Cytotoxic Activity against A549 Human Lung Cancer Cells and ADMET Analysis of New Pyrazole Derivatives
Abstract
1. Introduction
2. Results and Discussion
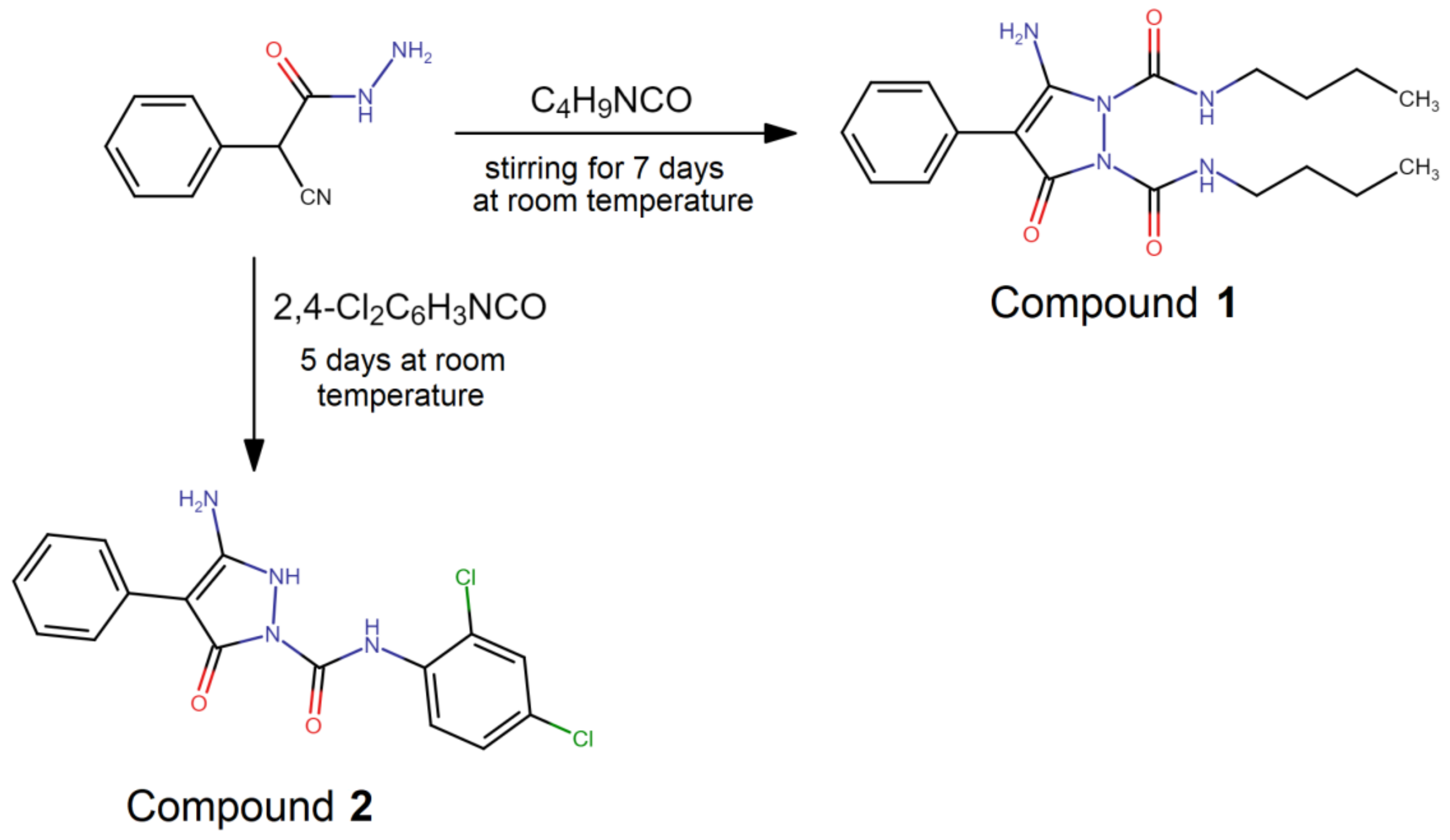
2.1. Synthesis
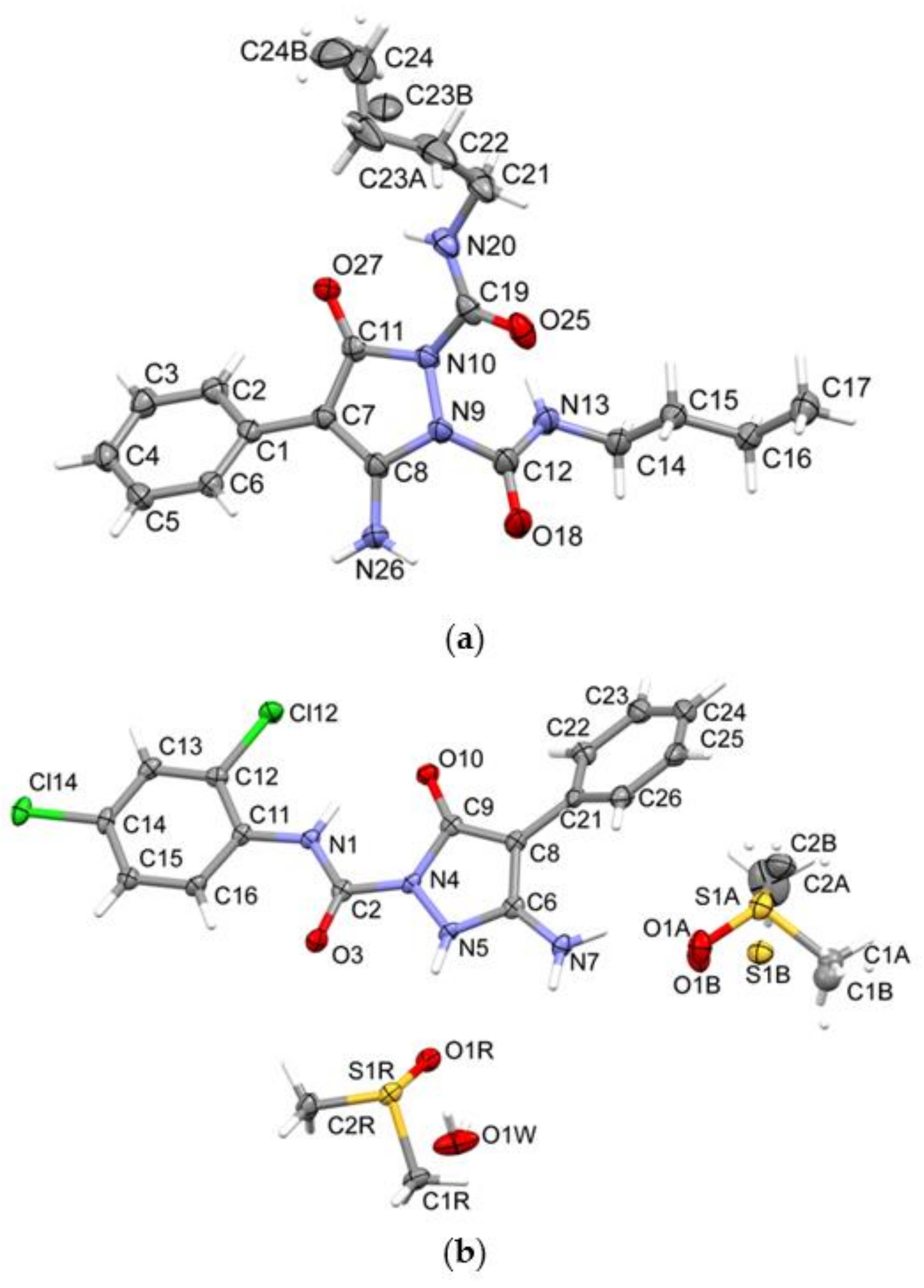
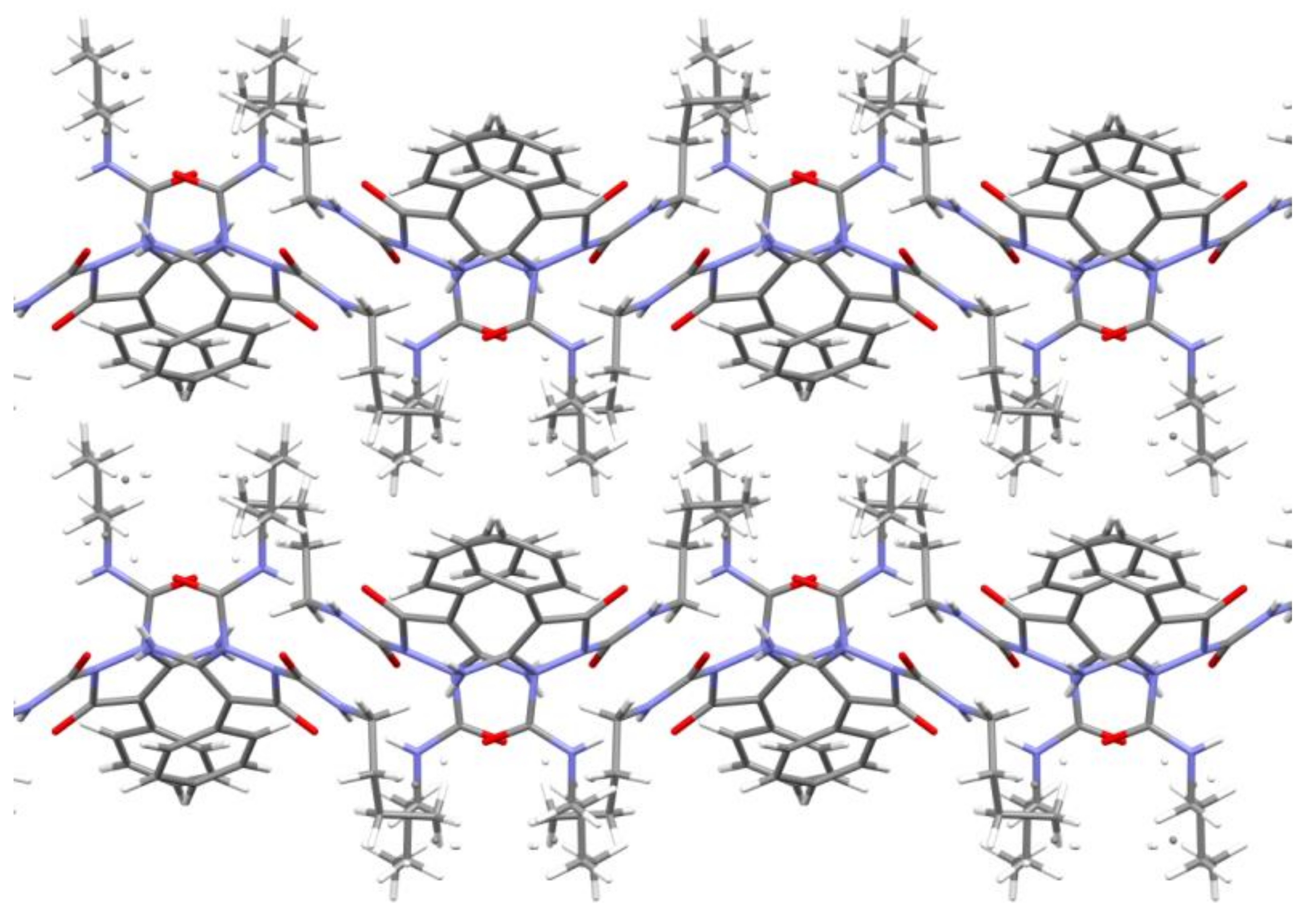
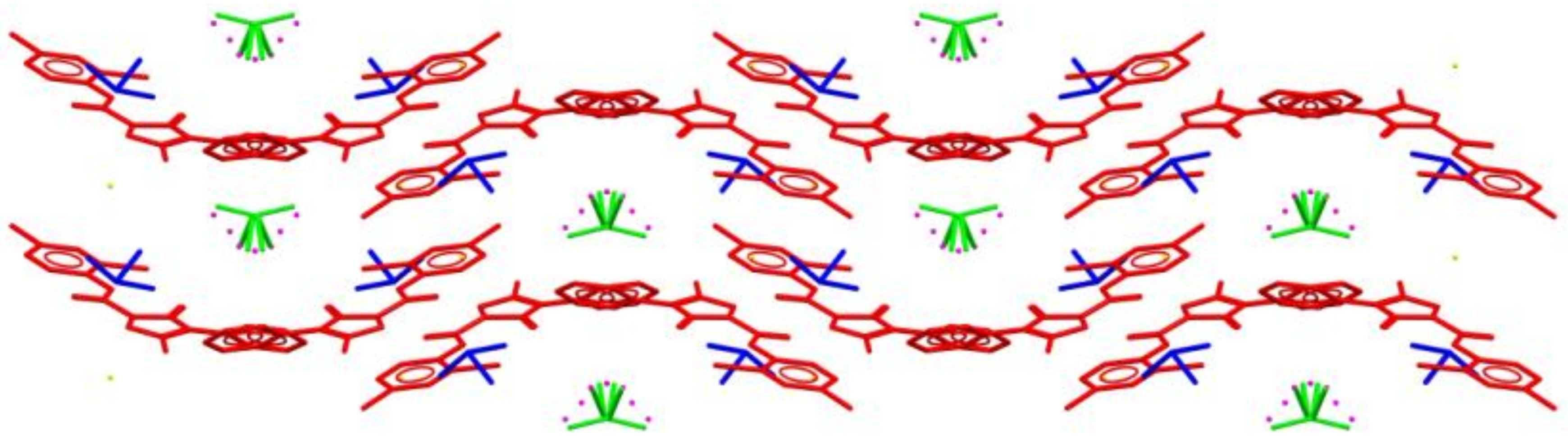
2.2. Crystal Structure Analysis
2.3. NMR Studies
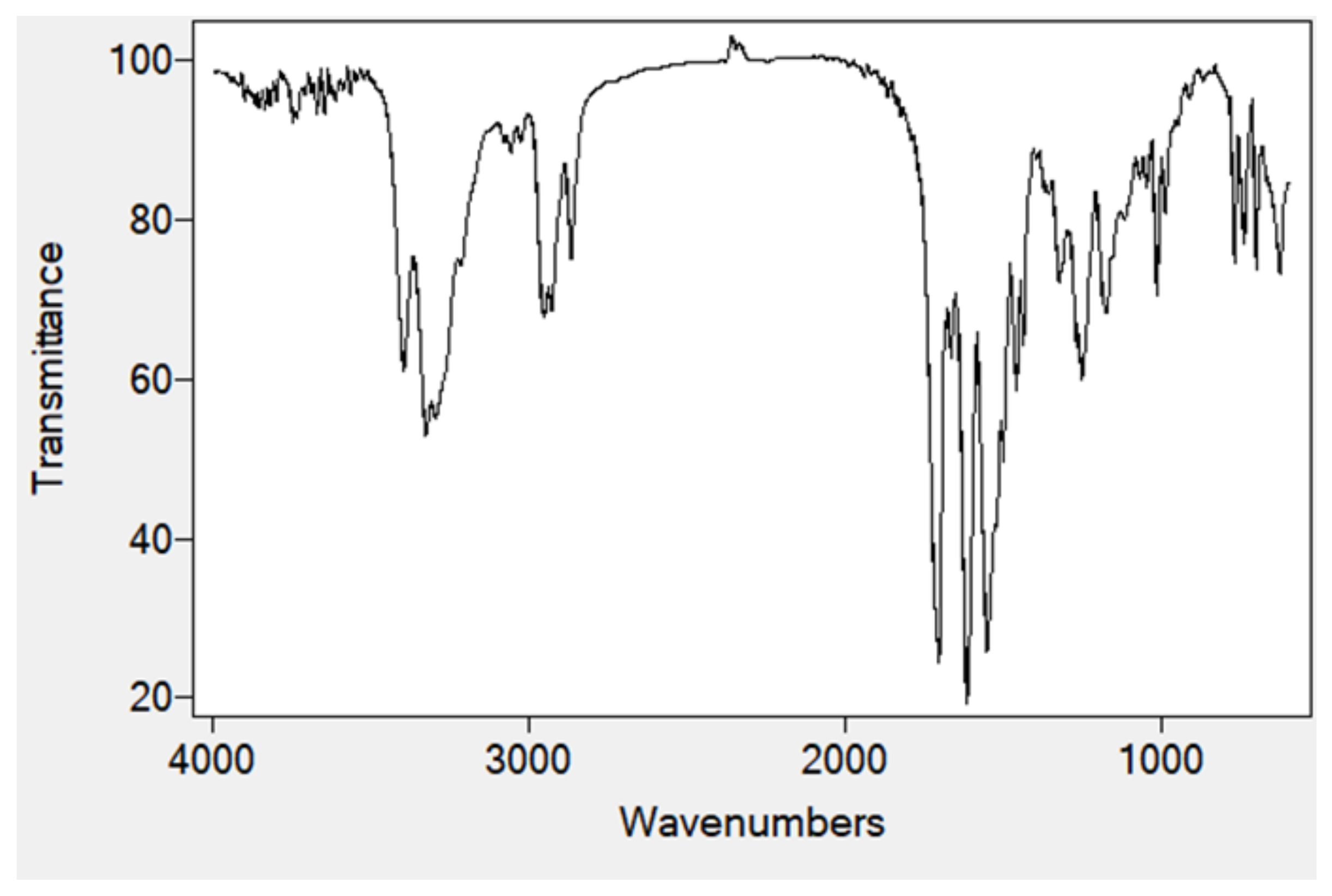
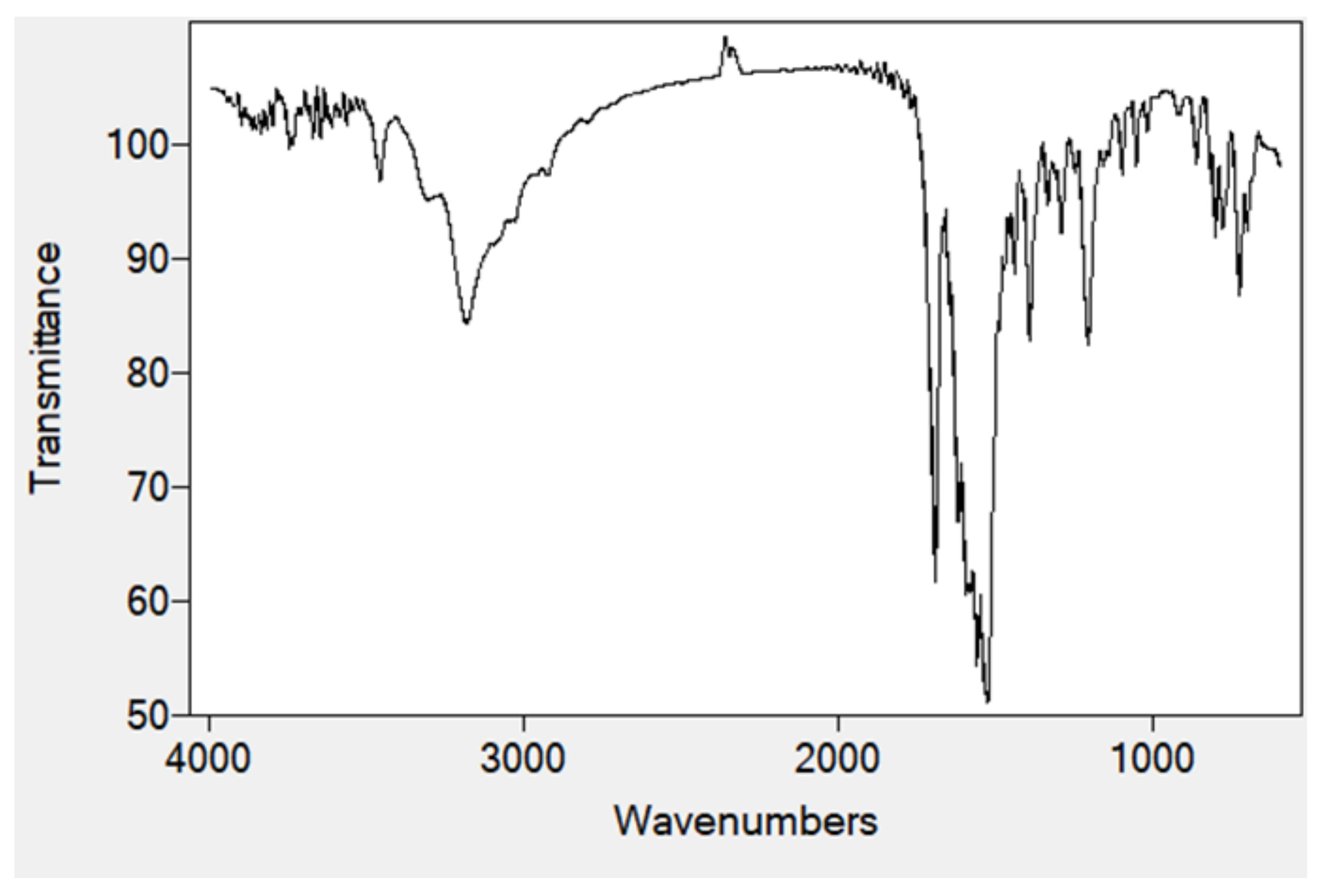
2.4. FTIR Spectra
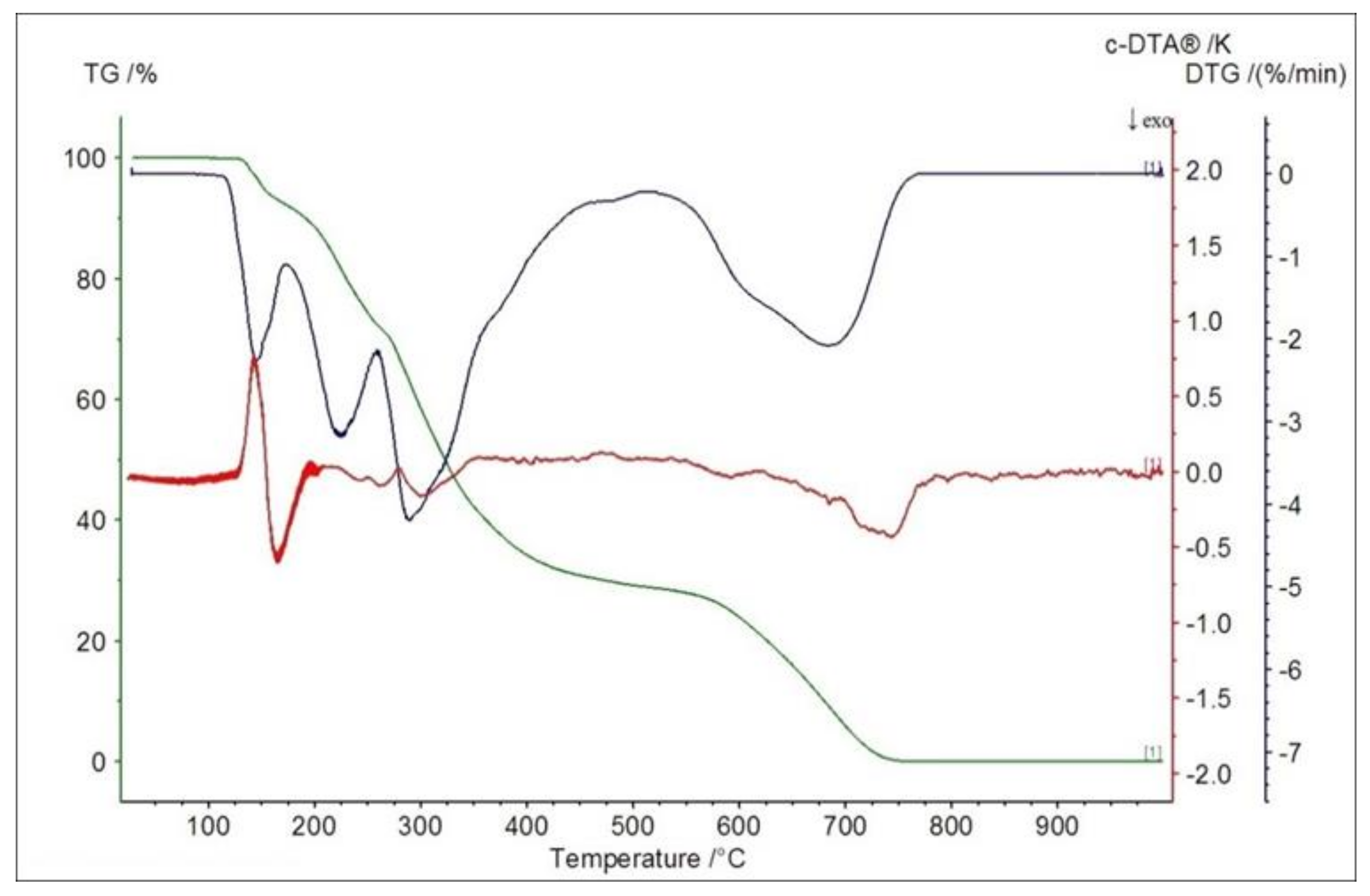
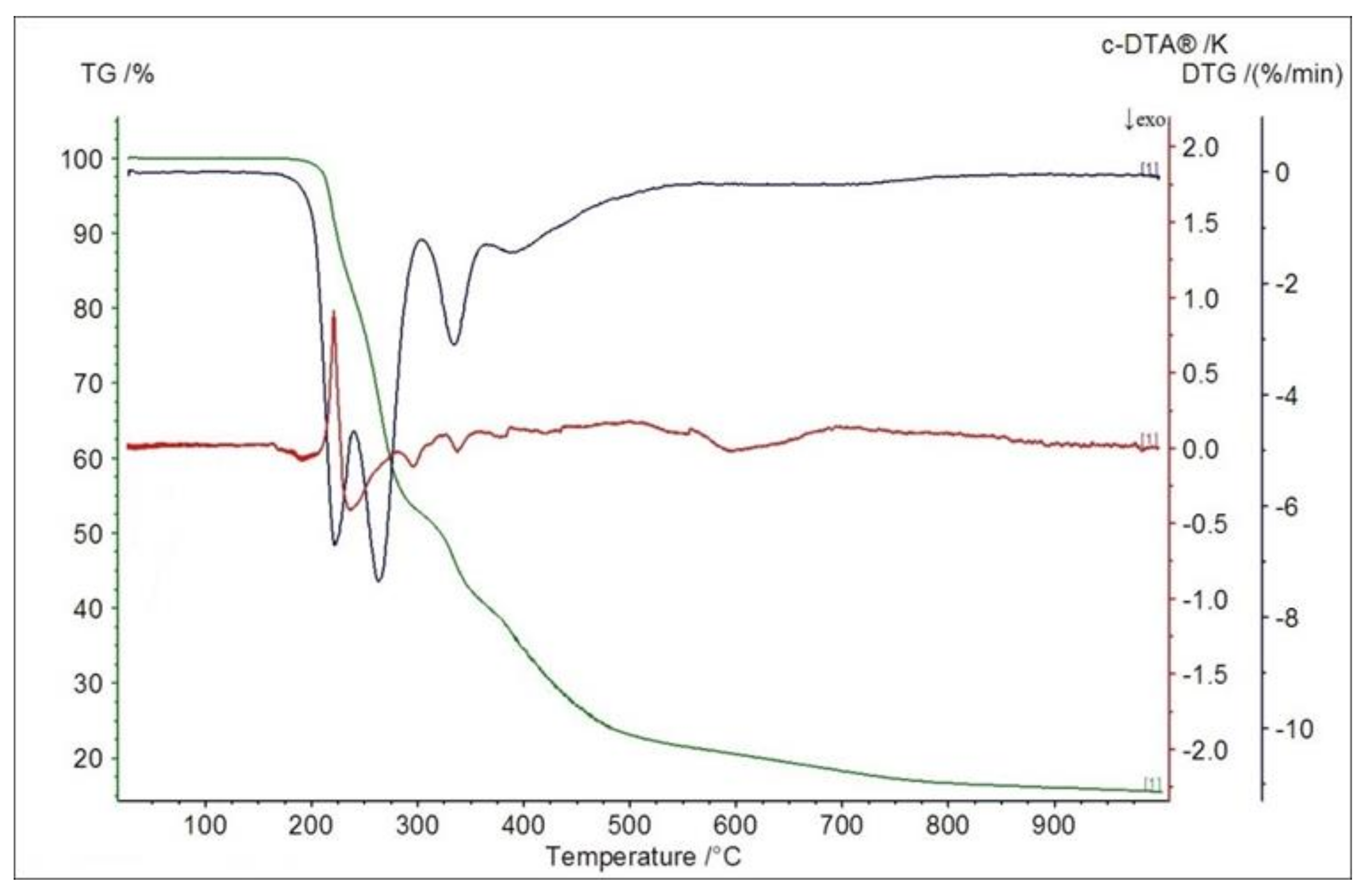
2.5. Thermogravimetric Studies in Air
2.6. Biological Assays
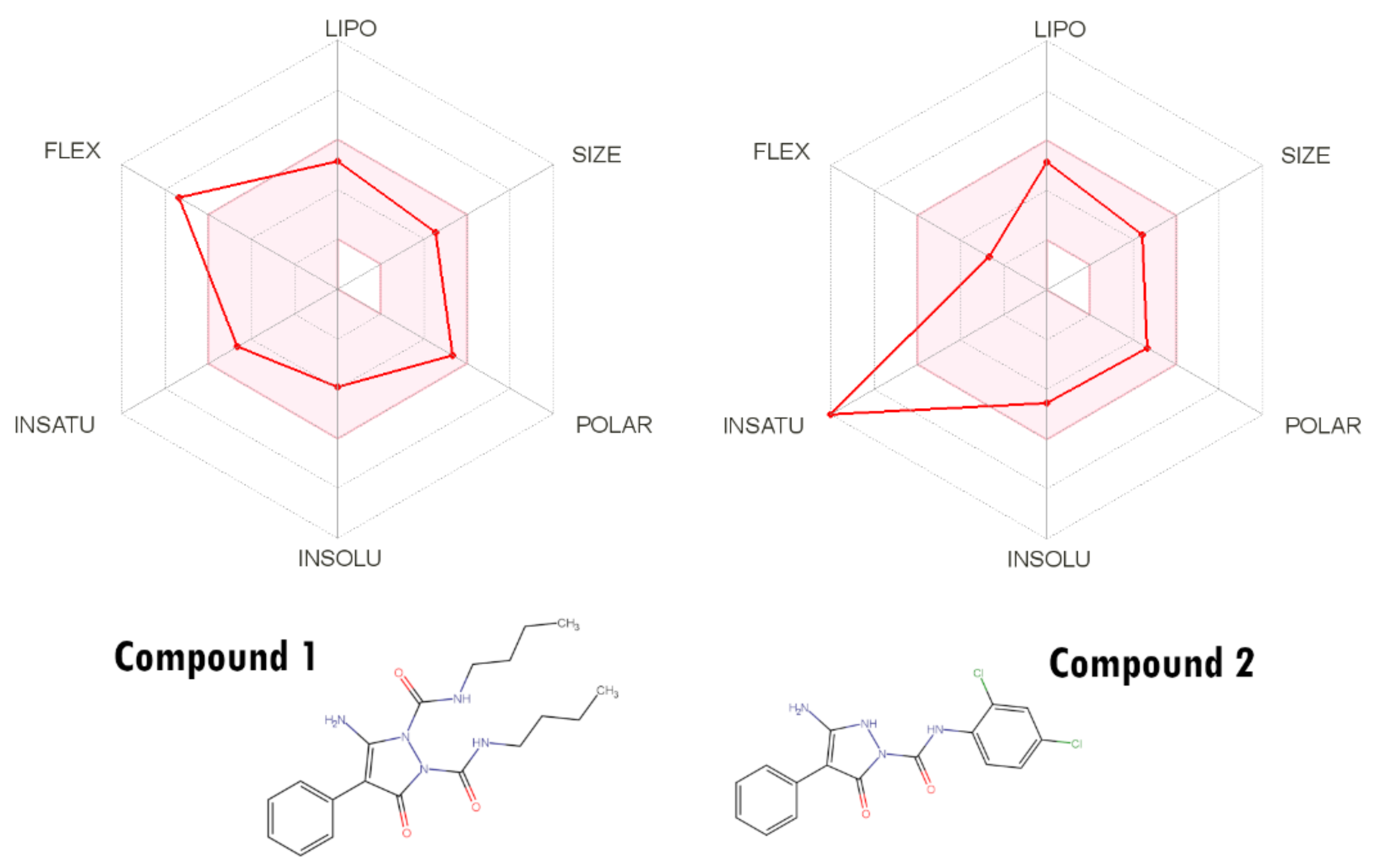
2.7. ADMET Analysis
3. Materials and Methods
3.1. Chemistry
3.2. Crystal Structure Determination
3.3. ADMET Analysis
3.4. Biological Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pitucha, M.; Rzymowska, J. Anticancer screening and structure activity relationship study of some semicarbazides and 1,2,4-Triazolin-5-ones. Lett. Drug Des. Discov. 2012, 9, 568–572. [Google Scholar] [CrossRef]
- Kumar, V.; Kaur, K.; Gupta, G.K.; Sharma, A.K. Pyrazole containing natural products: Synthetic preview and biological significance. Eur. J. Med. Chem. 2013, 69, 735–753. [Google Scholar] [CrossRef] [PubMed]
- Thawabteh, A.; Juma, S.; Bader, M.; Karaman, D.; Scrano, L.; Bufo, S.A.; Karaman, R. The biological activity of natural alkaloids against herbivores, cancerous cells and pathogens. Toxins 2019, 11, 656. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Xiao, Y.S.; Li, Z.; Xu, X.Y.; Qian, X.H. The thiazoylmethoxy modification on pyrazole oximes: Synthesis and insecticidal biological evaluation beyond acaricidal activity. Chin. Chem. Lett. 2014, 25, 1014–1016. [Google Scholar] [CrossRef]
- Saundane, A.R.; Verma, V.A.; Katkar, V.T. Synthesis and antimicrobial and antioxidant activities of some new 5-(2-Methyl-1 H -indol-3-yl)-1,3,4-oxadiazol-2-amine derivatives. J. Chem. 2013. [Google Scholar] [CrossRef]
- El-Borai, M.A.; Rizk, H.F.; Sadek, M.R.; El-Keiy, M.M. An eco-friendly synthesis of heterocyclic moieties condensed with pyrazole system under green conditions and their biological activity. Green Sustain. Chem. 2016, 6, 88–100. [Google Scholar] [CrossRef][Green Version]
- Yang, B.; Lu, Y.; Chen, C.J.; Cui, J.P.; Cai, M.S. The synthesis of 5-amino-4-arylazo-3-methyl-1H-pyrazoles and 5-aryl-3-methylpyrazole[3,4-e] [1–4] tetrazines. Dyes Pigment. 2009, 83, 144–147. [Google Scholar] [CrossRef]
- Sener, N.; Sener, I.; Yavuz, S.; Karci, F. Synthesis, absorption properties and biological evaluation of some novel disazo dyes derived from pyrazole derivatives. Asian J. Chem. 2015, 27, 3003–3012. [Google Scholar] [CrossRef]
- Abdel-Mohsen, S.A.; El-Emary, T.I. New pyrazolo [3,4-b] pyridines: Synthesis and antimicrobial Activity. Der Pharma Chem. 2018, 10, 44–51. [Google Scholar]
- El-Dean, A.M.K.; Zaki, R.M.; Geies, A.A.; Radwan, S.M.; Tolba, M.S. Synthesis and antimicrobial activity of new heterocyclic compounds containing thieno[3,2-c]coumarin and pyrazolo[4,3-c]coumarin frameworks. Russ. J. Bioorgan. Chem. 2013, 39, 553–564. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; Mcmahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Liu, X.H.; Qiao, L.; Zhai, Z.W.; Cai, P.P.; Cantrell, C.L.; Tan, C.X.; Weng, J.Q.; Han, L.; Wu, H.K. Novel 4-pyrazole carboxamide derivatives containing flexible chain motif: Design, synthesis and antifungal activity. Pest. Manag. Sci. 2019, 75, 2892–2900. [Google Scholar] [CrossRef]
- Yan, Z.; Liu, A.; Huang, M.; Liu, M.; Pei, H.; Huang, L.; Yi, H.; Liu, W.; Hu, A. Design, synthesis, DFT study and antifungal activity of the derivatives of pyrazolecarboxamide containing thiazole or oxazole ring. Eur. J. Med. Chem. 2018, 149, 170–181. [Google Scholar] [CrossRef]
- Ren, Z.L.; Liu, H.; Jiao, D.; Hu, H.T.; Wang, W.; Gong, J.X.; Wang, A.L.; Cao, H.Q.; Lv, X.H. Design, synthesis, and antifungal activity of novel cinnamon–pyrazole carboxamide derivatives. Drug Dev. Res. 2018, 79, 307–312. [Google Scholar] [CrossRef]
- Isyaku, Y.; Uzairu, A.; Uba, S. In silico studies of novel pyrazole-furan and pyrazole-pyrrole carboxamide as fungicides against Sclerotinia sclerotiorum. Beni Suef Univ. J. Basic Appl. Sci. 2020, 9. [Google Scholar] [CrossRef]
- Kaczor, A.A.; Wrobel, T.; Karczmarzyk, Z.; Wysocki, W.; Fruzinski, A.; Brodacka, M.; Matosiuk, D.; Pitucha, M. Structural studies on N-(1-naphthyl)-3-amino-5-oxo-4-phenyl-1hpyrazole-1-carboxamide with antibacterial activity. Lett. Organ. Chem. 2014, 11, 40–44. [Google Scholar] [CrossRef]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.-L. Patterns in hydrogen bonding: Functionality and graph set analysis in crystals. Angew. Chem. Int. Ed. Engl. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Bodenhausen, G.; Ruben, D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980, 69, 185–189. [Google Scholar] [CrossRef]
- Bax, A.; Summers, M.F. Proton and carbon-13 assignments from sensitivity-enhanced detection of heteronuclear multiple-bond connectivity by 2D multiple quantum NMR. J. Am. Chem. Soc. 1986, 108, 2093–2094. [Google Scholar] [CrossRef]
- Willker, W.; Leibfritz, D.; Kerssebaum, R.; Bermel, W. Gradient selection in inverse heteronuclear correlation spectroscopy. W. Magn. Reson. Chem. 1993, 31, 287–292. [Google Scholar] [CrossRef]
- Agilent Technologies UK. CrysAlisPro 1.171.39.33c; Rigaku Oxford Diffraction; Agilent Technologies UK Ltd.: Yarnton, UK, 2017. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; Mccabe, P.; Pidcock, E.; Rodriguez–Monge, L.; Taylor, R.; Van De Streek, J.; Wood, P.A. Mercury: Visualization and analysis of crystal structures. J. Appl. Cryst. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Westrip, S.P. publCIF: Software for editing, validating and formatting crystallographic information files. J. Appl. Cryst. 2010, 43, 920–925. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Yang, H.B.; Lou, C.F.; Sun, L.X.; Li, J.; Cai, Y.C.; Wang, Z.; Li, W.H.; Liu, G.X.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Plumb, J.A. Cell sensitivity assays: The MTT assay. In Cancer Cell Culture: Methods and Protocols; Langdon, S.P., Ed.; Humana Press: Totowa, NJ, USA, 2004; pp. 165–169. [Google Scholar]
- Girek, M.; Kłosiński, K.; Grobelski, B.; Pizzimenti, S.; Cucci, M.A.; Daga, M.; Barrera, G.; Pasieka, Z.; Czarnecka, K.; Szymański, P. Novel tetrahydroacridine derivatives with iodobenzoic moieties induce G0/G1 cell cycle arrest and apoptosis in A549 non-small lung cancer and HT-29 colorectal cancer cells. Mol. Cell. Biochem. 2019, 460, 123–150. [Google Scholar] [CrossRef] [PubMed]
- Mascaux, C.; Paesmans, M.; Berghmans, T.; Branle, F.; Lafitte, J.J.; Lemaitre, F.; Meert, A.P.; Vermylen, P.; Sculier, J.P. European Lung Cancer Working Party (ELCWP). A systematic review of the role of etoposide and cisplatin in the chemotherapy of small cell lung cancer with methodology assessment and meta-analysis. Lung Cancer 2000, 30, 23–36. [Google Scholar] [CrossRef]
| Compound | 1 | 2 |
|---|---|---|
| Chemical formula | C19H27N5O3 | C16H12Cl2N4O2·2(C2H6OS)·H2O |
| Mr | 373.45 | 537.47 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/c |
| Temperature (K) | 100 | 100 |
| a, b, c (Å) | 11.9591 (2), 16.2222 (2), 11.6359 (2) | 7.0863 (2), 25.7334 (8), 13.7460 (4) |
| β (°) | 117.695 (2) | 97.800 (3) |
| V (Å3) | 1998.78 (6) | 2483.45 (13) |
| Z | 4 | 4 |
| Radiation type | Cu Kα | Mo Kα |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 24,501, 4143, 3729 | 37,018, 7334, 5953 |
| Rint | 0.039 | 0.078 |
| (sin θ/λ)max (Å−1) | 0.635 | 0.736 |
| R[F2 > 2σ (F2)], wR(F2), S | 0.044, 0.119, 1.03 | 0.079, 0.203, 1.15 |
| No. of reflections | 4143 | 7334 |
| No. of parameters | 266 | 337 |
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N26—H26A⋯O25 i | 0.86 | 1.99 | 2.8055 (16) | 158 |
| N26—H26B⋯O18 | 0.86 | 2.11 | 2.7088 (17) | 127 |
| N20—H20⋯O27 | 0.86 | 2.00 | 2.6844 (15) | 136 |
| N13—H13⋯O27 ii | 0.86 | 2.00 | 2.8210 (15) | 160 |
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O10 | 0.86 | 1.97 | 2.689 (3) | 140 S(6) |
| N5—H5⋯S1R | 0.86 | 2.66 | 3.459 (3) | 155 |
| N5—H5⋯O1R | 0.86 | 2.26 | 2.799 (4) | 121 |
| N7—H7A⋯O1W i | 0.86 | 2.01 | 2.835 (4) | 160 |
| N7—H7B⋯O1Aa | 0.86 | 2.06 | 2.81 (4) | 146 |
| N7—H7B⋯O1Bb | 0.86 | 2.17 | 2.86 (3) | 137 |
| O1W—H1W1⋯O1R | 0.81 (2) | 2.02 (3) | 2.817 (4) | 166 (7) |
| O1W—H1W2⋯O3 ii | 0.81 (2) | 2.08 (3) | 2.867 (3) | 163 (6) |
| Compound | Cytotoxicity Activity EC50 [μM] |
|---|---|
| 1 | 613.22 +/−23.56 |
| 2 | 220.20+/−22.47 |
| etoposide | 451.47+/−18.27 * |
| Compound | Molecular Weight [g/mol] | LogP | pKa (Acid) | pKa (Base) | TPSA [Å2] | Molar Refractivity [m3/mol] |
|---|---|---|---|---|---|---|
| 1 | 373.45 | 2.83 | 11.70 | 3.90 | 111.15 | 106.29 |
| 2 | 363.20 | 3.17 | 9.60 | 3.48 | 92.91 | 95.59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czylkowska, A.; Szczesio, M.; Raducka, A.; Rogalewicz, B.; Kręcisz, P.; Czarnecka, K.; Szymański, P.; Pitucha, M.; Pawlak, T. Cytotoxic Activity against A549 Human Lung Cancer Cells and ADMET Analysis of New Pyrazole Derivatives. Int. J. Mol. Sci. 2021, 22, 6692. https://doi.org/10.3390/ijms22136692
Czylkowska A, Szczesio M, Raducka A, Rogalewicz B, Kręcisz P, Czarnecka K, Szymański P, Pitucha M, Pawlak T. Cytotoxic Activity against A549 Human Lung Cancer Cells and ADMET Analysis of New Pyrazole Derivatives. International Journal of Molecular Sciences. 2021; 22(13):6692. https://doi.org/10.3390/ijms22136692
Chicago/Turabian StyleCzylkowska, Agnieszka, Małgorzata Szczesio, Anita Raducka, Bartłomiej Rogalewicz, Paweł Kręcisz, Kamila Czarnecka, Paweł Szymański, Monika Pitucha, and Tomasz Pawlak. 2021. "Cytotoxic Activity against A549 Human Lung Cancer Cells and ADMET Analysis of New Pyrazole Derivatives" International Journal of Molecular Sciences 22, no. 13: 6692. https://doi.org/10.3390/ijms22136692
APA StyleCzylkowska, A., Szczesio, M., Raducka, A., Rogalewicz, B., Kręcisz, P., Czarnecka, K., Szymański, P., Pitucha, M., & Pawlak, T. (2021). Cytotoxic Activity against A549 Human Lung Cancer Cells and ADMET Analysis of New Pyrazole Derivatives. International Journal of Molecular Sciences, 22(13), 6692. https://doi.org/10.3390/ijms22136692







