed on 9 July 2021).
- Martins-Lopes, P.; Gomes, S.; Pereira, L.; Guedes-Pinto, H. Molecular markers for food traceability. Food Technol. Biotechnol. 2013, 51, 198–207. [Google Scholar]
- Scarano, D.; Rao, R. DNA markers for food products authentication. Diversity 2014, 6, 579–596. [Google Scholar] [CrossRef]
- Corrado, G. Advances in DNA typing in the agro-food supply chain. Trends Food Sci. Technol. 2016, 52, 80–89. [Google Scholar] [CrossRef]
- Abbas, O.; Zadravec, M.; Baeten, V.; Mikuš, T.; Lešić, T.; Vulić, A.; Prpić, J.; Jemeršić, L.; Pleadin, J. Analytical methods used for the authentication of food of animal origin. Food Chem. 2018, 246, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Wadood, S.A.; Boli, G.; Xiaowen, Z.; Hussain, I.; Yimin, W. Recent development in the application of analytical techniques for the traceability and authenticity of food of plant origin. Microchem. J. 2020, 152, 104295. [Google Scholar] [CrossRef]
- Lohumi, S.; Lee, S.; Lee, H.; Cho, B.K. A review of vibrational spectroscopic techniques for the detection of food authenticity and adulteration. Trends Food Sci. Technol. 2015, 46, 85–98. [Google Scholar] [CrossRef]
- Castro-Puyana, M.; Herrero, M. Metabolomics approaches based on mass spectrometry for food safety, quality and traceability. Trends Analyt. Chem. 2013, 52, 74–87. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, B.; Chen, G.; Chen, A.; Yang, S.; Ye, Z. Recent developments in application of stable isotope analysis on agro-product authenticity and traceability. Food Chem. 2014, 145, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Dymerski, T. Two-dimensional gas chromatography coupled with mass spectrometry in food analysis. Crit. Rev. Anal. Chem. 2018, 48, 252–278. [Google Scholar] [CrossRef]
- Esteki, M.; Shahsavari, Z.; Simal-Gandara, J. Food identification by high performance liquid chromatography fingerprinting and mathematical processing. Food Res. Int. 2019, 122, 303–317. [Google Scholar] [CrossRef]
- Nolvachai, Y.; Kulsing, C.; Marriott, P.J. Multidimensional gas chromatography in food analysis. Trends Analyt. Chem. 2017, 96, 124–137. [Google Scholar] [CrossRef]
- Medina, S.; Perestrelo, R.; Silva, P.; Pereira, J.A.M.; Câmara, J.S. Current trends and recent advances on food authenticity technologies and chemometric approaches. Trends Food Sci. Technol. 2019, 85, 163–176. [Google Scholar] [CrossRef]
- Wu, L.; Li, G.; Xu, X.; Zhu, L.; Huang, R.; Chen, X. Application of nano-ELISA in food analysis: Recent advances and challenges. Trends Analyt. Chem. 2019, 113, 140–156. [Google Scholar] [CrossRef]
- Ahmad, M.H.; Sahar, A.; Hitzmann, B. Fluorescence spectroscopy for the monitoring of food processes. In Measurement, Modeling and Automation in Advanced Food Processing; Hitzmann, B., Ed.; Springer: New York, NY, USA, 2017; Volume 161, pp. 121–151. [Google Scholar]
- Esteki, M.; Shahsavari, Z.; Simal-Gandara, J. Use of spectroscopic methods in combination with linear discriminant analysis for authentication of food products. Food Control 2018, 91, 100–112. [Google Scholar] [CrossRef]
- Xu, Y.; Zhong, P.; Jiang, A.; Shen, X.; Li, X.; Xu, Z.; Shen, Y.; Sun, Y.; Lei, H. Raman spectroscopy coupled with chemometrics for food authentication: A review. Trends Analyt. Chem. 2020, 131, 116017. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.R. The potentiality of NMR-based metabolomics in food science and food authentication assessment. Magn Reson Chem. 2019, 57, 558–578. [Google Scholar] [CrossRef]
- Rubert, J.; Zachariasova, M.; Hajslova, J. Advances in high-resolution mass spectrometry based on metabolomics studies for food—A review. Food Addit. Contam. Part A 2015, 32, 1685–1708. [Google Scholar] [CrossRef]
- Papapetros, S.; Louppis, A.; Kosma, I.; Kontakos, S.; Badeka, A.; Kontominas, M.G. Characterization and differentiation of botanical and geographical origin of selected popular sweet cherry cultivars grown in Greece. J. Food Compos. Anal. 2018, 72, 48–56. [Google Scholar] [CrossRef]
- Watanabe, E.; Miyake, S. Direct determination of neonicotinoid insecticides in an analytically challenging crop such as Chinese chives using selective ELISAs. J. Environ. Sci. Health Part B 2018, 53, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Hongsibsong, S.; Prapamontol, T.; Xu, T.; Hammock, B.D.; Wang, H.; Chen, Z.J.; Xu, Z.L. Monitoring of the organophosphate pesticide chlorpyrifos in vegetable samples from local markets in Northern Thailand by developed immunoassay. Int. J. Environ. Res. Public Health 2020, 17, 4723. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Li, R.; Jiang, Z.T.; Tang, S.H.; Wang, Y.; Shi, M.; Xiao, Y.Q.; Jia, B.; Lu, T.X.; Wang, H. Synchronous front-face fluorescence spectroscopy for authentication of the adulteration of edible vegetable oil with refined used frying oil. Food Chem. 2017, 217, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.; Kamal, G.M.; Wang, J.; Liu, H.; Zhang, G.; Hu, Z.; Anwar, F.; Du, H. 1H-NMRbased metabolomics for discrimination of rice from different geographical origins of China. J. Cereal Sci. 2017, 76, 243–252. [Google Scholar] [CrossRef]
- Longobardi, F.; Innamorato, V.; Di Gioia, A.; Ventrella, A.; Lippolis, V.; Logrieco, A.F.; Catucci, L.; Agostiano, A. Geographical origin discrimination of lentils (Lens culinaris Medik.) using 1H-NMR fingerprinting and multivariate statistical analyses. Food Chem. 2017, 237, 743–748. [Google Scholar] [CrossRef]
- Salazar, M.O.; Pisano, P.L.; Gonzalez Sierra, M.; Furlan, R.L.E. NMR and multivariate data analysis to assess traceability of argentine citrus. Microchem. J. 2018, 141, 264–270. [Google Scholar] [CrossRef]
- Drivelos, S.A.; Georgiou, C.A. Multi-element and multi-isotope-ratio analysis to determine the geographical origin of foods in the European Union. Trends Anal. Chem. 2012, 40, 38–51. [Google Scholar] [CrossRef]
- Mahne Opatić, A.; Nečemer, M.; Lojen, S.; Masten, J.; Zlatić, E.; Šircelj, H.; Stopar, D.; Vidrih, R. Determination of geographical origin of commercial tomato through analysis of stable isotopes, elemental composition and chemical markers. Food Control 2018, 89, 133–141. [Google Scholar] [CrossRef]
- Zambianchi, S.; Soffritti, G.; Stagnati, L.; Patrone, V.; Morelli, L.; Vercesi, A.; Busconi, M. Applicability of DNA traceability along the entire wine production chain in the real case of a large Italian cooperative winery. Food Control 2021, 124, 107929. [Google Scholar] [CrossRef]
- Chedid, E.; Rizou, M.; Kalaitzis, P. Application of high resolution melting combined with DNA-based markers for quantitative analysis of olive oil authenticity and adulteration. Food Chemistry X 2020, 6, 100082. [Google Scholar] [CrossRef]
- Gomes, S.; Breia, R.; Carvalho, T.; Carnide, V.; Martins-Lopes, P. Microsatellite high-resolution melting (SSR-HRM) to track olive genotypes: From field to olive oil. J. Food Sci. 2018, 83, 2415–2423. [Google Scholar] [CrossRef] [PubMed]
- di Rienzo, V.; Fanelli, V.; Miazzi, M.M.; Savino, V.; Pasqualone, A.; Summo, C.; Giannini, P.; Sabetta, W.; Montemurro, C. A reliable analytical procedure to discover table grape DNA adulteration in industrial wines and musts. Acta Hortic. 2017, 1188, 365–370. [Google Scholar] [CrossRef]
- Ben Ayed, R.; Rebai, A. Tunisian table olive oil traceability and quality using SNP genotyping and bioinformatics tools. BioMed Res. Int. 2019, 2019, 8291341. [Google Scholar] [CrossRef]
- Piarulli, L.; Savoia, M.A.; Taranto, F.; D’Agostino, N.; Sardaro, R.; Girone, S.; Gadaleta, S.; Fucili, V.; De Giovanni, C.; Montemurro, C.; et al. A robust DNA isolation protocol from filtered commercial olive oil for PCR-based fingerprinting. Foods 2019, 8, 462. [Google Scholar] [CrossRef] [Green Version]
- Pereira, L.; Gomes, S.; Barrias, S.; Fernandes, J.R.; Martins-Lopes, P. Applying high-resolution melting (HRM) technology to olive oil and wine authenticity. Food Res. Int. 2018, 103, 170–181. [Google Scholar] [CrossRef]
- Boccacci, P.; Chitarra, W.; Schneider, A.; Rolle, L.; Gambino, G. Single-nucleotide polymorphism (SNP) genotyping assays for the varietal authentication of ‘Nebbiolo’ musts and wines. Food Chem. 2020, 312, 126100. [Google Scholar] [CrossRef]
- Barrias, S.; Fernandes, J.R.; Eiras-Dias, J.E.; Brazão, J.; Martins-Lopes, P. Label free DNA-based optical biosensor as a potential system for wine authenticity. Food Chem. 2019, 270, 299–304. [Google Scholar] [CrossRef]
- Zhang, D.; Vega, F.E.; Infante, F.; Solano, W.; Johnson, E.S.; Meinhardt, L.W. Accurate differentiation of green beans of Arabica and Robusta coffee using nanofluidic array of Single Nucleotide Polymorphism (SNP) markers. J. AOAC Int. 2020, 103, 315–324. [Google Scholar] [CrossRef]
- Silletti, S.; Morello, L.; Gavazzi, F.; Gianì, S.; Braglia, L.; Breviario, D. Untargeted DNA-based methods for the authentication of wheat species and related cereals in food products. Food Chem. 2019, 271, 410–418. [Google Scholar] [CrossRef]
- Morcia, C.; Bergami, R.; Scaramagli, S.; Ghizzoni, R.; Carnevali, P.; Terzi, V. A Chip Digital PCR assay for quantification of common wheat contamination in pasta production chain. Foods 2020, 9, 911. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Gao, D.; Dong, J.; Chen, W.; Li, Z.; Wang, J.; Liu, J. Mass ratio quantitative detection for kidney bean in lotus seed paste using duplex droplet digital PCR and chip digital PCR. Anal. Bioanal. Chem. 2020, 412, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Swetha, V.P.; Parvathy, V.A.; Sheeja, T.E.; Sasikumar, B. Authentication of Myristica fragrans Houtt. using DNA barcoding. Food Control 2017, 73, 1010–1015. [Google Scholar] [CrossRef]
- Uncu, A.O. A trnH-psbA barcode genotyping assay for the detection of common apricot (Prunus armeniaca L.) adulteration in almond (Prunus dulcis Mill.). CyTA-J. Food 2020, 18, 187–194. [Google Scholar] [CrossRef] [Green Version]
- Lagiotis, G.; Stavridou, E.; Bosmali, I.; Osathanunkul, M.; Haider, N.; Madesis, P. Detection and quantification of cashew in commercial tea products using High Resolution Melting (HRM) analysis. J. Food Sci. 2020, 85, 1629–1634. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Jiang, G.; Huang, L.; Chen, C.; Sun, J.; Zhu, C. DNA barcoding coupled with high-resolution melting analysis for nut species and walnut milk beverage authentication. J. Sci. Food Agric. 2020, 100, 2372–2379. [Google Scholar] [CrossRef]
- Bosmali, I.; Lagiotis, G.; Stavridou, E.; Haider, N.; Osathanunkul, M.; Pasentsis, K.; Madesis, P. Novel authentication approach for coffee beans and the brewed beverage using a nuclear-based species-specific marker coupled with high resolution melting analysis. LWT-Food Sci. Technol. 2021, 137, 110336. [Google Scholar] [CrossRef]
- Valentini, P.; Galimberti, A.; Mezzasalma, V.; De Mattia, F.; Casiraghi, M.; Labra, M.; Pompa, P.P. DNA barcoding meets nanotechnology: Development of a universal colorimetric test for food authentication. Angew. Chem. Int. Ed. 2017, 56, 8094. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, M.; Yang, Y.; Jiang, L.; Liu, M.; Wang, B.; Wang, Y. Authentication of small berry fruit in fruit products by DNA barcoding method. J. Food Sci. 2018, 83, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, B.; Xiang, L.; Xiong, C.; Shi, Y.; Wu, L.; Meng, X.; Dong, G.; Xie, Y.; Sun, W. A novel onsite and visual molecular technique to authenticate saffron (Crocus sativus) and its adulterants based on recombinase polymerase amplification. Food Control 2019, 100, 117–121. [Google Scholar] [CrossRef]
- Zhao, M.; Shi, Y.; Wu, L.; Guo, L.; Liu, W.; Xiong, C.; Yan, S.; Sun, W.; Chen, S. Rapid authentication of the precious herb saffron by loop-mediated isothermal amplification (LAMP) based on internal transcribed spacer 2 (ITS2) sequence. Sci. Rep. 2016, 6, 25370. [Google Scholar] [CrossRef] [Green Version]
- Cibecchini, G.; Cecere, P.; Tumino, G.; Morcia, C.; Ghizzoni, R.; Carnevali, P.; Terzi, V.; Pompa, P.P. A fast, naked-eye assay for varietal traceability in the durum wheat production chain. Foods 2020, 9, 1691. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lu, X. Rapid pomegranate juice authentication using a simple sample-to-answer hybrid paper/polymer-based lab-on-a-chip device. ACS Sens. 2020, 5, 2168–2176. [Google Scholar] [CrossRef]
- Lo, Y.T.; Shaw, P.C. Application of next-generation sequencing for the identification of herbal products. Biotechnol. Adv. 2019, 37, 107450. [Google Scholar] [CrossRef]
- Raime, K.; Krjutškov, K.; Remm, M. Method for the identification of plant DNA in food using alignment-free analysis of sequencing reads: A case study on lupin. Front. Plant Sci. 2020, 11, 646. [Google Scholar] [CrossRef]
- Lee, H.J.; Koo, H.J.; Lee, J.; Lee, S.C.; Lee, D.Y.; Giang, V.N.L.; Kim, M.; Shim, H.; Park, J.Y. Yoo, K.O.; et al. Authentication of Zanthoxylum species based on integrated analysis of complete chloroplast genome sequences and metabolite profiles. J. Agric. Food Chem. 2017, 65, 10350–10359. [Google Scholar] [CrossRef]
- Kim, Y.; Shin, J.; Oh, D.R.; Kim, D.W.; Lee, H.S.; Choi, C. Complete chloroplast genome sequences of Vaccinium bracteatum Thunb., V. vitis-idaea L., and V. uliginosum L. (Ericaceae). Mitochondrial DNA B 2020, 5, 1843–1844. [Google Scholar] [CrossRef] [Green Version]
- Khansaritoreh, E.; Salmaki, Y.; Ramezani, E.; Akbari Azirani, T.; Keller, A.; Neumann, K.; Alizadeh, K.; Zarre, S.; Beckh, G.; Behling, H. Employing DNA metabarcoding to determine the geographical origin of honey. Heliyon 2020, 6, e05596. [Google Scholar] [CrossRef] [PubMed]
- Beltramo, C.; Cerutti, F.; Brusa, F.; Mogliotti, P.; Garrone, A.; Squadrone, S.; Acutis, P.L.; Peletto, S. Exploring the botanical composition of polyfloral and monofloral honeys through DNA metabarcoding. Food Control 2021, 128, 108175. [Google Scholar]
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Stagnati, L.; Soffritti, G.; Martino, M.; Bortolini, C.; Lanubile, A.; Busconi, M.; Marocco, A. Cocoa beans and liquor fingerprinting: A real case involving SSR profiling of CCN51 and “Nacional” varieties. Food Control 2020, 118, 107392. [Google Scholar] [CrossRef]
- Pinczinger, D.; von Reth, M.; Hanke, M.V.; Flachowsky, H. SSR fingerprinting of raspberry cultivars traded in Germany clearly showed that certainty about the genotype authenticity is a prerequisite for any horticultural experiment. Eur. J. Hortic. Sci. 2020, 85, 79–85. [Google Scholar] [CrossRef]
- Sabetta, W.; Miazzi, M.M.; di Rienzo, V.; Fanelli, V.; Pasqualone, A.; Montemurro, C. Development and application of protocols to certify the authenticity and traceability of Apulian typical products in olive sector. Riv. Ital. Delle Sostanze Grasse 2017, 94, 37–43. [Google Scholar]
- Verdone, M.; Rao, R.; Coppola, M.; Corrado, G. Identification of zucchini varieties in commercial food products by DNA typing. Food Control 2018, 84, 197–204. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Argiriou, A.; Tsaftaris, A. Microsatellite high resolution melting (SSR-HRM) analysis for authenticity testing of protected designation of origin (PDO) sweet cherry products. Food Control 2011, 22, 532–541. [Google Scholar] [CrossRef]
- Bosmali, I.; Ganopoulos, I.; Madesis, P.; Tsaftaris, A. Microsatellite and DNA-barcode regions typing combined with High Resolution Melting (HRM) analysis for food forensic uses: A case study on lentils (Lens culinaris). Food Res. Int. 2012, 46, 141–147. [Google Scholar] [CrossRef]
- di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Savino, V.; Pollastro, S.; Colucci, F.; Miccolupo, A.; Blanco, A.; Pasqualone, A.; Montemurro, C. An enhanced analytical procedure to discover table grape DNA adulteration in industrial musts. Food Control 2016, 60, 124–130. [Google Scholar] [CrossRef]
- Pasqualone, A.; Montemurro, C.; Grinn-Gofron, A.; Sonnante, G.; Blanco, A. Detection of soft wheat in semolina and durum wheat bread by analysis of DNA microsatellites. J. Agric. Food Chem. 2007, 55, 3312–3318. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, A.; Jin, X.; Pan, L. Technologies in individual animal identification and meat products traceability. Biotechnol. Biotechnol. Equip. 2020, 34, 48–57. [Google Scholar] [CrossRef]
- Marrano, A.; Birolo, G.; Prazzoli, M.L.; Lorenzi, S.; Valle, G.; Grando, M.S. SNP-discovery by RAD-sequencing in a germplasm collection of wild and cultivated grapevines (V. vinifera L.). PLoS ONE 2017, 12, e0170655. [Google Scholar] [CrossRef] [PubMed]
- Taranto, F.; D’Agostino, N.; Pavan, S.; Fanelli, V.; di Rienzo, V.; Sabetta, W.; Miazzi, M.M.; Zelasco, S.; Perri, E.; Montemurro, C. Single nucleotide polymorphism (SNP) diversity in an olive germplasm collection. Acta Hortic. 2018, 1199, 27–32. [Google Scholar] [CrossRef]
- Pavan, S.; Bardaro, N.; Fanelli, V.; Marcotrigiano, A.R.; Mangini, G.; Taranto, F.; Catalano, D.; Montemurro, C.; De Giovanni, C.; Lotti, C.; et al. Genotyping by Sequencing of cultivated lentil (Lens culinaris Medik.) highlights population structure in the Mediterranean gene pool associated with geographic patterns and phenotypic variables. Front. Genet. 2019, 10, 872. [Google Scholar] [CrossRef]
- Singh, R.; Iquebal, M.A.; Mishra, C.N.; Jaiswal, S.; Kumar, D.; Raghav, N.; Paul, S.; Sheoran, S.; Sharma, P.; Gupta, A.; et al. Development of model web-server for crop variety identification using throughput SNP genotyping data. Sci. Rep. 2019, 9, 5122. [Google Scholar] [CrossRef] [Green Version]
- Akpertey, A.; Padi, F.K.; Meinhardt, L.; Zhang, D. Effectiveness of Single Nucleotide Polymorphism markers in genotyping germplasm collections of Coffea canephora using KASP assay. Front. Plant Sci. 2021, 11, 612593. [Google Scholar] [CrossRef]
- Spaniolas, S.; Bazakos, C.; Tucker, G.A.; Bennett, M.J. Comparison of SNP-based detection assays for food analysis: Coffee authentication. J. AOAC Int. 2014, 97, 4. [Google Scholar] [CrossRef]
- Fang, W.; Meinhardt, L.W.; Mischke, S.; Bellato, C.M.; Motilal, L.; Zhang, D. Accurate determination of genetic identity for a single cacao bean, using molecular markers with a nanofluidic system, ensures cocoa authentication. J. Agric. Food Chem. 2014, 62, 481–487. [Google Scholar] [CrossRef]
- Fang, W.P.; Meinhardt, L.W.; Tan, H.W.; Zhou, L.; Mischke, S.; Zhang, D. Varietal identification of tea (Camellia sinensis) using nanofluidic array of single nucleotide polymorphism (SNP) markers. Hortic. Res. 2014, 1, 14035. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, R.; Paul, J.S.; Albrechtsen, A.; Song, Y.S. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 2011, 12, 443–451. [Google Scholar] [CrossRef]
- Galimberti, A.; De Mattia, F.; Losa, A.; Bruni, I.; Federici, S.; Casiraghi, M.; Martellos, S.; Labra, M. DNA barcoding as a new tool for food traceability. Food Res. Int. 2013, 50, 55–63. [Google Scholar] [CrossRef]
- Sonnante, G.; Montemurro, C.; Morgese, A.; Sabetta, W.; Blanco, A.; Pasqualone, A. DNA microsatellite region for a reliable quantification of ~soft wheat adulteration in durum wheat-based foodstuffs by real-time PCR. J. Agric. Food Chem. 2009, 57, 10199–1020411. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Arami, S.; Sato, M.; Haraguchi, H.; Kurimoto, Y.; Imai, S.; Tanaka, K.; Mano, J.; Furui, S.; Kitta, K. Development of methods to distinguish between durum/common wheat and common wheat in blended flour using PCR. J. Food Hyg. Soc. Jpn. 2012, 53, 195–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voorhuijzen, M.M.; van Dijk, J.P.; Prins, T.W.; Van Hoef, A.M.A.; Seyfarth, R.; Kok, E.J. Development of a multiplex DNA-based traceability tool for crop plant materials. Anal. Bioanal. Chem. 2012, 402, 693–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morisset, D.; Stebih, D.; Milavec, M.; Gruden, K.; Zel, J. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS ONE 2013, 8, e62583. [Google Scholar] [CrossRef] [Green Version]
- Low, H.; Chan, S.J.; Soo, G.H.; Ling, B.; Tan, E.L. Clarity™ digital PCR system: A novel platform for absolute quantification of nucleic acids. Anal. Bioanal. Chem. 2017, 409, 1869–1875. [Google Scholar] [CrossRef] [PubMed]
- Demeke, T.; Dobnik, D. Critical assessment of digital PCR for the detection and quantification of genetically modified organisms. Anal. Bioanal. Chem. 2018, 410, 4039–4050. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Simpson, D.J.; Gänzle, M.G. Detection of enterohaemorrhagic Escherichia coli in food by droplet digital PCR to detect simultaneous virulence factors in a single genome. Food Microbiol. 2020, 90, 103466. [Google Scholar] [CrossRef]
- Pierboni, E.; Rondini, C.; Torricelli, M.; Ciccone, L.; Tovo, G.R.; Mercuri, M.L.; Altissimi, S.; Haouet, N. Digital PCR for analysis of peanut and soybean allergens in foods. Food Control 2018, 92, 128–136. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; deWaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. Royal Soc. B-Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, X.; Wang, L.; Chen, X.; Pang, X.; Han, J. A nucleotide signature for the identification of American ginseng and its products. Front. Plant Sci. 2016, 18, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uncu, A.T.; Uncu, A.O. Plastid trnH-psbA intergenic spacer serves as a PCR-based marker to detect common grain adulterants of coffee (Coffea arabica L.). Food Control 2018, 91, 32–39. [Google Scholar] [CrossRef]
- Bosmali, I.; Ordoudi, S.A.; Tsimidou, M.Z.; Madesis, P. Greek PDO saffron authentication studies using species specific molecular markers. Food Res. Int. 2017, 100, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiang, L.; Zhang, Y.; Lai, X.; Xiong, C.; Li, J.; Su, Y.; Sun, W.; Chen, S. DNA barcoding based identification of Hippophae species and authentication of commercial products by high resolution melting analysis. Food Chem. 2018, 242, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Ballin, N.Z.; Onaindia, J.O.; Jawad, H.; Fernandez-Carazo, R.; Maquet, A. High-resolution melting of multiple barcode amplicons for plant species authentication. Food Control 2019, 105, 141–150. [Google Scholar] [CrossRef]
- Gong, L.; Qiu, X.H.; Huang, J.; Xu, W.; Bai, J.Q.; Zhang, J.; Su, H.; Xu, C.M.; Huang, Z.H. Constructing a DNA barcode reference library for southern herbs in China: A resource for authentication of southern Chinese medicine. PLoS ONE 2018, 13, e0201240. [Google Scholar]
- Kane, N.C.; Cronk, Q. Botany without borders, barcoding in focus. Mol. Ecol. 2008, 17, 5175–5176. [Google Scholar] [CrossRef]
- Kane, N.; Sveinsson, S.; Dempewolf, H.; Yang, J.Y.; Zhang, D.; Engels, J.M.M.; Cronk, Q. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot. 2012, 99, 320–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [Green Version]
- Leonardo, S.; Toldrà, A.; Campàs, M. Biosensors based on isothermal DNA amplification for bacterial detection in food safety and environmental monitoring. Sensors 2021, 21, 602. [Google Scholar] [CrossRef]
- Santiago-Felipe, S.; Tortajada-Genaro, L.A.; Puchades, R.; Maquieira, A. Recombinase polymerase and enzyme-linked immunosorbent assay as a DNA amplification-detection strategy for food analysis. Anal. Chim. Acta 2014, 811, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lowe, S.B.; Gooding, J.J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectron. 2014, 61, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.P.; Othman, S.; Lau, Y.L.; Radu, S.; Chee, H.Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2017, 124, 626–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Wang, C.; Ji, L.; Zhao, X.X.; Liu, M.; Zhang, D.; Shi, J. Loop mediated isothermal amplification (LAMP) assay for GMO detection: Recent progresses and future perspectives. Open Access Libr. J. 2015, 2, e1264. [Google Scholar]
- Wilkinson, M.J.; Szabo, C.; Ford, C.S.; Yarom, Y.; Croxford, A.E.; Camp, A.; Gooding, P. Replacing Sanger with Next Generation Sequencing to improve coverage and quality of reference DNA barcodes for plants. Sci. Rep. 2017, 7, 46040. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.R.; Cowley, M.J.; Davis, R.L. Next-Generation Sequencing and emerging technologies. Semin. Thromb. Hemost. 2019, 45, 661–673. [Google Scholar] [CrossRef]
- Ripp, F.; Krombholz, C.F.; Liu, Y.; Weber, M.; Schäfer, A.; Schmidt, B.; Köppel, R.; Hankeln, T. All-Food-Seq (AFS): A quantifiable screen for species in biological samples by deep DNA sequencing. BMC Genom. 2014, 15, 639. [Google Scholar] [CrossRef] [Green Version]
- Beck, K.L.; Haiminen, N.; Chambliss, D.; Edlund, S.; Kunitomi, M.; Huang, B.C.; Kong, N.; Ganesan, B.; Baker, R.; Markwell, P.; et al. Monitoring the microbiome for food safety and quality using deep shotgun sequencing. NPJ Sci Food 2021, 5, 3. [Google Scholar] [CrossRef]
- Haynes, E.; Jimenez, E.; Pardo, M.A.; Helyar, S.J. The future of NGS (Next Generation Sequencing) analysis in testing food authenticity. Food Control 2019, 101, 134–143. [Google Scholar] [CrossRef]
- Haiminen, N.; Edlund, S.; Chambliss, D.; Kunitomi, M.; Weimer, B.C.; Ganesan, B.; Baker, R.; Markwell, P.; Davis, M.; Huang, C.; et al. Food authentication from shotgun sequencing reads with an application on high protein powders. NPJ Sci Food 2019, 3, 24. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Ramachandran, P.; Wen, J.; Duke, J.A.; Metzman, H.; McLaughlin, W.; Ottesen, A.R.; Timme, R.E.; Handy, S.M. Development of a reference standard library of chloroplast genome sequences, GenomeTrakrCP. Planta Med. 2017, 83, 1420–1430. [Google Scholar] [CrossRef]
- Bruno, A.; Sandionigi, A.; Agostinetto, G.; Bernabovi, L.; Frigerio, J.; Casiraghi, M.; Labra, M. Food tracking perspective: DNA metabarcoding to identify plant composition in complex and processed food products. Genes 2019, 10, 248. [Google Scholar] [CrossRef] [Green Version]
- Speranskaya, A.S.; Khafizov, K.; Ayginin, A.A.; Krinitsina, A.A.; Omelchenko, D.O.; Nilova, M.V.; Severova, E.E.; Samokhina, E.N.; Shipulin, G.A.; Logacheva, M.D. Comparative analysis of Illumina and Ion Torrent high-throughput sequencing platforms for identification of plant components in herbal teas. Food Control 2018, 93, 315–324. [Google Scholar] [CrossRef]
- Frigerio, J.; Gorini, T.; Galimberti, A.; Bruni, I.; Tommasi, N.; Mezzasalma, V.; Labra, M. DNA barcoding to trace Medicinal and Aromatic Plants from the field to the food supplement. J. Appl. Bot. Food Qual. 2019, 92, 33–38. [Google Scholar]
- Banchi, E.; Ametrano, C.G.; Greco, S.; Stanković, D.; Muggia, L.; Pallavicini, A. PLANiTS: A curated sequence reference dataset for plant ITS DNA metabarcoding. Database 2020, 2020, baz155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Utzeri, V.J.; Ribani, A.; Schiavo, G.; Bertolini, F.; Bovo, S.; Fontanesi, L. Application of next generation semiconductor based sequencing to detect the botanical composition of monofloral, polyfloral and honeydew honey. Food Control 2018, 86, 342–349. [Google Scholar] [CrossRef]
- Gostel, M.R.; Zúñiga, J.D.; Kress, W.J.; Funk, V.A.; Puente-Lelievre, C. Microfluidic Enrichment Barcoding (MEBarcoding): A new method for high throughput plant DNA barcoding. Sci. Rep. 2020, 10, 8701. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chen, F.; Jiang, L.; Li, S.; Zhang, J. Development of an efficient method to extract DNA from refined soybean oil. Food Anal. Methods 2021, 14, 196–207. [Google Scholar] [CrossRef]
- Pereira, L.; Guedes-Pinto, H.; Martins-Lopes, P. An enhanced method for Vitis vinifera L. DNA extraction from wines. Am. J. Enol. Vitic. 2011, 62, 4. [Google Scholar] [CrossRef] [Green Version]
- Bojang, K.P.; Kuna, A.; Pushpavalli, S.N.C.V.L.; Sarkar, S.; Sreedhar, M. Evaluation of DNA extraction methods for molecular traceability in cold pressed, solvent extracted and refined groundnut oils. J. Food Sci. Technol. 2021, 1–7. [Google Scholar] [CrossRef]
- Soares, S.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I. Improving DNA isolation from honey for the botanical origin identification. Food Control 2015, 48, 130–136. [Google Scholar] [CrossRef] [Green Version]
- López-Calleja, I.M.; de la Cruz, S.; Martín, R.; González, I.; García, T. Duplex real-time PCR method for the detection of sesame (Sesamum indicum) and flaxseed (Linum usitatissimum) DNA in processed food products. Food Addit. Contam. Part A 2015, 32, 1772–1785. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Gupta, V.; Misra, A.; Modi, D.; Pandey, B. Potential of molecular markers in plant biotechnology. Plant Omics 2009, 4, 141–162. [Google Scholar]
- Jagadeesan, B.; Gerner-Smidt, P.; Allard, M.W.; Leuillet, S.; Winkler, A.; Xiao, Y.; Chaffron, S.; Van Der Vossen, J.; Tang, S.; Katase, M.; et al. The use of next generation sequencing for improving food safety: Translation into practice. Food Microbiol. 2019, 79, 96–115. [Google Scholar] [CrossRef]
- Galimberti, A.; Bruno, A.; Mezzasalma, V.; De Mattia, F.; Bruni, I.; Labra, M. Emerging DNA-based technologies to characterize food ecosystems. Food Res. Int. 2015, 69, 424–433. [Google Scholar] [CrossRef]
- Besse, P.; Da Silva, D.; Grisoni, M. Plant DNA barcoding principles and limits: A case study in the Genus Vanilla. In Molecular Plant Taxonomy; Besse, P., Ed.; Springer: New York, NY, USA, 2021; Volume 2222, pp. 131–148. [Google Scholar]
| Analytical Method | Food Products | References |
|---|---|---|
| Vibrational spectroscopic techniques | Different agri-food products | Lohumi et al. [14] |
| Mass spectrometry techniques | Different agri-food products | Castro-Puyana and Herrero [15] |
| Stable isotope analysis | Cereals, wine, and vegetable oils | Zhao et al. [16] |
| Gas chromatography coupled with mass spectrometry | Wine, hazelnuts, barley, terebinth, olive oil, coffee, vegetables, and fruits | Dymerski [17] |
| HPLC | Olive oil, coffee, tea, wine, juice, fruit, nuts | Esteki et al. [18] |
| Gas chromatography | Wine, chocolate, coffee, saffron, vegetable oil, fruit | Nolvachai et al. [19] |
| Spectroscopic and spectrometric techniques | Wine, vegetable oils, coffee, wheat, nuts, rice, vegetables and fruits | Medina et al. [20] |
| ELISA | Different agri-food products | Wu et al. [21] |
| Fluorescence spectroscopy | Vegetable oils, cereals, vegetables and fruits | Ahmad et al. [22] |
| Spectroscopic techniques | Vegetable oils, coffee, wine, fruit juice | Esteki et al. [23] |
| Raman spectroscopy | Olive oil, coffee, wine, rice | Xu et al. [24] |
| NMR | Balsamic vinegar, saffron, coffee, tomato | Consonni and Cagliani [25] |
| Mass spectrometry techniques | Wine, fruit juice, olive oil, beer, coffee | Rubert et al. [26] |
| Spectroscopic and spectrometric techniques | Different agri-food products | Wadood et al. [13] |
| Technique | Agri-Food Product | Detected Species | References |
|---|---|---|---|
| SSR/capillary electrophoresis | Grapes, must, and wine | Grapevine (Vitis vinifera L.) | [36] |
| SSR/HRM and SNP/HRM | Olive oil | Olive (Olea europea L.) | [37] |
| SSR/HRM | Olive oil | Olive (Olea europea L.) | [38] |
| SSR/capillary electrophoresis and SSR/HRM | Must and wine | Grapevine (Vitis vinifera L.) | [39] |
| SNP/PCR-RFLP | Olive oil | Olive (Olea europea L.) | [40] |
| SSR/capillary electrophoresis and SNP/Sanger sequencing | Extra virgin olive oil | Olive (Olea europea L.) | [41] |
| SNP/HRM | Must and wine | Grapevine (Vitis vinifera L.) | [42] |
| TaqMan SNP Genotyping Assay | Must and wine | Grapevine (Vitis vinifera L.) | [43] |
| SNP/biosensor | Must and wine | Grapevine (Vitis vinifera L.) | [44] |
| SNP/nanofluidic array | Coffee beans | Coffee (Coffea arabica L. and Coffea canephora Pierre ex. A. Froehner). | [45] |
| Species-specific primer PCR/sequencing | Flour, pasta, bread, and cookies | Common wheat (Triticum aestivum L.) | [46] |
| Species-specific primer/digital PCR | Flour and pasta | Common wheat (Triticum aestivum L.) | [47] |
| Species-specific primer/digital PCR | Lotus seed paste | White kidney bean (Phaseolus vulgaris L.). | [48] |
| DNA barcoding/sequencing | Nutmeg mace | Nutmeg tree (Myristica fragrans Houtt) | [49] |
| DNA barcoding/capillary electrophoresis | Almond oil and almond kernels | Almond (Prunus dulcis Mill.) | [50] |
| Bar-HRM | Tea products | Tea (Camellia sinensis L.) | [51] |
| Bar-HRM | Nut species and walnut milk beverage | Walnut (Juglans regia L.), pecan (Carya illinoensis K. Koch), hickory (Carya cathayensis Sarg.), and peanut (Arachis hypogaea L.) | [52] |
| Bar-HRM | Raw seeds and ground coffee | Coffee (Coffea arabica L. and Coffea canephora Pierre ex. A. Froehner). | [53] |
| DNA barcoding/NanoTracer strategy | Saffron powder | Saffron (Crocus sativus L.) | [54] |
| DNA barcoding/sequencing | Berry fruit and fruit juice | Different taxa | [55] |
| RPA-LFD | Saffron powder | Saffron (Crocus sativus L.) | [56] |
| DNA barcoding/LAMP | Saffron powder | Saffron (Crocus sativus L.) | [57] |
| LAMP | Durum wheat products | Durum wheat variety Aureo (Triticum turgidum var. durum L.) | [58] |
| DNA barcoding/LAMP | Fruit juice | Pomegranate (Punica granatum L.), Apple (Malus domestica (Suckow) Borkh.), and grape (Vitis vinifera L.) | [59] |
| Whole metagenome sequencing | Different herbal products | Different taxa | [60] |
| Whole metagenome sequencing | Lupin seed, flour, and cookies | Lupin (Lupinus spp.) | [61] |
| Whole chloroplast genome sequencing | Dried fruit | Different species of aromatic trees (Zanthoxylum spp.) | [62] |
| Whole chloroplast genome sequencing | Berry fruit | Different berry species (Vaccinium spp.) | [63] |
| DNA metabarcoding | Honey | Different taxa | [64] |
| DNA metabarcoding | Honey | Different taxa | [65] |
| Agri-Food Matrices | Method | Reference |
|---|---|---|
| Must and wine | CTAB-based method/post-extraction purification | di Rienzo et al. [39] |
| Extra virgin olive oil | Hexane-based method | Piarulli et al. [41] |
| Nutmeg mace | SDS-based method | Swetha et al. [49] |
| Fruit juice | Filtration device | Hu and Lu [59] |
| Soybean oil | CTAB-based method | Xia et al. [122] |
| Wine | CTAB-based method | Pereira et al. [123] |
| Groundnut oil | DNA enrichment/CTAB-based method | Bojang et al. [124] |
| Honey | CTAB-based method | Soares et al. [125] |
| Sesame and flaxseed | SDS-based method/post-extraction purification | López-Calleja et al. [126] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanelli, V.; Mascio, I.; Miazzi, M.M.; Savoia, M.A.; De Giovanni, C.; Montemurro, C. Molecular Approaches to Agri-Food Traceability and Authentication: An Updated Review. Foods 2021, 10, 1644. https://doi.org/10.3390/foods10071644
Fanelli V, Mascio I, Miazzi MM, Savoia MA, De Giovanni C, Montemurro C. Molecular Approaches to Agri-Food Traceability and Authentication: An Updated Review. Foods. 2021; 10(7):1644. https://doi.org/10.3390/foods10071644
Chicago/Turabian StyleFanelli, Valentina, Isabella Mascio, Monica Marilena Miazzi, Michele Antonio Savoia, Claudio De Giovanni, and Cinzia Montemurro. 2021. "Molecular Approaches to Agri-Food Traceability and Authentication: An Updated Review" Foods 10, no. 7: 1644. https://doi.org/10.3390/foods10071644
APA StyleFanelli, V., Mascio, I., Miazzi, M. M., Savoia, M. A., De Giovanni, C., & Montemurro, C. (2021). Molecular Approaches to Agri-Food Traceability and Authentication: An Updated Review. Foods, 10(7), 1644. https://doi.org/10.3390/foods10071644









