Genome-Wide Identification of Soybean ABC Transporters Relate to Aluminum Toxicity
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of Soybean ABC Transporters
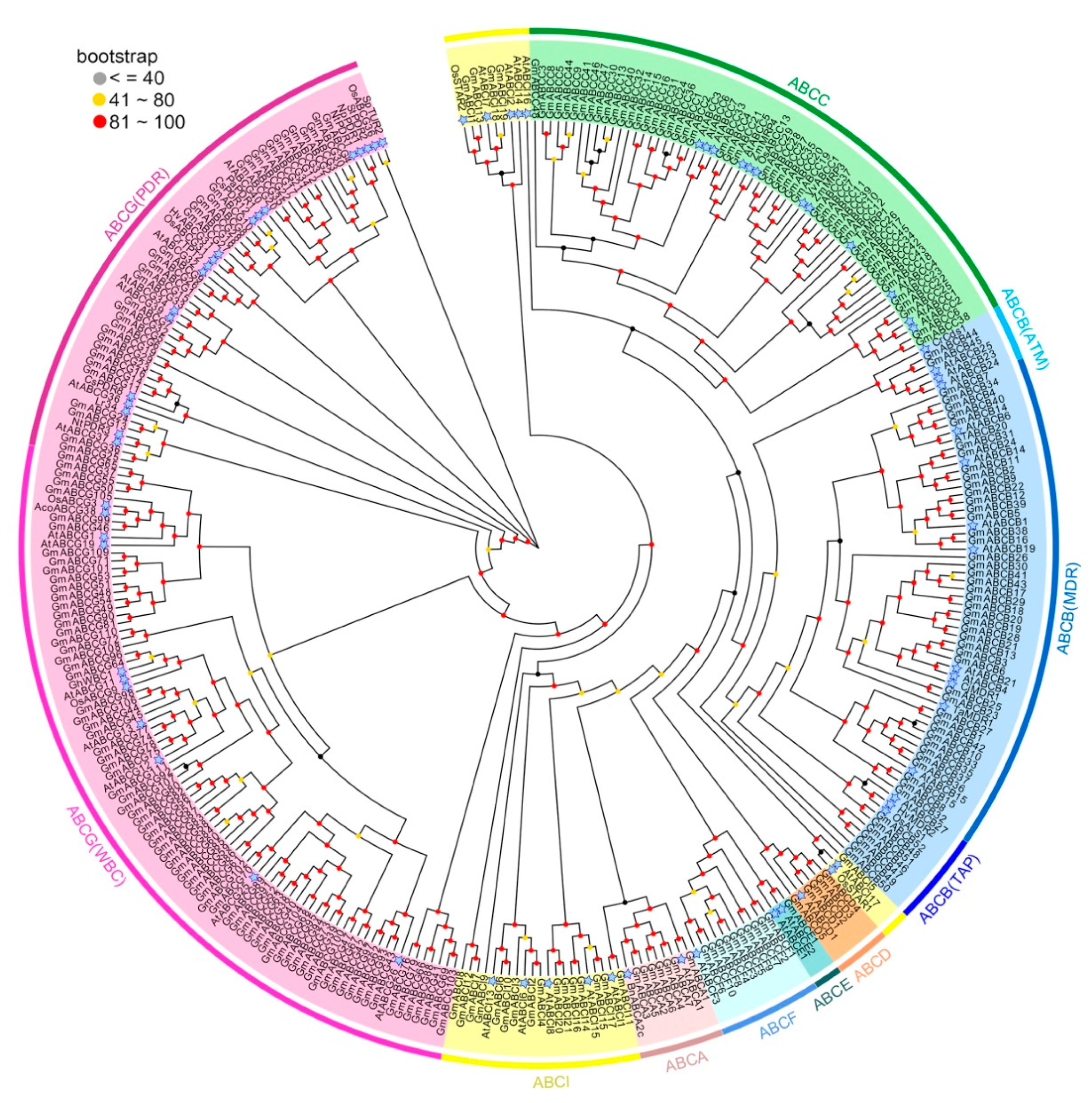
2.2. Phylogenetic Analysis of the GmABC Family
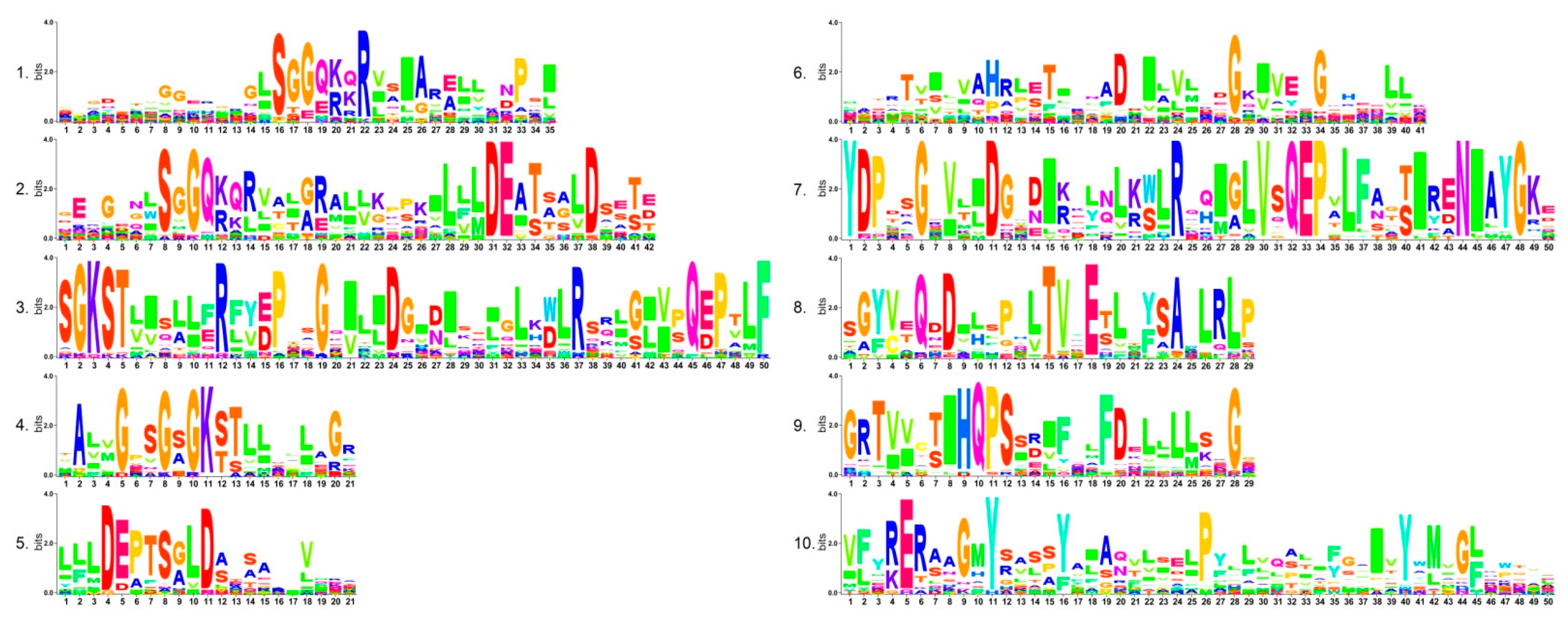
2.3. Gene Structure and Conserved Motif Analysis
2.4. Chromosome Distribution
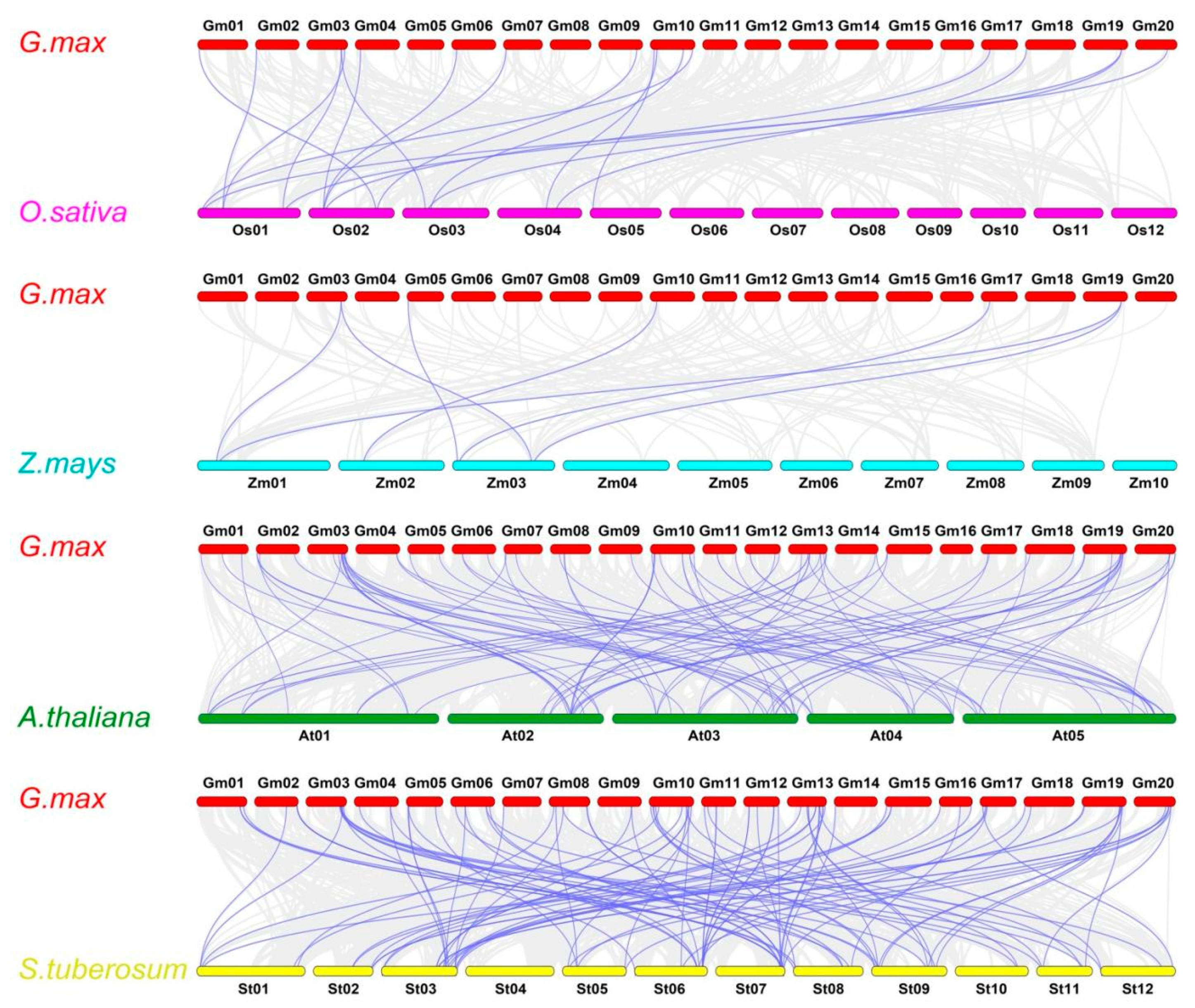
2.5. Gene Duplication and Synteny Analysis
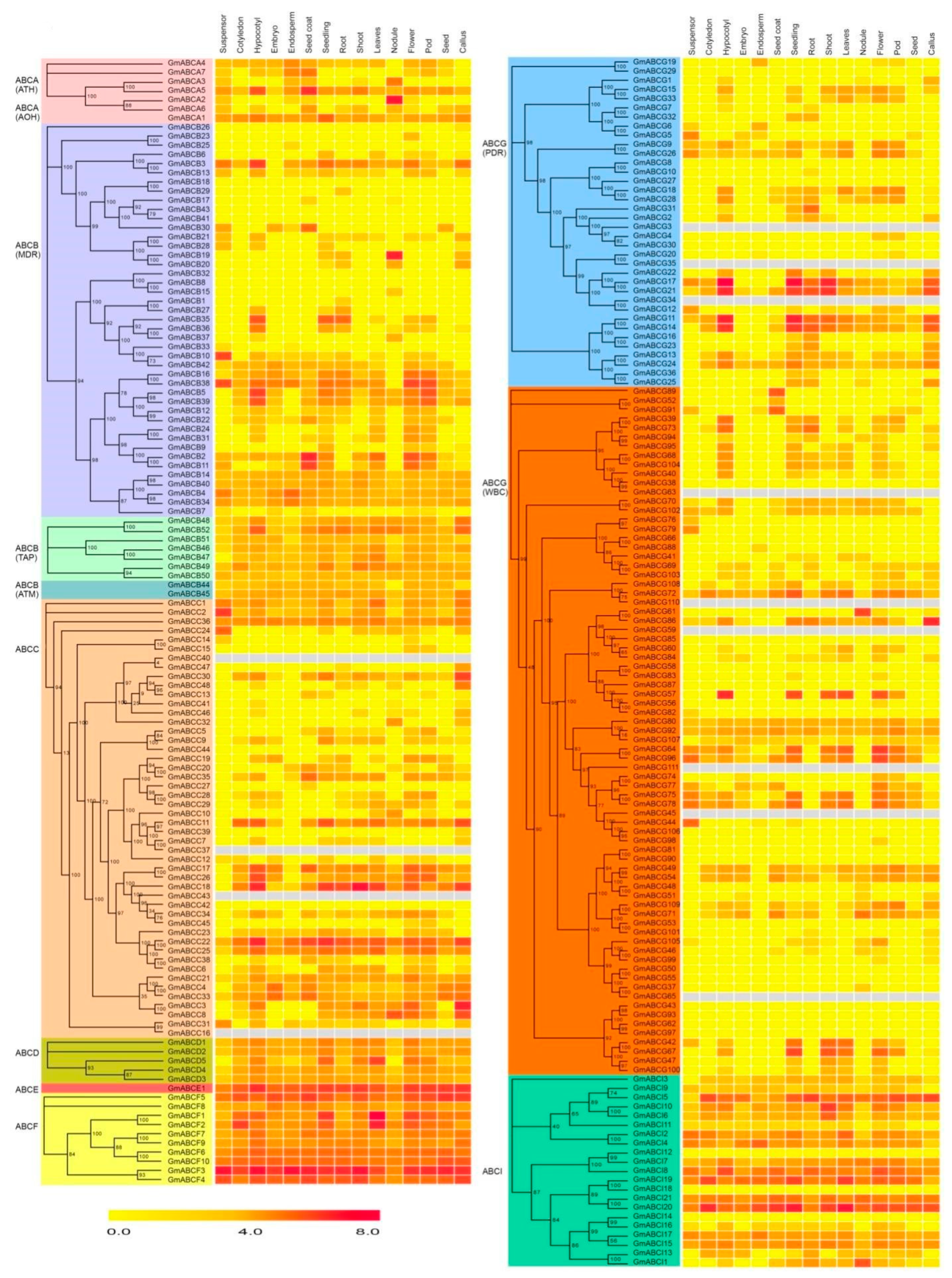
2.6. Expression Pattern of GmABC Genes in Different Tissues
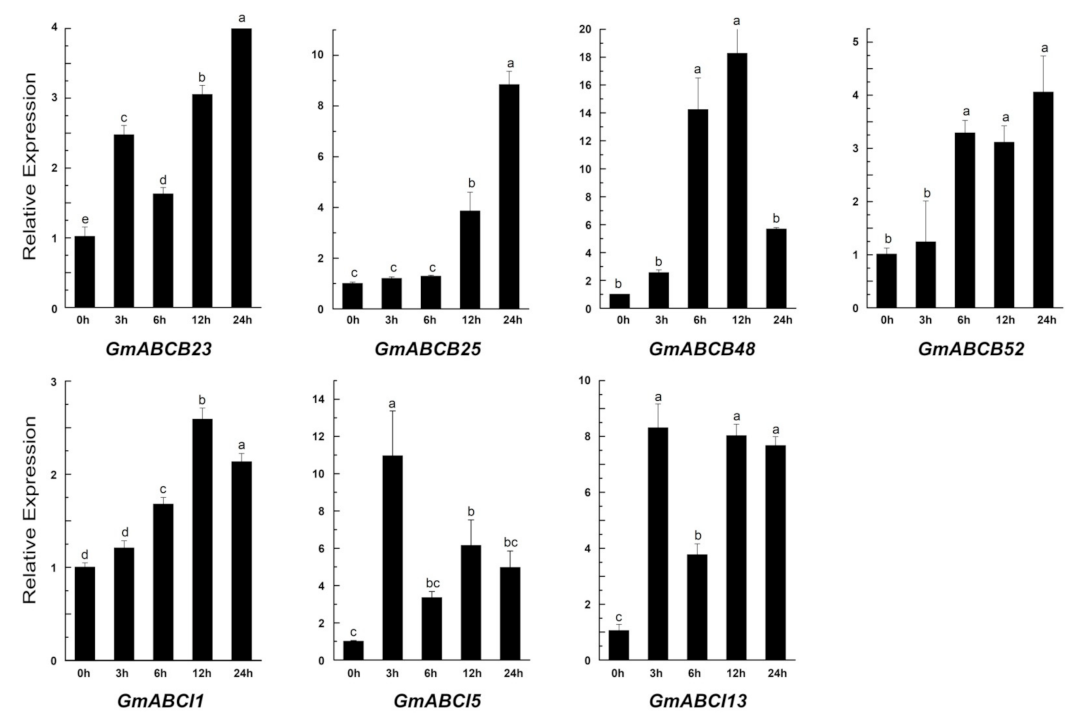
2.7. Expression of GmABC Genes in Response to Al Toxicity
3. Discussion
4. Materials and Methods
4.1. Identification of Putative ABC Proteins in Soybean
4.2. Gene Structure, Conversed Motif, and Phylogeny Analysis
4.3. Chromosomal Locations, Gene Duplication, and Gene Collinearity Analysis
4.4. Expression Pattern Analysis of the GmABC Genes
4.5. Plant Materials and QRT-PCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Theodoulou, F.L.; Kerr, L.D. ABC transporter research: Going strong 40 years on. Biochem. Soc. Trans. 2015, 43, 1033–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higgins, C.F.; Linton, K.J. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004, 11, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xu, J.; Zhang, D.B. Molecular evolution, expression and functional network prediction analysis of ABC transporter gene family in Arabidopsis thaliana. Plant Physiol. J. 2012, 48, 1151–1166. [Google Scholar]
- Nguyen, V.N.; Moon, S.; Jung, K.H. Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J. Plant Physiol. 2014, 171, 1276–1288. [Google Scholar] [CrossRef]
- Zhang, X.D.; Zhao, K.X.; Yang, Z.M. Identification of genomic ATP binding cassette (ABC) transporter genes and Cd-responsive ABCs in Brassica napus. Gene 2018, 664, 139–151. [Google Scholar] [CrossRef]
- Ofori, P.A.; Mizuno, A.; Suzuki, M.; Martinoia, E.; Reuscher, S.; Aoki, K.; Shibata, D.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854. [Google Scholar] [CrossRef]
- Schaedler, T.A.; Thornton, J.D.; Kruse, I.; Schwarzländer, M.; Meyer, A.J.; van Veen, H.W.; Balk, J. A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly. J. Biol. Chem. 2014, 289, 23264–23274. [Google Scholar] [CrossRef] [Green Version]
- Davies, T.G.E.; Theodoulou, F.L.; Hallahan, D.L.; Hallahan, D.L.; Forde, B.G. Cloning and characterization of a novel p-glycoprotein homologue from barley. Gene 1997, 19, 195–202. [Google Scholar] [CrossRef]
- Shitan, N.; Bazin, I.; Dan, K.; Obata, K.; Kigawa, K.; Ueda, K.; Sato, F.; Forestier, C.; Yazaki, K. Involvement of CjMDR1, a plant multidrugresistance-type ATP-binding cassette protein, in alkaloid transport in Coptis japonica. Proc. Natl. Acad. Sci. USA 2003, 100, 751–756. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Song, W.-Y.; Ko, D.; Eom, Y.; Hansen, T.H.; Schiller, M.; Lee, T.G.; Martinoia, E.; Lee, Y. The phytochelatin transporters ATABCC1 and ATABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012, 69, 278–288. [Google Scholar] [CrossRef]
- Bessire, M.; Borel, S.; Fabre, G.; Carraca, L.; Efremova, N.; Yephremov, A.; Cao, Y.; Jetter, R.; Jacquat, A.C.; Metraux, J.P.; et al. A member of the pleiotropic drug resistance family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 2011, 23, 1958–1970. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Komatsuda, T.; Ma, J.F.; Nawrath, C.; Pourkheirandish, M.; Tagiri, A.; Hu, Y.G.; Sameri, M.; Li, X.; Zhao, X.; et al. An ATP-binding cassette subfamily G full transporter is essential for the retention of leaf water in both wild barley and rice. Proc. Natl. Acad. Sci. USA 2011, 108, 12354–12359. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.; Lu, Y.; Zhang, X.; Yang, G.; Chao, D.; Wang, Z.; Shi, M.; Chen, J.; Chao, D.-Y.; Li, R.; et al. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 2019, 70, 5909–5918. [Google Scholar] [CrossRef]
- Sasaki, T.; Ezaki, B.; Matsumoto, H. A gene encoding mutidrug resistance (MDR)-like protein is induced by aluminium and inhibitors of calcium flux in wheat. Plant Cell Physiol. 2002, 43, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Hanikenne, M.; Motte, P.; Wu, M.C.S.; Wang, T.; Loppes, R.; Matagne, R.F. A mitochondrial half-size ABC transporter is involved in cadmium tolerance in Chlamydomonas reinhardtii. Plant Cell Environ. 2005, 28, 863–873. [Google Scholar] [CrossRef]
- Gaillard, S.; Jacquet, H.; Vavasseur, A.; Leonhardt, N.; Forestier, C. AtMRP6/AtABCC6, an ATP-binding cassette transporter gene expressed during early steps of seedling development and up-regulated by cadmium in Arabidopsis thaliana. BMC Plant Biol. 2008, 8, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunetti, P.; Zanella, L.; De Paolis, A.; Di Litta, D.; Cecchetti, V.; Falasca, G.; Barbieri, M.; Altamura, M.M.; Costantino, P.; Cardarelli, M. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot 2015, 66, 3815–3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, J.; Xu, P.; Xiang, C. Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. Plant J. 2012, 69, 782–791. [Google Scholar] [CrossRef]
- Khare, D.; Choi, H.; Huh, S.U.; Bassin, B.; Kim, J.; Martinoia, E.; Sohn, K.H.; Paek, K.-H.; Lee, Y. Arabidopsis ABCG34 contributes to defense against necrotrophic pathogens by mediating the secretion of camalexin. Proc. Natl. Acad. Sci. USA 2017, 114, E5712–E5720. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Fu, S.; Huang, J.; Li, L.; Long, Y.; Wei, Q.; Wang, Z.; Chen, Z.; Xia, J. The tonoplast-localized transporter OsABCC9 is involved in cadmium tolerance and accumulation in rice. Plant Sci. 2021, 307, 110894. [Google Scholar] [CrossRef]
- Gupta, N.; Gaurav, S.S.; Kumar, A. Molecular basis of aluminium toxicity in plants: A Review. Am. J. Plant Sci. 2013, 4, 21–37. [Google Scholar] [CrossRef] [Green Version]
- Nunes-Nesi, A.; Brito, D.S.; Inostroza-Blancheteau, C.; Fernie, A.R.; Araújo, W.L. The complex role of mitochondrial metabolism in plant aluminum resistance. Trends Plant Sci. 2014, 19, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 2007, 264, 225–252. [Google Scholar] [PubMed]
- Yokosho, K.; Yamaji, N.; Mitani-Ueno, N.; Shen, R.F.; Ma, J.F. An Aluminum-Inducible IREG gene is required for internal detoxification of aluminum in buckwheat. Plant Cell Physiol. 2016, 57, 1169–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Fujii-Kashino, M.; Ma, J.F. Functional characterization of two half-size ABC transporter genes in aluminium-accumulating buckwheat. New Phytol. 2017, 215, 1080–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Ji, F.; Zhang, Y.; Hou, J.; Liu, W.; Huang, J.; Liang, W. Interactions between hydrogen sulphide and nitric oxide regulate two soybean citrate transporters during the alleviation of aluminium toxicity. Plant Cell Environ. 2019, 42, 2340–2356. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.R.; Tyerman, S.D.; Sasaki, T.; Furuichi, T.; Yamamoto, Y.; Zhang, W.H.; Delhaize, E. The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J. Exp. Bot. 2011, 62, 9–20. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Piñeros, M.A.; Li, X.; Yang, H.; Liu, Y.; Murphy, A.S.; Kochian, L.V.; Liu, D. An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol. Plant 2017, 10, 244–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wang, Z.; Zheng, Z.; Dong, J.; Song, L.; Sui, L.; Nussaume, L.; Desnos, T.; Liu, D. Genetic dissection of Fe-dependent signaling in root developmental responses to phosphate deficiency. Plant Physiol. 2019, 179, 300–316. [Google Scholar] [CrossRef] [Green Version]
- Tokizawa, M.; Kobayashi, Y.; Saito, T.; Kobayashi, M.; Iuchi, S.; Nomoto, M.; Tada, Y.; Yamamoto, Y.Y.; Koyama, H. Sensitive to proton rhizotoxicity1, calmodulin binding transcription activator2, and other transcription factors are involved in aluminum-activated malate transporter1 expression. Plant Physiol. 2015, 167, 991–1003. [Google Scholar] [CrossRef] [Green Version]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447–1458. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef] [Green Version]
- Che, J.; Yamaji, N.; Yokosho, K.; Shen, R.F.; Ma, J.F. Two genes encoding a bacterial-type ATP-binding cassette transporter are implicated in aluminum tolerance in buckwheat. Plant Cell Physiol. 2018, 59, 2502–2511. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.M.; Lou, H.Q.; Jin, J.F.; Chen, W.W.; Wan, J.X.; Fan, W.; Yang, J.L. A half-type ABC transporter FeSTAR1 regulates Al resistance possibly via UDP-glucose-based hemicelluloses metabolism and Al binding. Plant Soil 2018, 432, 303–314. [Google Scholar] [CrossRef]
- Delhaize, E.; Ma, J.F.; Ryan, P.R. Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci. 2012, 17, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Chen, Z.; Ma, J.F. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 2012, 69, 857–867. [Google Scholar] [CrossRef]
- Xu, J.M.; Wang, Z.Q.; Jin, J.F.; Chen, W.W.; Fan, W.; Zheng, S.J.; Yang, J.L. Festar2 interacted by festar1 alters its subcellular location and regulates al tolerance in buckwheat. Plant Soil 2019, 436, 489–501. [Google Scholar] [CrossRef]
- Artigas Ramírez, M.D.; Silva, J.D.; Ohkama-Ohtsu, N.; Yokoyama, T. In vitro rhizobia response and symbiosis process under aluminum stress. Can. J. Microbiol. 2018, 64, 511–526. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cjeng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Sun, Q.; Zhang, G.; Wu, T.; Zhang, X.; Xu, X.; Han, Z.; Wang, Y. Genome-wide identification and characterization of ABC transporters in nine rosaceae species identifying MdABCG28 as a possible cytokinin transporter linked to dwarfing. Int. J. Mol. Sci. 2019, 20, 5783. [Google Scholar] [CrossRef] [Green Version]
- Garcia, O.; Bouige, P.; Forestier, C.; Dassa, E. Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J. Mol. Biol. 2004, 343, 249–265. [Google Scholar] [CrossRef]
- Pang, K.; Li, Y.; Liu, M.; Meng, Z.; Yu, Y. Inventory and general analysis of the ATP-binding cassette (ABC) gene superfamily in maize (Zea mays L.). Gene 2013, 526, 411–428. [Google Scholar] [CrossRef]
- Chen, W.W.; Xu, J.M.; Jin, J.F.; Lou, H.Q.; Fan, W.; Yang, J.L. Genome-wide transcriptome analysis reveals conserved and distinct molecular mechanisms of Al resistance in buckwheat (Fagopyrum esculentum Moench) leaves. Int. J. Mol. Sci. 2017, 18, 1859. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Wickett, N.J.; Saravanaraj, A.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Sotis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Ding, X.; Lian, T.; Wang, M.; Tong, Y.; Liang, D.; An, Q.; Sun, S.; Jackson, S.A.; Liu, B.; et al. The effects of gene duplication modes on the evolution of regulatory divergence in wild and cultivated soybean. Front. Genet. 2020, 11, 601003. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Duan, W.; Lyu, S.; Li, Y.; Hou, X. Genome-wide identification, evolution, and expression analysis of the ATP-binding cassette transporter gene family in Brassica rapa. Front. Plant Sci. 2017, 8, 349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, N.; You, F.M.; Datla, R.; Ravichandran, S.; Jia, B.; Cloutier, S. Genome-wide identification of ATP binding cassette (ABC) transporter and heavy metal associated (HMA) gene families in flax (Linum usitatissimum L.). BMC Genom. 2020, 21, 722. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Jin, J.; Sarojam, R.; Ramachandran, S. A comprehensive survey on the terpene synthase gene family provides new insight into its evolutionary patterns. Genome Biol. Evol. 2019, 11, 2078–2098. [Google Scholar] [CrossRef] [Green Version]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Schwager, E.E.; Sharma, P.P.; Clarke, T.; Leite, D.J.; Wierschin, T.; Pechmann, M.; Akiyama-Oda, Y.; Esposito, L.; Bechsgaard, J.; Bilde, T.; et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Zhikai, L.; Schnable, J.C. Functional divergence between subgenomes and gene pairs after whole genome duplications. Mol. Plant 2018, 11, 388–397. [Google Scholar]
- Clark, J.W.; Donoghue, P.C. Whole-genome duplication and plant macroevolution. Trends Plant Sci. 2018, 23, 933–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segraves, K.A. The effects of genome duplications in a community context. New Phytol. 2017, 215, 57–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, N.; Fatima, F.; Haider, M.S.; Shazadee, H.; Liu, Z.; Zheng, T.; Fang, J. Genome-wide identification and expression profiling of the polygalacturonase (PG) and pectin methylesterase (PME) genes in grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2019, 20, 3180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shazadee, H.; Khan, N.; Wang, J.; Wang, C.; Zeng, J.; Huang, Z.; Wang, X. Identification and expression profiling of protein phosphatases (PP2C) gene family in Gossypium hirsutum L. Int. J. Mol. Sci. 2019, 20, 1395. [Google Scholar] [CrossRef] [Green Version]
- Die, J.V.; Gil, J.; Millán, T. Genome-wide identification of the auxin response factor gene family in Cicer arietinum. BMC Genom. 2018, 19, 301. [Google Scholar] [CrossRef] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Goodman, C.D.; Casati, P.; Walbot, V. A Multidrug resistance–Associated protein involved in anthocyanin transport in Zea mays. Plant Cell 2004, 16, 1812–1826. [Google Scholar] [CrossRef] [Green Version]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC transporters. Arab. Book 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mentewab, A.; Stewart, C.N., Jr. Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat. Biotechnol. 2005, 23, 1177–1180. [Google Scholar] [CrossRef] [PubMed]
- Borghi, L.; Kang, J.; Ko, D.; Lee, Y.; Martinoia, E. The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 2015, 43, 924–930. [Google Scholar] [CrossRef]
- Geisler, M.; Aryal, B.; Di Donato, M.; Hao, P. A Critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 2017, 58, 1601–1614. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, Y.; Zhao, L.; Hou, Z.; Yan, M.; Hu, B.; Liu, Y.; Azam, S.M.; Zhang, Z.; Rahman, Z.U.; et al. Genome-wide identification and expression profiling of ATP-binding cassette (ABC) transporter gene family in pineapple (Ananas comosus) reveal the role of AcABCG38 in pollen development. Front. Plant Sci. 2017, 8, 2150. [Google Scholar] [CrossRef]
- Xu, J.M.; Fan, W.; Jin, J.F.; Lou, H.Q.; Chen, W.W.; Yang, J.L.; Zheng, S.J. Transcriptome analysis of Al-induced genes in buckwheat (Fagopyrum esculentum Moench) root apex: New insight into Al toxicity and resistance mechanisms in an Al ac-cumulating species. Front Plant Sci. 2017, 8, 1141. [Google Scholar] [CrossRef] [Green Version]
- Agrahari, R.K.; Kobayashi, Y.; Borgohain, P.; Panda, S.K.; Koyama, H. Aluminum-specific upregulation of GmALS3 in the shoots of soybeans: A potential biomarker for managing soybean production in acidic soil regions. Agronomy 2020, 10, 1228. [Google Scholar] [CrossRef]
- Bernard, D.G.; Cheng, Y.; Zhao, Y.; Balk, J. An allelic mutant series of ATM3 reveals its key role in the biogenesis of cytosolic iron-sulfur proteins in Arabidopsis. Plant Physiol. 2009, 151, 590–602. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elisabeth, G.; Alexandre, G.; Christine, H.; Ivan, I.; Appel, R.D.; Amos, B. Expasy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for in-teractive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995, 3, 21–29. [Google Scholar]
- De Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Jiang, H.Y.; Zhou, L.Y.; Deng, L.; Lin, Y.X.; Peng, X.J.; Yan, H.; Cheng, B. Molecular evolution of the HD-ZIP I gene family in legume genomes. Gene 2014, 533, 218–228. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize class III peroxidase gene family reveals a con-served subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Machado, F.B.; Moharana, K.C.; Almeida-Silva, F.; Gazara, R.; Pedrosa-Silva, F.; Coelho, F.S.; Grativol, C.; Venancio, T.M. Systematic analysis of 1298 RNA-Seq samples and construction of a comprehensive soybean (Glycine max) expression atlas. Plant J. 2020, 103, 1894–1909. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Li, X.; Chen, X.; Guo, Y.; Liang, W.; Wang, H. Genome-Wide Identification of Soybean ABC Transporters Relate to Aluminum Toxicity. Int. J. Mol. Sci. 2021, 22, 6556. https://doi.org/10.3390/ijms22126556
Huang J, Li X, Chen X, Guo Y, Liang W, Wang H. Genome-Wide Identification of Soybean ABC Transporters Relate to Aluminum Toxicity. International Journal of Molecular Sciences. 2021; 22(12):6556. https://doi.org/10.3390/ijms22126556
Chicago/Turabian StyleHuang, Junjun, Xiaoyu Li, Xin Chen, Yaru Guo, Weihong Liang, and Huahua Wang. 2021. "Genome-Wide Identification of Soybean ABC Transporters Relate to Aluminum Toxicity" International Journal of Molecular Sciences 22, no. 12: 6556. https://doi.org/10.3390/ijms22126556





