Cancer Stem Cells as a Source of Drug Resistance in Bone Sarcomas
Abstract
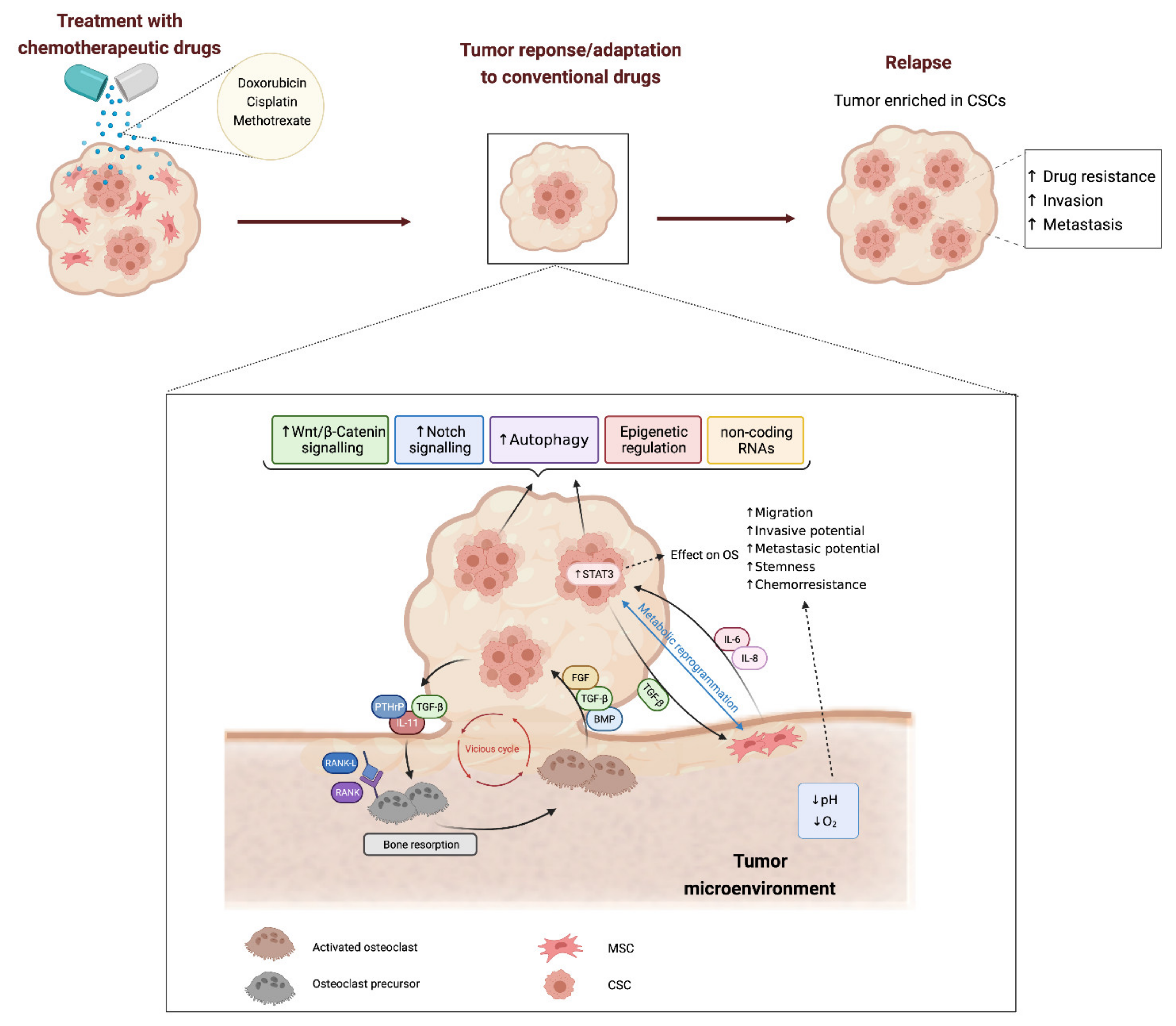
:1. Introduction: Cell Heterogeneity and Cancer Stem Cells in Bone Sarcomas
2. Mechanisms Involved in Cancer Stem Cell-Mediated Drug-Resistance
2.1. Stemness-Related Signalling Pathways
2.2. Regulation of Metabolism
2.3. Autophagy
2.4. Epigenetic Regulation of Stemness in Bone Sarcoma Stem Cells
2.5. Non-Coding RNAs
3. Influence of Tumor Microenvironment in CSCs and Drug Resistance in Osteosarcoma
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heymann, D. (Ed.) Bone Cancer, 3rd ed.; Academic Press: London, UK, 2021. [Google Scholar]
- The WHO Classification of Tumours Editorial Board. WHO Classification of Tumours: Soft Tissue and Bone Tumours, 5th ed.; IARC Press: Lyon, France, 2020. [Google Scholar]
- Kovac, M.; Blattmann, C.; Ribi, S.; Smida, J.; Mueller, N.S.; Engert, F.; Castro-Giner, F.; Weischenfeldt, J.; Kovacova, M.; Krieg, A.; et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat. Commun. 2015, 6, 8940. [Google Scholar] [CrossRef]
- Sayles, L.C.; Breese, M.R.; Koehne, A.L.; Leung, S.G.; Lee, A.G.; Liu, H.-Y.; Spillinger, A.; Shah, A.T.; Tanasa, B.; Straessler, K.; et al. Genome-Informed Targeted Therapy for Osteosarcoma. Cancer Discov. 2019, 9, 46–63. [Google Scholar] [CrossRef] [Green Version]
- Smida, J.; Xu, H.; Zhang, Y.; Baumhoer, D.; Ribi, S.; Kovac, M.; Von Luettichau, I.; Bielack, S.; O’Leary, V.B.; Leib-Mösch, C.; et al. Genome-wide analysis of somatic copy number alterations and chromosomal breakages in osteosarcoma. Int. J. Cancer 2017, 141, 816–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speetjens, F.M.; de Jong, Y.; Gelderblom, H.; Bovée, J.V. Molecular oncogenesis of chondrosarcoma: Impact for targeted treatment. Curr. Opin. Oncol. 2016, 28, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Tarpey, P.S.; Behjati, S.; Cooke, S.L.; Van Loo, P.; Wedge, D.; Pillay, N.; Marshall, J.; O’Meara, S.; Davies, H.; Nik-Zainal, S.; et al. Frequent mutation of the major cartilage collagen gene COL2A1 in chondrosarcoma. Nat. Genet. 2013, 45, 923–926. [Google Scholar] [CrossRef]
- Grünewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; De Álava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Prim. 2018, 4, 5. [Google Scholar] [CrossRef]
- Casali, P.G.; Bielack, S.; Abecassis, N.; Aro, H.; Bauer, S.; Biagini, R.; Bonvalot, S.; Boukovinas, I.; Bovee, J.V.M.G.; Brennan, B.; et al. Bone sarcomas: ESMO-PaedCan-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv79–iv95. [Google Scholar] [CrossRef]
- Grünewald, T.G.; Alonso, M.; Avnet, S.; Banito, A.; Burdach, S.; Cidre-Aranaz, F.; Di Pompo, G.; Distel, M.; Dorado-Garcia, H.; Garcia-Castro, J.; et al. Sarcoma treatment in the era of molecular medicine. EMBO Mol. Med. 2020, 12, e11131. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.D.; Lizardo, M.M.; Reed, D.R.; Hingorani, P.; Glover, J.; Allen-Rhoades, W.; Fan, T.; Khanna, C.; Sweet-Cordero, E.A.; Cash, T.; et al. Provocative questions in osteosarcoma basic and translational biology: A report from the Children’s Oncology Group. Cancer 2019, 125, 3514–3525. [Google Scholar] [CrossRef] [PubMed]
- Valery, P.C.; Laversanne, M.; Bray, F. Bone cancer incidence by morphological subtype: A global assessment. Cancer Causes Control. 2015, 26, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.B. Age-specific bone tumour incidence rates are governed by stem cell exhaustion influencing the supply and demand of progenitor cells. Mech. Ageing Dev. 2014, 139, 31–40. [Google Scholar] [CrossRef]
- Diomede, F.; Marconi, G.D.; Fonticoli, L.; Pizzicanella, J.; Merciaro, I.; Bramanti, P.; Mazzon, E.; Trubiani, O. Functional Relationship between Osteogenesis and Angiogenesis in Tissue Regeneration. Int. J. Mol. Sci. 2020, 21, 3242. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage Potential of Adult Human Mesenchymal Stem Cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [Green Version]
- Abarrategi, A.; Tornin, J.; Cruzado, L.M.; Hamilton, A.; Martinez-Campos, E.; Rodrigo, J.P.; González, M.V.; Baldini, N.; Garcia-Castro, J.; Rodriguez, R. Osteosarcoma: Cells-of-Origin, Cancer Stem Cells, and Targeted Therapies. Stem Cells Int. 2016, 2016, 3631764. [Google Scholar] [CrossRef] [Green Version]
- Boehme, K.A.; Schleicher, S.B.; Traub, F.; Rolauffs, B. Chondrosarcoma: A Rare Misfortune in Aging Human Cartilage? The Role of Stem and Progenitor Cells in Proliferation, Malignant Degeneration and Therapeutic Resistance. Int. J. Mol. Sci. 2018, 19, 311. [Google Scholar] [CrossRef] [Green Version]
- Jacques, C.; Renema, N.; Ory, B.; Walkley, C.R.; Grigoriadis, A.E.; Heymann, D. Murine Models of Bone Sarcomas. Adv. Struct. Saf. Stud. 2019, 1914, 331–342. [Google Scholar] [CrossRef]
- Lu, C.; Venneti, S.; Akalin, A.; Fang, F.; Ward, P.; DeMatteo, R.G.; Intlekofer, A.M.; Chen, C.; Ye, J.; Hameed, M.; et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013, 27, 1986–1998. [Google Scholar] [CrossRef] [Green Version]
- Rey, V.; Menendez, S.T.; Estupiñan, O.; Rodriguez, A.; Santos, L.; Tornin, J.; Martinez-Cruzado, L.; Castillo, D.; Ordoñez, G.R.; Costilla, S.; et al. New Chondrosarcoma Cell Lines with Preserved Stem Cell Properties to Study the Genomic Drift During In Vitro/In Vivo Growth. J. Clin. Med. 2019, 8, 455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, R.; García-Castro, J.; Trigueros, C.; Arranz, M.G.; Menéndez, P. Multipotent Mesenchymal Stromal Cells: Clinical Applications and Cancer Modeling. Adv. Exp. Med. Biol. 2012, 741, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Rosu-Myles, M.; Aráuzo-Bravo, M.; Horrillo, A.; Pan, Q.; Gonzalez-Rey, E.; Delgado, M.; Menendez, P. Human Bone Marrow Stromal Cells Lose Immunosuppressive and Anti-inflammatory Properties upon Oncogenic Transformation. Stem Cell Rep. 2014, 3, 606–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, R.; Rubio, R.; Menendez, P. Modeling sarcomagenesis using multipotent mesenchymal stem cells. Cell Res. 2011, 22, 62–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riggi, N.; Cironi, L.; Provero, P.; Suvà, M.-L.; Kaloulis, K.; Garcia-Echeverria, C.; Hoffmann, F.; Trumpp, A.; Stamenkovic, I. Development of Ewing’s Sarcoma from Primary Bone Marrow–Derived Mesenchymal Progenitor Cells. Cancer Res. 2005, 65, 11459–11468. [Google Scholar] [CrossRef] [Green Version]
- Tirode, F.; Laud-Duval, K.; Prieur, A.; Delorme, B.; Charbord, P.; Delattre, O. Mesenchymal Stem Cell Features of Ewing Tumors. Cancer Cell 2007, 11, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Von Levetzow, C.; Jiang, X.; Gwye, Y.; Von Levetzow, G.; Hung, L.; Cooper, A.; Hsu, J.H.-R.; Lawlor, E.R. Modeling Initiation of Ewing Sarcoma in Human Neural Crest Cells. PLoS ONE 2011, 6, e19305. [Google Scholar] [CrossRef] [Green Version]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Hatina, J.; Kripnerova, M.; Houfkova, K.; Pesta, M.; Kuncova, J.; Sana, J.; Slaby, O.; Rodriguez, R. Sarcoma Stem Cell Heterogeneity. Adv. Exp. Med. Biol. 2019, 1123, 95–118. [Google Scholar] [CrossRef]
- Lytle, N.K.; Barber, A.G.; Reya, T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer 2018, 18, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, H.K.; Tellez-Gabriel, M.; Heymann, D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017, 386, 189–195. [Google Scholar] [CrossRef]
- Tornin, J.; Hermida-Prado, F.; Padda, R.S.; Gonzalez, M.V.; Alvarez-Fernandez, C.; Rey, V.; Martinez-Cruzado, L.; Estupi-nan, O.; Menendez, S.T.; Fernandez-Nevado, L.; et al. FUS-CHOP Promotes Invasion in Myxoid Liposarcoma through a SRC/FAK/RHO/ROCK-Dependent Pathway. Neoplasia 2018, 20, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.-N.; Lv, Y.-F.; Guo, Q.-N. Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett. 2016, 370, 268–274. [Google Scholar] [CrossRef] [Green Version]
- Honoki, K.; Fujii, H.; Kubo, A.; Kido, A.; Mori, T.; Tanaka, Y.; Tsujiuchi, T. Possible involvement of stem-like populations with elevated ALDH1 in sarcomas for chemotherapeutic drug resistance. Oncol. Rep. 2010, 24, 501–505. [Google Scholar] [CrossRef]
- Cruzado, L.M.; Tornin, J.; Santos, L.; Rodriguez, A.; García-Castro, J.; Morís, F.; Rodriguez, R. Aldh1 Expression and Activity Increase During Tumor Evolution in Sarcoma Cancer Stem Cell Populations. Sci. Rep. 2016, 6, 27878. [Google Scholar] [CrossRef] [Green Version]
- Roundhill, E.A.; Jabri, S.; Burchill, S.A. ABCG1 and Pgp identify drug resistant, self-renewing osteosarcoma cells. Cancer Lett. 2019, 453, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Vallette, F.M.; Olivier, C.; Lézot, F.; Oliver, L.; Cochonneau, D.; Lalier, L.; Cartron, P.-F.; Heymann, D. Dormant, quiescent, tolerant and persister cells: Four synonyms for the same target in cancer. Biochem. Pharmacol. 2019, 162, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Alfranca, A.; Cruzado, L.M.; Tornin, J.; Abarrategi, A.; Amaral, T.; De Alava, E.; Menendez, P.; Garcia-Castro, J.; Rodriguez, R. Bone microenvironment signals in osteosarcoma development. Cell. Mol. Life Sci. 2015, 72, 3097–3113. [Google Scholar] [CrossRef] [PubMed]
- David, E.; Blanchard, F.; Heymann, M.-F.; De Pinieux, G.; Gouin, F.; Rédini, F.; Heymann, D. The Bone Niche of Chondrosarcoma: A Sanctuary for Drug Resistance, Tumour Growth and also a Source of New Therapeutic Targets. Sarcoma 2011, 2011, 932451. [Google Scholar] [CrossRef] [PubMed]
- Izadpanah, S.; Shabani, P.; Aghebati-Maleki, A.; Baghbanzadeh, A.; Fotouhi, A.; Bisadi, A.; Baradaran, B.; Aghebati-Maleki, L. Prospects for the involvement of cancer stem cells in the pathogenesis of osteosarcoma. J. Cell. Physiol. 2020, 235, 4167–4182. [Google Scholar] [CrossRef]
- Menendez, S.T.; Rey, V.; Martinez-Cruzado, L.; Gonzalez, M.V.; Morales-Molina, A.; Santos, L.; Blanco, V.; Alvarez, C.; Estupiñan, O.; Allonca, E.; et al. SOX2 Expression and Transcriptional Activity Identifies a Subpopulation of Cancer Stem Cells in Sarcoma with Prognostic Implications. Cancers 2020, 12, 964. [Google Scholar] [CrossRef]
- Adhikari, A.S.; Agarwal, N.; Wood, B.M.; Porretta, C.; Ruiz, B.; Pochampally, R.; Iwakuma, T. CD117 and Stro-1 Identify Osteosarcoma Tumor-Initiating Cells Associated with Metastasis and Drug Resistance. Cancer Res. 2010, 70, 4602–4612. [Google Scholar] [CrossRef] [Green Version]
- Awad, O.; Yustein, J.T.; Shah, P.; Gul, N.; Katuri, V.; O’Neill, A.; Kong, Y.; Brown, M.L.; Toretsky, J.A.; Loeb, D.M. High ALDH Activity Identifies Chemotherapy-Resistant Ewing’s Sarcoma Stem Cells That Retain Sensitivity to EWS-FLI1 Inhibition. PLoS ONE 2010, 5, e13943. [Google Scholar] [CrossRef] [Green Version]
- Honoki, K.; Fujii, H.; Tsujiuchi, T.; Kido, A.; Yoshitani, K.; Takakura, Y. Sphere-forming stem-like cell populations with drug resistance in human sarcoma cell lines. Int. J. Oncol. 2009, 34, 1381–1386. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, T.; Katsuda, T.; Hagiwara, K.; Kosaka, N.; Yoshioka, Y.; Takahashi, R.-U.; Takeshita, F.; Kubota, D.; Kondo, T.; Ichikawa, H.; et al. Clinical Relevance and Therapeutic Significance of MicroRNA-133a Expression Profiles and Functions in Malignant Osteosarcoma-Initiating Cells. Stem Cells 2014, 32, 959–973. [Google Scholar] [CrossRef] [Green Version]
- Greco, N.; Schott, T.; Mu, X.; Rothenberg, A.; Voigt, C.; McGough, R.L., III; Goodman, M.; Huard, J.; Weiss, K.R. ALDH Activity Correlates with Metastatic Potential in Primary Sarcomas of Bone. J. Cancer Ther. 2014, 05, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Hotfilder, M.; Mallela, N.; Seggewiß, J.; Dirksen, U.; Korsching, E. Defining a Characteristic Gene Expression Set Responsible for Cancer Stem Cell-Like Features in a Sub-Population of Ewing Sarcoma Cells CADO-ES1. Int. J. Mol. Sci. 2018, 19, 3908. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Gwye, Y.; Russell, D.; Cao, C.; Douglas, D.; Hung, L.; Kovar, H.; Triche, T.J.; Lawlor, E.R. CD133 expression in chemo-resistant Ewing sarcoma cells. BMC Cancer 2010, 10, 116. [Google Scholar] [CrossRef] [Green Version]
- Martins-Neves, S.R.; Lopes, A.O.; do Carmo, A.; Paiva, A.A.; Simoes, P.C.; Abrunhosa, A.J.; Gomes, C.M. Therapeutic implications of an enriched cancer stem-like cell population in a human osteosarcoma cell line. BMC Cancer 2012, 12, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.-X.; Liao, G.-J.; Liu, K.-G.; Jian, H. Endosialin-expressing bone sarcoma stem-like cells are highly tumor-initiating and invasive. Mol. Med. Rep. 2015, 12, 5665–5670. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Li, X.; Si, M.; Liu, T.; Li, J. CD271+ Osteosarcoma Cells Display Stem-Like Properties. PLoS ONE 2014, 9, e98549. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Teng, J.-S. Increased multi-drug resistance and reduced apoptosis in osteosarcoma side population cells are crucial factors for tumor recurrence. Exp. Ther. Med. 2016, 12, 81–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Yan, M.; Zhang, R.; Li, J.; Luo, Z. Side population cells isolated from human osteosarcoma are enriched with tumor-initiating cells. Cancer Sci. 2011, 102, 1774–1781. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, R.; Yan, M.; Ye, Z.; Liang, W.; Luo, Z. Detection and characterization of side population in Ewing’s sarcoma SK-ES-1 cells in vitro. Biochem. Biophys. Res. Commun. 2010, 391, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Ying, M.; Liu, G.; Shimada, H.; Ding, W.; May, W.A.; He, Q.; Adams, G.B.; Wu, L. Human osteosarcoma CD49f−CD133+ cells: Impaired in osteogenic fate while gain of tumorigenicity. Oncogene 2013, 32, 4252–4263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Liu, S.; Zhang, C.; Zhang, B.; Simões, B.M.; Eyre, R.; Liang, Y.; Yan, H.; Wu, Z.; Guo, W.; et al. Enrichment of human osteosarcoma stem cells based on hTERT transcriptional activity. Oncotarget 2013, 4, 2326–2338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Zhao, Q.; Sun, H.; Yin, L.; Wu, J.; Xu, J.; He, T.; Yang, C.; Liang, C. Defective autophagy leads to the suppression of stem-like features of CD271+ osteosarcoma cells. J. Biomed. Sci. 2016, 23, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Li, Y.; Kuang, M.; Wang, X.; Jia, Q.; Cao, J.; Hu, J.; Wu, S.; Wang, Z.; Xiao, J. The CD24+ cell subset promotes invasion and metastasis in human osteosarcoma. EBioMedicine 2020, 51, 102598. [Google Scholar] [CrossRef] [Green Version]
- Di Fiore, R.; Guercio, A.; Puleio, R.; Di Marco, P.; Drago-Ferrante, R.; D’Anneo, A.; De Blasio, A.; Carlisi, D.; Di Bella, S.; Pentimalli, F.; et al. Modeling human osteosarcoma in mice through 3AB-OS cancer stem cell xenografts. J. Cell. Biochem. 2012, 113, 3380–3392. [Google Scholar] [CrossRef]
- Di Fiore, R.; Santulli, A.; Ferrante, R.D.; Giuliano, M.; De Blasio, A.; Messina, C.; Pirozzi, G.; Tirino, V.; Tesoriere, G.; Vento, R. Identification and expansion of human osteosarcoma-cancer-stem cells by long-term 3-aminobenzamide treatment. J. Cell. Physiol. 2009, 219, 301–313. [Google Scholar] [CrossRef]
- Li, Y.; Xian, M.; Yang, B.; Ying, M.; He, Q. Inhibition of KLF4 by Statins Reverses Adriamycin-Induced Metastasis and Cancer Stemness in Osteosarcoma Cells. Stem Cell Rep. 2017, 8, 1617–1629. [Google Scholar] [CrossRef] [Green Version]
- Martins-Neves, S.R.; Paiva-Oliveira, D.I.; Wijers-Koster, P.M.; Abrunhosa, A.J.; Fontes-Ribeiro, C.; Bovée, J.V.; Cleton-Jansen, A.-M.; Gomes, C.M. Chemotherapy induces stemness in osteosarcoma cells through activation of Wnt/β-catenin signaling. Cancer Lett. 2016, 370, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.-L.; Liang, Y.; Xie, X.-B.; Yin, J.-Q.; Zou, C.-Y.; Zhao, Z.-Q.; Shen, J.-N.; Wang, J. Enrichment of osteosarcoma stem cells by chemotherapy. Chin. J. Cancer 2011, 30, 426–432. [Google Scholar] [CrossRef] [Green Version]
- Tsuchida, R.; Das, B.; Yeger, H.; Koren, G.; Shibuya, M.; Thorner, P.S.; Baruchel, S.; Malkin, D. Cisplatin treatment increases survival and expansion of a highly tumorigenic side-population fraction by upregulating VEGF/Flt1 autocrine signaling. Oncogene 2008, 27, 3923–3934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zeng, L.; Liang, C.; Zan, R.; Ji, W.; Zhang, Z.; Wei, Y.; Tu, S.; Dong, Y. Integrated analysis of transcriptome-wide m6A methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics 2019, 11, 1693–1715. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Fan, Z.; Fang, S.; Yang, J.; Gao, T.; Simões, B.M.; Eyre, R.; Guo, W.; Clarke, R.B. Cisplatin selects for stem-like cells in osteosarcoma by activating Notch signaling. Oncotarget 2016, 7, 33055–33068. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Song, X.; Xu, D.; Tiek, D.; Goenka, A.; Wu, B.; Sastry, N.; Hu, B.; Cheng, S.-Y. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics 2020, 10, 8721–8743. [Google Scholar] [CrossRef] [PubMed]
- Martins-Neves, S.R.; Paiva-Oliveira, D.I.; Fontes-Ribeiro, C.; Bovée, J.V.; Cleton-Jansen, A.-M.; Gomes, C.M. IWR-1, a tankyrase inhibitor, attenuates Wnt/β-catenin signaling in cancer stem-like cells and inhibits in vivo the growth of a subcutaneous human osteosarcoma xenograft. Cancer Lett. 2018, 414, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Rainusso, N.; Lee, Y.-C.; Dawson, B.; Coarfa, C.; Han, R.; Larson, J.L.; Shuck, R.; Kurenbekova, L.; Yustein, J.T. Tegavivint and the β-Catenin/ALDH Axis in Chemotherapy-Resistant and Metastatic Osteosarcoma. J. Natl. Cancer Inst. 2019, 111, 1216–1227. [Google Scholar] [CrossRef]
- Feng, J.; Lan, R.; Cai, G.; Lin, J. TREX1 suppression imparts cancer-stem-cell-like characteristics to CD133- osteosarcoma cells through the activation of E2F4 signaling. Int. J. Clin. Exp. Pathol. 2019, 12, 1134–1153. [Google Scholar] [PubMed]
- Liu, L.; Wang, T.; Yang, X.; Xu, C.; Liao, Z.; Wang, X.; Su, D.; Li, Y.; Zhou, H.; Qiu, X.; et al. MTNR1B loss promotes chordoma recurrence by abrogating melatonin-mediated β-catenin signaling repression. J. Pineal Res. 2019, 67, e12588. [Google Scholar] [CrossRef]
- Dai, G.; Deng, S.; Guo, W.; Yu, L.; Yang, J.; Zhou, S.; Gao, T. Notch pathway inhibition using DAPT, a γ-secretase inhibitor (GSI), enhances the antitumor effect of cisplatin in resistant osteosarcoma. Mol. Carcinog. 2019, 58, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Xia, K.; Gao, T.; Chen, J.; Zhang, Z.; Sun, X.; Simões, B.M.; Eyre, R.; Fan, Z.; Guo, W.; et al. The Notch Pathway Promotes Osteosarcoma Progression through Activation of Ephrin Reverse Signaling. Mol. Cancer Res. 2019, 17, 2383–2394. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Ma, K.; Li, W.-Y. IL-6 Promotes Cancer Stemness and Oncogenicity in U2OS and MG-63 Osteosarcoma Cells by Upregulating the OPN-STAT3 Pathway. J. Cancer 2019, 10, 6511–6525. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ma, K.; Li, W.-Y. Cinobufagin Suppresses The Characteristics Of Osteosarcoma Cancer Cells By Inhibiting The IL-6-OPN-STAT3 Pathway. Drug Des. Dev. Ther. 2019, 13, 4075–4090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, X.-T.; Li, Y.-L.; Zhang, Y.-Q.; Xu, T.; Lu, B.; Fang, L.; Gao, J.-Q.; Yu, L.-S.; Zhu, D.-F.; Yang, B.; et al. KLF4 functions as an oncogene in promoting cancer stem cell-like characteristics in osteosarcoma cells. Acta Pharmacol. Sin. 2019, 40, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, P.; Liu, Q.; Wang, J.; Yan, F.; Duan, L.; Lin, F.; Lian, Z. KLF8 promotes cancer stem cell-like phenotypes in osteosarcoma through miR-429-SOX2 signaling. Neoplasma 2020, 67, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Hosain, S.B.; Khiste, S.K.; Uddin, M.; Vorubindi, V.; Ingram, C.; Zhang, S.; Hill, R.A.; Gu, X.; Liu, Y.-Y. Inhibition of glucosylceramide synthase eliminates the oncogenic function of p53 R273H mutant in the epithelial-mesenchymal transition and induced pluripotency of colon cancer cells. Oncotarget 2016, 7, 60575–60592. [Google Scholar] [CrossRef] [Green Version]
- Di Fiore, R.; Marcatti, M.; Drago-Ferrante, R.; D’Anneo, A.; Giuliano, M.; Carlisi, D.; De Blasio, A.; Querques, F.; Pastore, L.; Tesoriere, G.; et al. Mutant p53 gain of function can be at the root of dedifferentiation of human osteosarcoma MG63 cells into 3AB-OS cancer stem cells. Bone 2014, 60, 198–212. [Google Scholar] [CrossRef] [Green Version]
- Guerzoni, C.; Fiori, V.; Terracciano, M.; Manara, M.C.; Moricoli, D.; Pasello, M.; Sciandra, M.; Nicoletti, G.; Gellini, M.; Dominici, S.; et al. CD99 Triggering in Ewing Sarcoma Delivers a Lethal Signal through p53 Pathway Reactivation and Cooperates with Doxorubicin. Clin. Cancer Res. 2015, 21, 146–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, C.; Martins-Neves, S.R.; Paiva-Oliveira, D.; Oliveira, V.E.; Fontes-Ribeiro, C.; Gomes, C.M. Sensitizing osteosarcoma stem cells to doxorubicin-induced apoptosis through retention of doxorubicin and modulation of apoptotic-related proteins. Life Sci. 2015, 130, 47–56. [Google Scholar] [CrossRef]
- Stuart, J.A.; Brown, M.F. Energy, quiescence and the cellular basis of animal life spans. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 2006, 143, 12–23. [Google Scholar] [CrossRef]
- Dasgupta, A.; Trucco, M.; Rainusso, N.; Bernardi, R.J.; Shuck, R.; Kurenbekova, L.; Loeb, D.M.; Yustein, J.T. Metabolic modulation of Ewing sarcoma cells inhibits tumor growth and stem cell properties. Oncotarget 2017, 8, 77292–77308. [Google Scholar] [CrossRef] [Green Version]
- Shang, D.; Wu, J.; Guo, L.; Xu, Y.; Liu, L.; Lu, J. Metformin increases sensitivity of osteosarcoma stem cells to cisplatin by inhibiting expression of PKM2. Int. J. Oncol. 2017, 50, 1848–1856. [Google Scholar] [CrossRef]
- Chen, X.; Hu, C.; Zhang, W.; Shen, Y.; Wang, J.; Hu, F.; Yu, P. Metformin inhibits the proliferation, metastasis, and cancer stem-like sphere formation in osteosarcoma MG63 cells in vitro. Tumor Biol. 2015, 36, 9873–9883. [Google Scholar] [CrossRef]
- Zhao, B.; Luo, J.; Wang, Y.; Zhou, L.; Che, J.; Wang, F.; Peng, S.; Zhang, G.; Shang, P. Metformin Suppresses Self-Renewal Ability and Tumorigenicity of Osteosarcoma Stem Cells via Reactive Oxygen Species-Mediated Apoptosis and Autophagy. Oxidative Med. Cell. Longev. 2019, 2019, 9290728. [Google Scholar] [CrossRef]
- Camuzard, O.; Trojani, M.-C.; Santucci-Darmanin, S.; Pagnotta, S.; Breuil, V.; Carle, G.; Pierrefite-Carle, V. Autophagy in Osteosarcoma Cancer Stem Cells Is Critical Process which Can Be Targeted by the Antipsychotic Drug Thioridazine. Cancers 2020, 12, 3675. [Google Scholar] [CrossRef]
- Wang, W.; Chen, D.; Zhu, K. SOX2OT variant 7 contributes to the synergistic interaction between EGCG and Doxorubicin to kill osteosarcoma via autophagy and stemness inhibition. J. Exp. Clin. Cancer Res. 2018, 37, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Burdach, S.; Plehm, S.; Unland, R.; Borkhardt, A.; Staege, M.; Müller-Tidow, C.; Richter, G.H. Epigenetic maintenance of stemness and malignancy in peripheral neuroectodermal tumors by EZH2. Cell Cycle 2009, 8, 1991–1996. [Google Scholar] [CrossRef]
- Richter, G.H.S.; Plehm, S.; Fasan, A.; Rössler, S.; Unland, R.; Bennani-Baiti, I.M.; Hotfilder, M.; Löwel, D.; von Luettichau, I.; Mossbrugger, I.; et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 5324–5329. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, S.; Hersey, J.M.; Mellor, P.; Dai, W.; Santos-Silva, A.; Liber, D.; Luk, L.; Titley, I.; Carden, C.P.; Box, G.; et al. Ovarian Cancer Stem Cell–Like Side Populations Are Enriched Following Chemotherapy and Overexpress EZH2. Mol. Cancer Ther. 2011, 10, 325–335. [Google Scholar] [CrossRef] [Green Version]
- Suvà, M.-L.; Riggi, N.; Janiszewska, M.; Radovanovic, I.; Provero, P.; Stehle, J.-C.; Baumer, K.; Le Bitoux, M.-A.; Marino, D.; Cironi, L.; et al. EZH2 Is Essential for Glioblastoma Cancer Stem Cell Maintenance. Cancer Res. 2009, 69, 9211–9218. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.; Shen, J.; Gao, Y.; Zhou, Y.; Yu, Z.; Hornicek, F.; Kan, Q.; Duan, Z. Overexpression of EZH2 is associated with the poor prognosis in osteosarcoma and function analysis indicates a therapeutic potential. Oncotarget 2016, 7, 38333–38346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.; He, C.; Wu, Z.; Li, F.; Xiao, J. Histone methyltransferase SETD2 regulates osteosarcoma cell growth and chemosensitivity by suppressing Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 2018, 502, 382–388. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Liu, C.; Wang, L.; Sun, Y.; Jiang, Y.; Hao, Y. Histone methyltransferase NSD2 regulates apoptosis and chemosensitivity in osteosarcoma. Cell Death Dis. 2019, 10, 65. [Google Scholar] [CrossRef] [Green Version]
- Pozzi, V.; Salvolini, E.; Lucarini, G.; Salvucci, A.; Campagna, R.; Rubini, C.; Sartini, D.; Emanuelli, M. Cancer stem cell enrichment is associated with enhancement of nicotinamide N-methyltransferase expression. IUBMB Life 2020, 72, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; He, Y.; He, J.; Wang, L.; Liu, Z.; Yang, J.; Gao, Z.; Lu, G.; Zou, C.; Zhao, W. Epigenetic Profiling Identifies LIF as a Super-enhancer-Controlled Regulator of Stem Cell–like Properties in Osteosarcoma. Mol. Cancer Res. 2020, 18, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef]
- Wang, T.; Wang, D.; Zhang, L.; Yang, P.; Wang, J.; Liu, Q.; Yan, F.; Lin, F. The TGFβ-miR-499a-SHKBP1 pathway induces resistance to EGFR inhibitors in osteosarcoma cancer stem cell-like cells. J. Exp. Clin. Cancer Res. 2019, 38, 226. [Google Scholar] [CrossRef] [Green Version]
- Di Fiore, R.; Drago-Ferrante, R.; Pentimalli, F.; Di Marzo, D.; Forte, I.M.; D’Anneo, A.; Carlisi, D.; De Blasio, A.; Giuliano, M.; Tesoriere, G.; et al. MicroRNA-29b-1 impairs in vitro cell proliferation, self-renewal and chemoresistance of human osteosarcoma 3AB-OS cancer stem cells. Int. J. Oncol. 2014, 45, 2013–2023. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Jin, H.; Xu, C.-X.; Sun, B.; Song, Z.-G.; Bi, W.-Z.; Wang, Y. miR-382 Inhibits Osteosarcoma Metastasis and Relapse by Targeting Y Box-Binding Protein 1. Mol. Ther. 2015, 23, 89–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lü, J.; Song, G.; Tang, Q.; Yin, J.; Zou, C.; Zhao, Z.; Xie, X.; Xu, H.; Huang, G.; Wang, J.; et al. MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1. Oncogene 2017, 36, 231–241. [Google Scholar] [CrossRef]
- Zou, Y.; Huang, Y.; Yang, J.; Wu, J.; Luo, C. miR-34a is downregulated in human osteosarcoma stem-like cells and promotes invasion, tumorigenic ability and self-renewal capacity. Mol. Med. Rep. 2017, 15, 1631–1637. [Google Scholar] [CrossRef] [Green Version]
- Marino, M.T.; Grilli, A.; Baricordi, C.; Manara, M.C.; Ventura, S.; Pinca, R.S.; Bellenghi, M.; Calvaruso, M.; Mattia, G.; Donati, D.; et al. Prognostic significance of miR-34a in Ewing sarcoma is associated with cyclin D1 and ki-67 expression. Ann. Oncol. 2014, 25, 2080–2086. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Chen, Y.; Chen, S.; Zheng, C.; Hu, J.; Luo, S. MiR-19a regulates the cell growth and apoptosis of osteosarcoma stem cells by targeting PTEN. Tumor Biol. 2017, 39, 1010428317705341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, H.; Luo, S.; Wang, Y.; Liu, C.; Piao, Z.; Xu, M.; Guan, W.; Li, Q.; Zou, H.; Tan, Q.-Y.; et al. miR-135b Stimulates Osteosarcoma Recurrence and Lung Metastasis via Notch and Wnt/β-Catenin Signaling. Mol. Ther. Nucleic Acids 2017, 8, 111–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Huang, W.; Sun, W.; Zheng, B.; Wang, C.; Luo, Z.; Wang, J.; Yan, W. LncRNA MALAT1 Promotes Cancer Metastasis in Osteosarcoma via Activation of the PI3K-Akt Signaling Pathway. Cell. Physiol. Biochem. 2018, 51, 1313–1326. [Google Scholar] [CrossRef]
- Jiang, N.; Wang, X.; Xie, X.; Liao, Y.; Liu, N.; Liu, J.; Miao, N.; Shen, J.; Peng, T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017, 405, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Lu, C.; Qu, X.; Li, P.; Chen, K.; Shan, L.; Zhu, X. LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and drug resistance via the miR-134-5p/MBTD1 axis. Aging 2019, 11, 8374–8385. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Ji, S.; Yuan, Q.; Zhou, H. Downregulated Long Non-Coding RNA MSC-AS1 Inhibits Osteosarcoma Progression and Increases Sensitivity to Cisplatin by Binding to MicroRNA-142. Med. Sci. Monit. 2020, 26, e921594. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A. Circular RNAs and their participation in stemness of cancer. Med. Oncol. 2020, 37, 42. [Google Scholar] [CrossRef]
- Kun-Peng, Z.; Xiao-Long, M.; Chun-Lin, Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int. J. Biol. Sci. 2018, 14, 321–330. [Google Scholar] [CrossRef]
- Kun-Peng‡, Z.; Xiao-Long‡, M.; Lei‡, Z.; Chun-Lin, Z.; Jian-Ping, H.; Tai-Cheng, Z. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics 2018, 10, 1327–1346. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Y.; Zhang, X.; Shen, J.; Xu, R.; Liu, Y.; Yu, X. Circular RNA hsa_circ_0000073 contributes to osteosarcoma cell proliferation, migration, invasion and methotrexate resistance by sponging miR-145-5p and miR-151-3p and upregulating NRAS. Aging 2020, 12, 14157–14173. [Google Scholar] [CrossRef] [PubMed]
- Yanbin, Z.; Jing, Z. CircSAMD4A accelerates cell proliferation of osteosarcoma by sponging miR-1244 and regulating MDM2 mRNA expression. Biochem. Biophys. Res. Commun. 2019, 516, 102–111. [Google Scholar] [CrossRef]
- Dong, L.; Qu, F. CircUBAP2 promotes SEMA6D expression to enhance the cisplatin resistance in osteosarcoma through sponging miR-506-3p by activating Wnt/β-catenin signaling pathway. J. Mol. Histol. 2020, 51, 329–340. [Google Scholar] [CrossRef]
- Heymann, M.-F.; Lézot, F.; Heymann, D. The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell. Immunol. 2019, 343, 103711. [Google Scholar] [CrossRef]
- Corre, I.; Verrecchia, F.; Crenn, V.; Redini, F.; Trichet, V. The Osteosarcoma Microenvironment: A Complex but Targetable Ecosystem. Cells 2020, 9, 976. [Google Scholar] [CrossRef] [Green Version]
- Le Nail, L.-R.; Brennan, M.; Rosset, P.; Deschaseaux, F.; Piloquet, P.; Pichon, O.; Le Caignec, C.; Crenn, V.; Layrolle, P.; Hérault, O.; et al. Comparison of Tumor- and Bone Marrow-Derived Mesenchymal Stromal/Stem Cells from Patients with High-Grade Osteosarcoma. Int. J. Mol. Sci. 2018, 19, 707. [Google Scholar] [CrossRef] [Green Version]
- Rédini, F.; Heymann, D. Bone Tumor Environment as a Potential Therapeutic Target in Ewing Sarcoma. Front. Oncol. 2015, 5, 279. [Google Scholar] [CrossRef] [Green Version]
- Rubio, R.; Abarrategi, A.; Garcia-Castro, J.; Cruzado, L.M.; Suarez, C.; Tornin, J.; Santos, L.; Astudillo, A.; Colmenero, I.; Mulero, F.; et al. Bone Environment is Essential for Osteosarcoma Development from Transformed Mesenchymal Stem Cells. Stem Cells 2014, 32, 1136–1148. [Google Scholar] [CrossRef]
- Adamski, J.; Price, A.; Dive, C.; Makin, G. Hypoxia–Induced Cytotoxic Drug Resistance in Osteosarcoma Is Independent of HIF-1Alpha. PLoS ONE 2013, 8, e65304. [Google Scholar] [CrossRef] [Green Version]
- Roncuzzi, L.; Pancotti, F.; Baldini, N. Involvement of HIF-1α activation in the doxorubicin resistance of human osteosarcoma cells. Oncol. Rep. 2014, 32, 389–394. [Google Scholar] [CrossRef] [Green Version]
- Scholten, D.J., II; Timmer, C.M.; Peacock, J.D.; Pelle, D.W.; Williams, B.; Steensma, M.R. Down Regulation of Wnt Signaling Mitigates Hypoxia-Induced Chemoresistance in Human Osteosarcoma Cells. PLoS ONE 2014, 9, e111431. [Google Scholar] [CrossRef] [Green Version]
- Birru, B.; Durthi, C.P.; Kacham, S.; Pola, M.; Rajulapati, S.B.; Parcha, S.R.; Kamal, M.A. Stem Cells in Tumour Microenvironment Aid in Prolonged Survival Rate of Cancer Cells and Developed Drug Resistance: Major Challenge in Osteosarcoma Treatment. Curr. Drug Metab. 2020, 21, 44–52. [Google Scholar] [CrossRef]
- Cortini, M.; Avnet, S.; Baldini, N. Mesenchymal stroma: Role in osteosarcoma progression. Cancer Lett. 2017, 405, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, G.; Chen, R.; Hua, Y.; Cai, Z. Mesenchymal stem cells in the osteosarcoma microenvironment: Their biological properties, influence on tumor growth, and therapeutic implications. Stem Cell Res. Ther. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trubiani, O.; Marconi, G.D.; Pierdomenico, S.D.; Piattelli, A.; Diomede, F.; Pizzicannella, J. Human Oral Stem Cells, Biomaterials and Extracellular Vesicles: A Promising Tool in Bone Tissue Repair. Int. J. Mol. Sci. 2019, 20, 4987. [Google Scholar] [CrossRef] [Green Version]
- Vallabhaneni, K.C.; Hassler, M.-Y.; Abraham, A.; Whitt, J.; Mo, Y.-Y.; Atfi, A.; Pochampally, R. Mesenchymal Stem/Stromal Cells under Stress Increase Osteosarcoma Migration and Apoptosis Resistance via Extracellular Vesicle Mediated Communication. PLoS ONE 2016, 11, e0166027. [Google Scholar] [CrossRef] [PubMed]
- Vallabhaneni, K.C.; Penfornis, P.; Dhule, S.; Guillonneau, F.; Adams, K.V.; Mo, Y.Y.; Xu, R.; Liu, Y.; Watabe, K.; Vemuri, M.C.; et al. Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget 2015, 6, 4953–4967. [Google Scholar] [CrossRef] [Green Version]
- Avril, P.; Duteille, F.; Ridel, P.; Heymann, M.-F.; De Pinieux, G.; Rédini, F.; Blanchard, F.; Heymann, D.; Trichet, V.; Perrot, P. Opposite Effects of Soluble Factors Secreted by Adipose Tissue on Proliferating and Quiescent Osteosarcoma Cells. Plast. Reconstr. Surg. 2016, 137, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Tu, B.; Zhu, J.; Liu, S.; Wang, L.; Fan, Q.; Hao, Y.; Fan, C.; Tang, T.-T. Mesenchymal stem cells promote osteosarcoma cell survival and drug resistance through activation of STAT3. Oncotarget 2016, 7, 48296–48308. [Google Scholar] [CrossRef] [Green Version]
- Avril, P.; Le Nail, L.-R.; Brennan, M.Á.; Rosset, P.; De Pinieux, G.; Layrolle, P.; Heymann, D.; Perrot, P.; Trichet, V. Mesenchymal stem cells increase proliferation but do not change quiescent state of osteosarcoma cells: Potential implications according to the tumor resection status. J. Bone Oncol. 2016, 5, 5–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, S.; Jiang, L.; Shen, L.; Xiong, Z. Role of microRNA-92a in metastasis of osteosarcoma cells in vivo and in vitro by inhibiting expression of TCF21 with the transmission of bone marrow derived mesenchymal stem cells. Cancer Cell Int. 2019, 19, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietrovito, L.; Leo, A.; Gori, V.; Lulli, M.; Parri, M.; Becherucci, V.; Piccini, L.; Bambi, F.; Taddei, M.L.; Chiarugi, P. Bone marrow-derived mesenchymal stem cells promote invasiveness and transendothelial migration of osteosarcoma cells via a mesenchymal to amoeboid transition. Mol. Oncol. 2018, 12, 659–676. [Google Scholar] [CrossRef] [PubMed]
- Cortini, M.; Massa, A.; Avnet, S.; Bonuccelli, G.; Baldini, N. Tumor-Activated Mesenchymal Stromal Cells Promote Osteosarcoma Stemness and Migratory Potential via IL-6 Secretion. PLoS ONE 2016, 11, e0166500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baglio, S.R.; Lagerweij, T.; Pérez-Lanzón, M.; Ho, X.D.; Léveillé, N.; Melo, S.; Cleton-Jansen, A.-M.; Jordanova, E.S.; Roncuzzi, L.; Greco, M.; et al. Blocking Tumor-Educated MSC Paracrine Activity Halts Osteosarcoma Progression. Clin. Cancer Res. 2017, 23, 3721–3733. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Han, X.-G.; Tu, B.; Wang, M.-Q.; Qiao, H.; Zhang, S.-H.; Fan, Q.-M.; Tang, T.-T. CXCR1/Akt signaling activation induced by mesenchymal stem cell-derived IL-8 promotes osteosarcoma cell anoikis resistance and pulmonary metastasis. Cell Death Dis. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Zhang, R.-M.; Tang, T.; Yu, H.-M.; Yao, X.-D. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem. Biophys. Res. Commun. 2018, 507, 260–266. [Google Scholar] [CrossRef]
- Zhao, W.; Qin, P.; Zhang, D.; Cui, X.; Gao, J.; Yu, Z.; Chai, Y.; Wang, J.; Li, J. Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging 2019, 11, 9581–9596. [Google Scholar] [CrossRef]
- Bian, Z.-Y.; Fan, Q.-M.; Li, G.; Xu, W.-T.; Tang, T. Human mesenchymal stem cells promote growth of osteosarcoma: Involvement of interleukin-6 in the interaction between human mesenchymal stem cells and Saos-2. Cancer Sci. 2010, 101, 2554–2560. [Google Scholar] [CrossRef] [PubMed]
- Tu, B.; Du, L.; Fan, Q.-M.; Tang, Z.; Tang, T.-T. STAT3 activation by IL-6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett. 2012, 325, 80–88. [Google Scholar] [CrossRef]
- Tu, B.; Peng, Z.-X.; Fan, Q.-M.; Du, L.; Yan, W.; Tang, T.-T. Osteosarcoma cells promote the production of pro-tumor cytokines in mesenchymal stem cells by inhibiting their osteogenic differentiation through the TGF-β/Smad2/3 pathway. Exp. Cell Res. 2014, 320, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Bonuccelli, G.; Avnet, S.; Grisendi, G.; Salerno, M.; Granchi, D.; Dominici, M.; Kusuzaki, K.; Baldini, N. Role of mesenchymal stem cells in osteosarcoma and metabolic reprogramming of tumor cells. Oncotarget 2014, 5, 7575–7588. [Google Scholar] [CrossRef] [PubMed]
- Avnet, S.; Chano, T.; Massa, A.; Bonuccelli, G.; Lemma, S.; Falzetti, L.; Grisendi, G.; Dominici, M.; Baldini, N. Acid microen-vironment promotes cell survival of human bone sarcoma through the activation of cIAP proteins and NF-kappaB pathway. Am. J. Cancer Res. 2019, 9, 1127–1144. [Google Scholar]
- Avnet, S.; Di Pompo, G.; Chano, T.; Errani, C.; Ibrahim-Hashim, A.; Gillies, R.J.; Donati, D.M.; Baldini, N. Cancer-associated mesenchymal stroma fosters the stemness of osteosarcoma cells in response to intratumoral acidosis via NF-κB activation. Int. J. Cancer 2017, 140, 1331–1345. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.; Wilding, C.; Jones, R.L.; Huang, P.H. Proteomic research in sarcomas—Current status and future opportunities. Semin. Cancer Biol. 2020, 61, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Mao, S.; Lin, H.; Lin, J.-M.; Lin, J. Comparative proteomics of cancer stem cells in osteosarcoma using ultra-high-performance liquid chromatography and Orbitrap Fusion mass spectrometer. Talanta 2018, 178, 362–368. [Google Scholar] [CrossRef]
- Tellez-Gabriel, M.; Heymann, M.-F.; Heymann, D. Circulating Tumor Cells as a Tool for Assessing Tumor Heterogeneity. Theranostics 2019, 9, 4580–4594. [Google Scholar] [CrossRef]
- Tellez-Gabriel, M.; Ory, B.; Lamoureux, F.; Heymann, M.-F.; Heymann, D. Tumour Heterogeneity: The Key Advantages of Single-Cell Analysis. Int. J. Mol. Sci. 2016, 17, 2142. [Google Scholar] [CrossRef]

| Method of CSC Isolation/Characterization | Bone Sarcoma Models | Drug Resistance | Ref. | |
|---|---|---|---|---|
| Drug | Fold Resistance * | |||
| Sphere formation | MNNG/HOS (OS) | DOX CIS MTX | 2.67 2.23 8.33 | [49] |
| MG63 (OS), HTB166 (EWS) | DOX CIS | N/Q N/Q | [44] | |
| CD133 | Human primary, Saos2, MG63, U2OS, HOS, MNNG/HOS, 143B (OS) | DOX CIS MTX | N/Q N/Q N/Q | [45] |
| Human primary, STA-ET8.2, TC71, A4573, 5838 and other 5 lines (EWS) | DOX ETO VNC | N/Q N/Q N/Q | [48] | |
| STRO-1/CD177 | 318-1, K7M2 (OS) | DOX | 1.47, 1.73 | [42] |
| CD271 | Saos2, U2OS, MNNG/HOS (OS) | CIS | 2.16, 1.42, 1.65 | [51] |
| Saos2, MNNG/HOS (OS) | CIS EPR | N/Q N/Q | [57] | |
| CD24 | Human primary, MG63, MNNG/HOS, U2OS, OSC228 (OS) | CIS EPR | N/Q N/Q | [58] |
| CD49f | UT2, TTC606 (OS) | CIS IDR PTX | 6.56, >1.74 1.77, NA 2.10, NA | [55] |
| Side population | OS1, OS2, OS5 (Human Primary, OS) | DOX CIS MTX | 1.33, 1.8, 1.67 1.09, 2.01, 1.07 1.46, 1.43, 0.94 | [53] |
| Human primary (OS) | DOX CIS MTX | N/Q N/Q N/Q | [50] | |
| OS65 (OS) | ETO 5-FU CIS PTX GEM OXP | N/Q N/Q N/Q N/Q N/Q N/Q | [52] | |
| SK-ES-1 (EWS) | DOX CIS | 1.64 1.92 | [54] | |
| CADO-ES1 (EWS) | DOX CIS ETO | N/Q N/Q N/Q | [47] | |
| ALDH activity | Human primary (OS/CDS/EWS) | DOX DSF | N/Q N/Q | [46] |
| Human primary, TC71, MHH-ES, SK-ES-1, A4573 (EWS) | DOX ETO | N/Q N/Q | [43] | |
| hTERT | Human primary, MG63, MNNG/HOS, 143B (OS) | DOX | N/Q | [56] |
| Bone Sarcoma Models | Drug Resistance Induction | Characterization of CSC Phenotype | Ref. | ||
|---|---|---|---|---|---|
| Drug | Method | Fold Resistance * | |||
| MG63 (OS) | 3AB | Exposure to 3AB 5 mM for 100 days | N/Q | Sphere formation, pluripot. factors, ABC transporters, CSC markers, MSC features, tumorigenic growth | [59,60] |
| U2OS, MG63 (OS) | MTX | Exposure to MTX 100 or 300 ng/mL for 5 days | N/Q | Sphere formation, SP, CSC markers, tumorigenic growth | [63] |
| HOS (OS) | CIS | Exposure to CIS 2 µM for 3 days | N/Q | SP, sphere formation, pluripot. factors, VEGF signaling, tumorigenic growth | [64] |
| 143B, U2OS (OS) | CIS | Exposure to CIS 5 µM for 24 h | 1.54, 1.75 | Sphere formation, MSC features, pluripot. factors, CSC markers, NOTCH signaling, tumorigenic growth | [66] |
| HOS, MG63, MHN, MNNG/HOS, OHS, U2OS (OS) | DOX MTX CIS | Exposure to DOX (0.91, 0.31, 0.92, 0.46, 0.52, 0.92 µM), MTX (8, 20, 22, 22, 5, 7 nM) or CIS (3.72, 7.55, 15.87, 3.3, 4.17, 12.48 µM) for 24 h | N/Q N/Q N/Q | ALDH activity, pluripot. factors, ABC transporters, WNT/β-Catenin signaling, tumorigenic growth | [62] |
| Human primary, U2OS, KHOS/NP (OS) | DOX | Exposure to DOX (50, 100, 100 nM) for 24 h | N/Q | Sphere formation, CSC markers, migration, tumorigenic growth | [61] |
| HOS, MG63, U2OS (OS) | DOX | Exposure to DOX (14, 28, 28 nM) for 6 months | 4.0, 4.0, 6.0 | Multidrug resistance, ABC transporters, migration | [36] |
| MG63 (OS) | DOX | Exposure to increasing doses of DOX (from 2.5 to 1000 ng/mL) | 10.53 | Multidrug resistance, pluripot. factors, CSC markers, sphere formation, tumorigenic growth | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menéndez, S.T.; Gallego, B.; Murillo, D.; Rodríguez, A.; Rodríguez, R. Cancer Stem Cells as a Source of Drug Resistance in Bone Sarcomas. J. Clin. Med. 2021, 10, 2621. https://doi.org/10.3390/jcm10122621
Menéndez ST, Gallego B, Murillo D, Rodríguez A, Rodríguez R. Cancer Stem Cells as a Source of Drug Resistance in Bone Sarcomas. Journal of Clinical Medicine. 2021; 10(12):2621. https://doi.org/10.3390/jcm10122621
Chicago/Turabian StyleMenéndez, Sofía T., Borja Gallego, Dzohara Murillo, Aida Rodríguez, and René Rodríguez. 2021. "Cancer Stem Cells as a Source of Drug Resistance in Bone Sarcomas" Journal of Clinical Medicine 10, no. 12: 2621. https://doi.org/10.3390/jcm10122621
APA StyleMenéndez, S. T., Gallego, B., Murillo, D., Rodríguez, A., & Rodríguez, R. (2021). Cancer Stem Cells as a Source of Drug Resistance in Bone Sarcomas. Journal of Clinical Medicine, 10(12), 2621. https://doi.org/10.3390/jcm10122621






