Omega-3 PUFAs Suppress IL-1β-Induced Hyperactivity of Immunoproteasomes in Astrocytes
Abstract
1. Introduction
2. Results
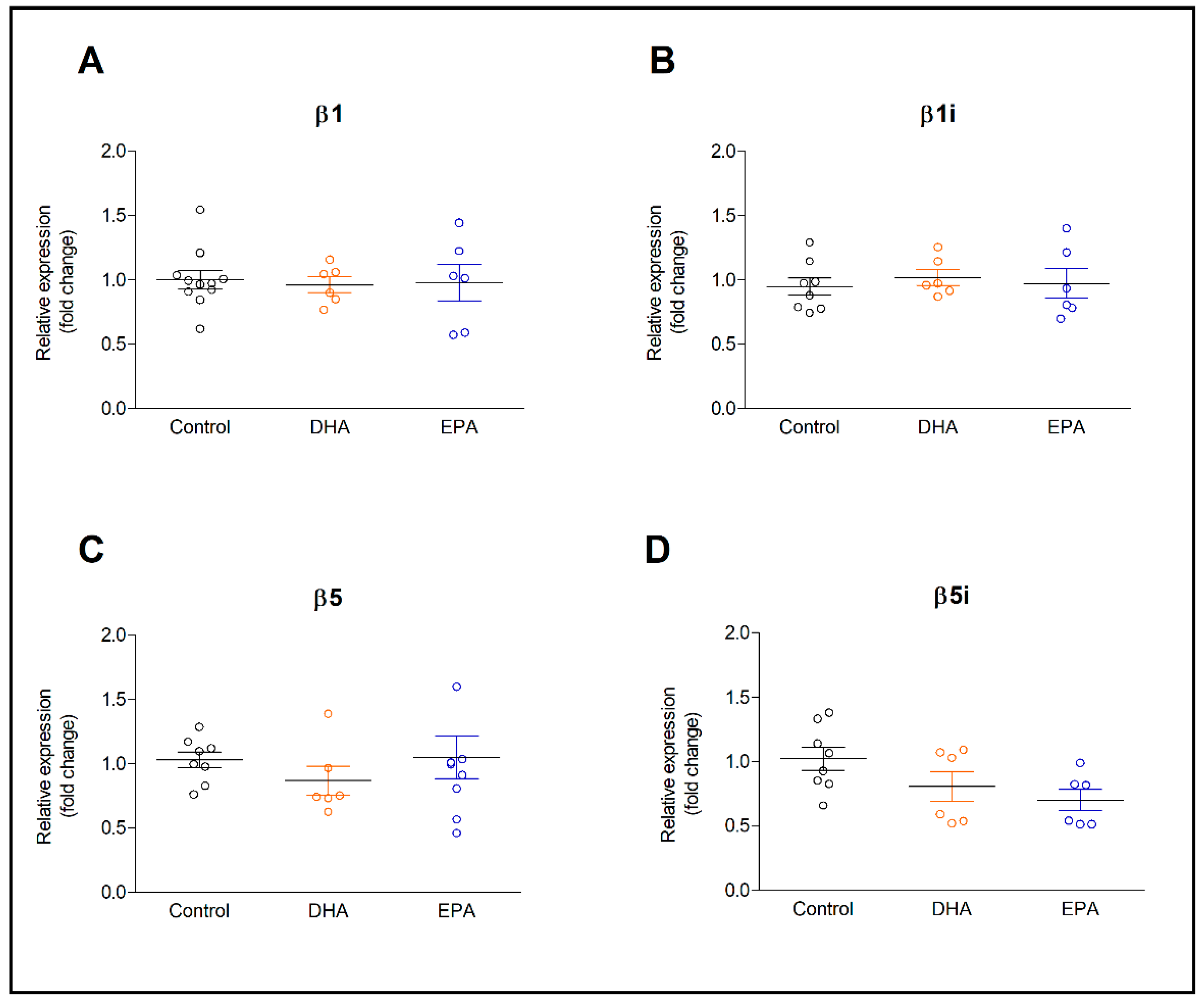
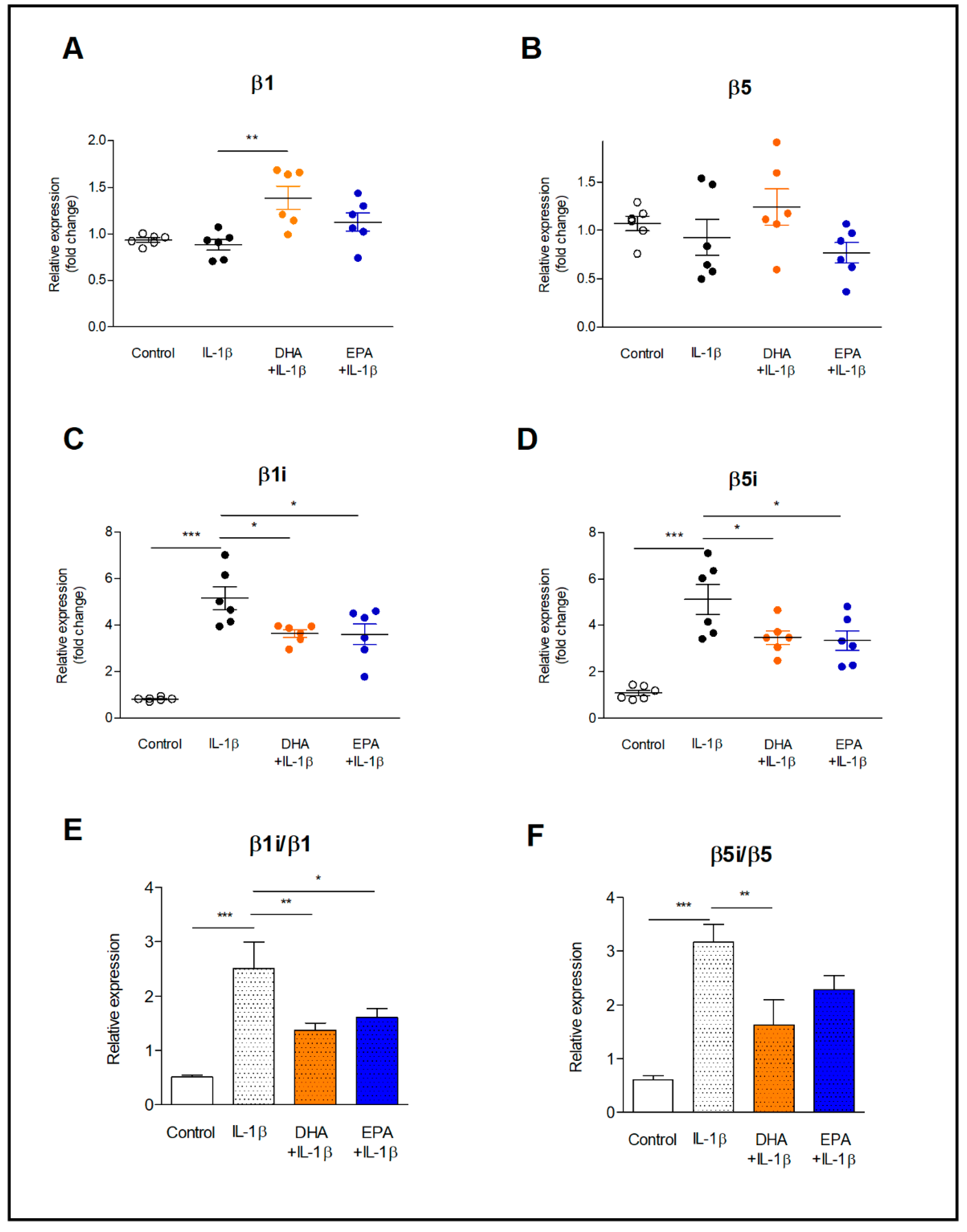
2.1. Expression of Constitutive and Inducible Proteasome Subunits in Resting and IL-1β-Activated Astrocytes
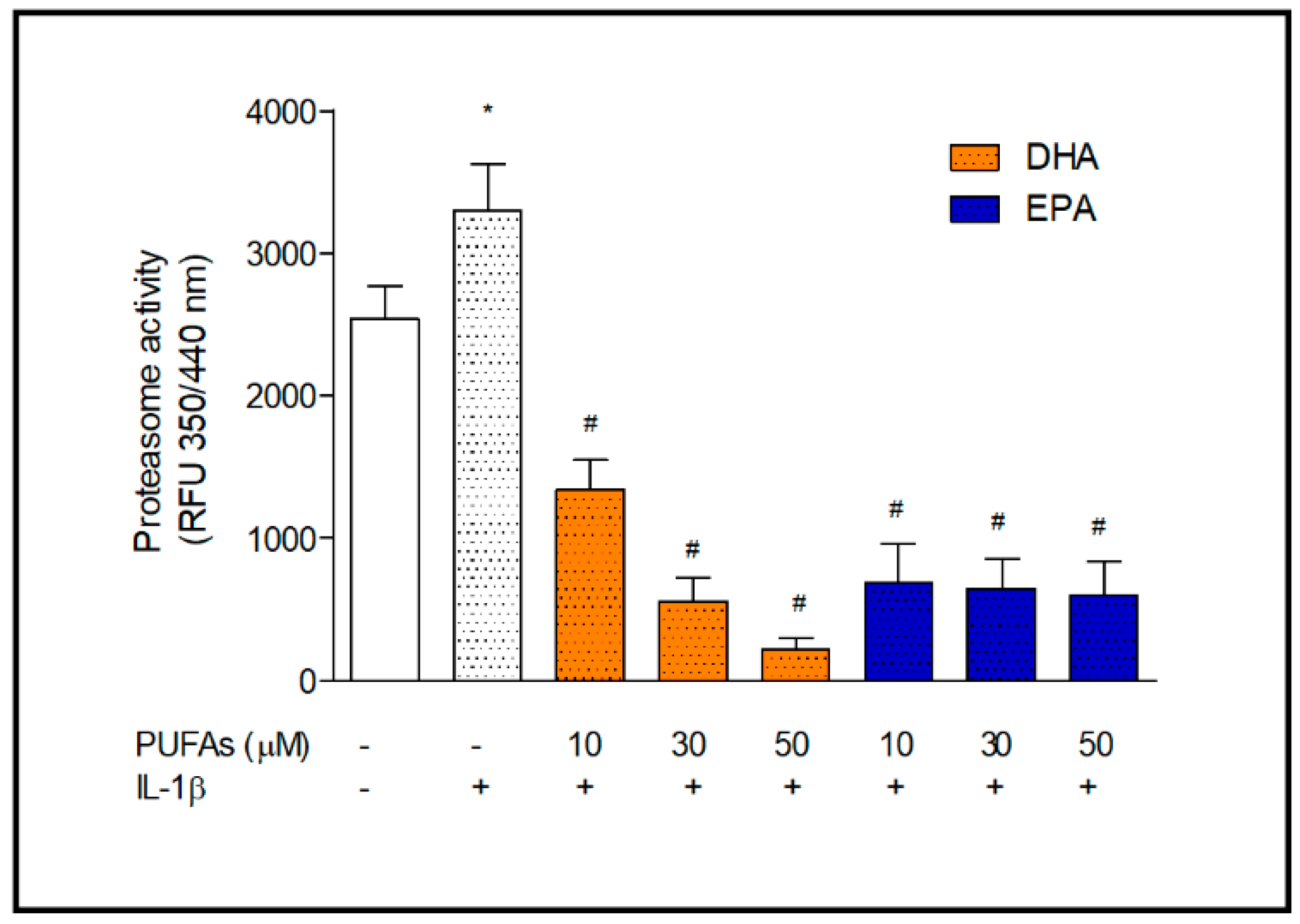
2.2. Proteasome Activity
2.3. C/EBPs Transcription Factors Activation
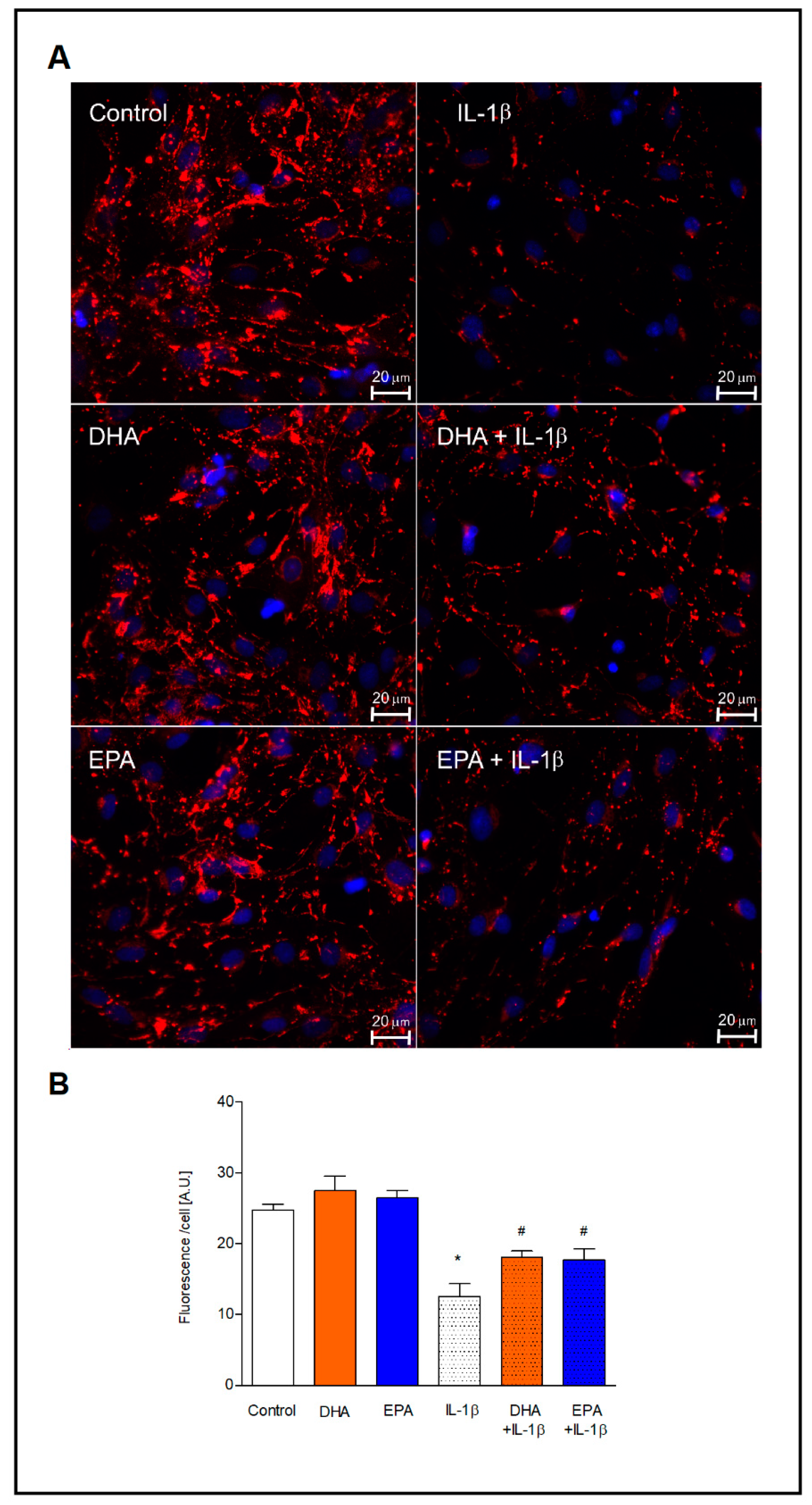
2.4. Expression of Connexin 43
3. Discussion
4. Materials and Methods
4.1. Culture of Primary Astrocytes and Treatment
4.2. RNA Isolation and Gene Expression Analysis
4.3. Proteasome Activity Assay
4.4. Determination of C/EBPs Activation
4.5. Immunofluorescence Microscopy
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krüger, E.; Kloetzel, P.M. Immunoproteasomes at the interface of innate and adaptive immune responses: Two faces of one enzyme. Curr. Opin. Immunol. 2012, 24, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012, 109, 75–112. [Google Scholar]
- Orre, M.; Kamphuis, W.; Dooves, S.; Kooijman, L.; Chan, E.T.; Kirk, C.J.; Dimayuga Smith, V.; Koot, S.; Mamber, C.; Jansen, A.H.; et al. Reactive glia show increased immunoproteasome activity in Alzheimer’s disease. Brain 2013, 136, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Ferrington, D.A.; Hussong, S.A.; Roehrich, H.; Kapphahn, R.J.; Kavanaugh, S.M.; Heuss, N.D.; Gregerson, D.S. Immunoproteasome responds to injury in the retina and brain. J. Neurochem. 2008, 106, 158–169. [Google Scholar] [CrossRef]
- Volterra, A.; Steinhäuser, C. Glial modulation of synaptic transmission in the hippocampus. Glia 2004, 47, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Parpura, V.; Scemes, E.; Spray, D.C. Mechanisms of glutamate release from astrocytes: Gap junction “hemichannels”, purinergic receptors and exocytotic release. Neurochem. Int. 2004, 45, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Liberto, C.M.; Albrecht, P.J.; Herx, L.M.; Yong, V.W.; Levison, S.W. Pro-regenerative properties of cytokine-activated astrocytes. J. Neurochem. 2004, 89, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, J.L.; Xu, L.; Foo, L.C.; Nouri, N.; Zhou, L.; Giffard, R.G.; Barres, B.A. Genomic analysis of reactive astrogliosis. J. Neurosci. 2012, 32, 6391–6410. [Google Scholar] [CrossRef] [PubMed]
- Zgórzyńska, E.; Stulczewski, D.; Dziedzic, B.; Su, K.P.; Walczewska, A. Docosahexaenoic fatty acid reduces the pro-inflammatory response induced by IL-1β in astrocytes through inhibition of NF-κB and AP-1 transcription factor activation. BMC Neurosci. 2021, 22, 4. [Google Scholar] [CrossRef]
- Zgórzyńska, E.; Dziedzic, B.; Gorzkiewicz, A.; Stulczewski, D.; Bielawska, K.; Su, K.P.; Walczewska, A. Omega-3 polyunsaturated fatty acids improve the antioxidative defense in rat astrocytes via an Nrf2-dependent mechanism. Pharmacol. Rep. 2017, 69, 935–942. [Google Scholar] [CrossRef]
- Giaume, C.; Naus, C.C. Connexins, gap junctions, and glia. WIREs Membr. Transp. Signal. 2013, 2, 133–142. [Google Scholar] [CrossRef]
- Giaume, C.; Koulakoff, A.; Roux, L.; Holcman, D.; Rouach, N. Astroglial networks: A step further in neuroglial and gliovascular interactions. Nat. Rev. Neurosci. 2010, 11, 87–99. [Google Scholar] [CrossRef]
- Antonietta Ajmone-Cat, M.; Lavinia Salvatori, M.; De Simone, R.; Mancini, M.; Biagioni, S.; Bernardo, A.; Cacci, E.; Minghetti, L. Docosahexaenoic acid modulates inflammatory and antineurogenic functions of activated microglial cells. J. Neurosci. Res. 2012, 90, 575–587. [Google Scholar] [CrossRef]
- Lu, D.Y.; Tsao, Y.Y.; Leung, Y.M.; Su, K.P. Docosahexaenoic acid suppresses neuroinflammatory responses and induces heme oxygenase-1 expression in BV-2 microglia: Implications of antidepressant effects for ω-3 fatty acids. Neuropsychopharmacology 2010, 35, 2238–2248. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.; Calderon, F.; Wen, Z.; Kim, H.Y. Docosahexaenoic acid: A positive modulator of Akt signaling in neuronal survival. Proc. Natl. Acad. Sci. USA 2005, 102, 10858–10863. [Google Scholar] [CrossRef]
- Stillwell, W.; Wassall, S.R. Docosahexaenoic acid: Membrane properties of a unique fatty acid. Chem. Phys. Lipids 2003, 26, 1–27. [Google Scholar] [CrossRef]
- Farooqui, A.A. n-3 fatty acid-derived lipid mediators in the brain: New weapons against oxidative stress and inflammation. Curr. Med. Chem. 2012, 19, 532–543. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef]
- Aki, M.; Shimbara, N.; Takashina, M.; Akiyama, K.; Kagawa, S.; Tamura, T.; Tanahashi, N.; Yoshimura, T.; Tanaka, K.; Ichihara, A. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J. Biochem. 1994, 115, 257–269. [Google Scholar] [CrossRef]
- Coux, O.; Tanaka, K.; Goldberg, A.L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996, 65, 801–847. [Google Scholar] [CrossRef]
- Basler, M.; Moebius, J.; Elenich, L.; Groettrup, M.; Monaco, J.J. An altered T cell repertoire in MECL-1-deficient mice. J. Immunol. 2006, 176, 6665–6672. [Google Scholar] [CrossRef]
- Drews, O.; Wildgruber, R.; Zong, C.; Sukop, U.; Nissum, M.; Weber, G.; Gomes, A.V.; Ping, P. Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol. Cell. Proteom. 2007, 6, 2021–2031. [Google Scholar] [CrossRef]
- Baldovino, S.; Piccinini, M.; Anselmino, A.; Ramondetti, C.; Rinaudo, M.T.; Costanzo, P.; Sena, L.M.; Roccatello, D. Structural and functional properties of proteasomes purified from the human kidney. J. Nephrol. 2006, 19, 710–716. [Google Scholar]
- Noda, C.; Tanahashi, N.; Shimbara, N.; Hendil, K.B.; Tanaka, K. Tissue distribution of constitutive proteasomes, immunoproteasomes, and PA28 in rats. Biochem. Biophys. Res. Commun. 2000, 277, 348–354. [Google Scholar] [CrossRef]
- Gimsa, U.; Mitchison, N.A.; Brunner-Weinzierl, M.C. Immune privilege as an intrinsic CNS property: Astrocytes protect the CNS against T-cell-mediated neuroinflammation. Mediat. Inflamm. 2013, 2013, 320519. [Google Scholar] [CrossRef] [PubMed]
- Moritz, K.E.; McCormack, N.M.; Abera, M.B.; Viollet, C.; Yauger, Y.J.; Sukumar, G.; Dalgard, C.L.; Burnett, B.G. The role of the immunoproteasome in interferon-γ-mediated microglial activation. Sci. Rep. 2017, 7, 9365. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Mundt, S.; Muchamuel, T.; Moll, C.; Jiang, J.; Groettrup, M.; Kirk, C.J. Inhibition of the immunoproteasome ameliorates experimental autoimmune encephalomyelitis. EMBO Mol. Med. 2014, 6, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Hernández, M.; Hernández, F.; Martín-Aparicio, E.; Gómez-Ramos, P.; Morán, M.A.; Castaño, J.G.; Ferrer, I.; Avila, J.; Lucas, J.J. Neuronal induction of the immunoproteasome in Huntington’s disease. J. Neurosci. 2003, 23, 11653–11661. [Google Scholar] [CrossRef]
- Mishto, M.; Bellavista, E.; Santoro, A.; Stolzing, A.; Ligorio, C.; Nacmias, B.; Spazzafumo, L.; Chiappelli, M.; Licastro, F.; Sorbi, S.; et al. Immunoproteasome and LMP2 polymorphism in aged and Alzheimer’s disease brains. Neurobiol. Aging. 2006, 27, 54–66. [Google Scholar] [CrossRef]
- Gavilán, M.P.; Castaño, A.; Torres, M.; Portavella, M.; Caballero, C.; Jiménez, S.; García-Martínez, A.; Parrado, J.; Vitorica, J.; Ruano, D. Age-related increase in the immunoproteasome content in rat hippocampus: Molecular and functional aspects. J. Neurochem. 2009, 108, 260–272. [Google Scholar] [CrossRef]
- Seifert, U.; Bialy, L.P.; Ebstein, F.; Bech-Otschir, D.; Voigt, A.; Schröter, F.; Prozorovski, T.; Lange, N.; Steffen, J.; Rieger, M.; et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 2010, 142, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, A.; Maekawa, Y.; Uehara, H.; Izumi, K.; Kawachi, I.; Nishizawa, M.; Toyoshima, Y.; Takahashi, H.; Standley, D.M.; Tanaka, K.; et al. A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. J. Clin. Investig. 2011, 121, 4150–4160. [Google Scholar] [CrossRef] [PubMed]
- Bö, L.; Mörk, S.; Kong, P.A.; Nyland, H.; Pardo, C.A.; Trapp, B.D. Detection of MHC class II-antigens on macrophages and microglia, but not on astrocytes and endothelia in active multiple sclerosis lesions. J. Neuroimmunol. 1994, 51, 135–146. [Google Scholar] [CrossRef]
- Kort, J.J.; Kawamura, K.; Fugger, L.; Weissert, R.; Forsthuber, T.G. Efficient presentation of myelin oligodendrocyte glycoprotein peptides but not protein by astrocytes from HLA-DR2 and HLA-DR4 transgenic mice. J. Neuroimmunol. 2006, 173, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Rostami, J.; Fotaki, G.; Sirois, J.; Mzezewa, R.; Bergström, J.; Essand, M.; Healy, L.; Erlandsson, A. Astrocytes have the capacity to act as antigen-presenting cells in the Parkinson’s disease brain. J. Neuroinflammation 2020, 17, 119. [Google Scholar] [CrossRef]
- Krumm, B.; Xiang, Y.; Deng, J. Structural biology of the IL-1 superfamily: Key cytokines in the regulation of immune and inflammatory responses. Protein Sci. 2014, 23, 526–538. [Google Scholar] [CrossRef]
- James, A.B.; Conway, A.M.; Morris, B.J. Regulation of the neuronal proteasome by Zif268 (Egr1). J. Neurosci. 2006, 26, 1624–1634. [Google Scholar] [CrossRef]
- Zanelli, E.; Zhou, P.; Cao, H.; Smart, M.K.; David, C.S. Genomic organization and tissue expression of the mouse proteasome gene Lmp-7. Immunogenetics 1993, 38, 400–407. [Google Scholar] [CrossRef]
- Lekstrom-Himes, J.; Xanthopoulos, K.G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 1998, 273, 28545–28548. [Google Scholar] [CrossRef]
- Vinson, C.R.; Hai, T.; Boyd, S.M. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: Prediction and rational design. Genes Dev. 1993, 7, 1047–1058. [Google Scholar] [CrossRef]
- Xu, H.; Fu, J.; Ha, S.W.; Ju, D.; Zheng, J.; Li, L.; Xie, Y. The CCAAT box-binding transcription factor NF-Y regulates basal expression of human proteasome genes. Biochim. Biophys. Acta 2012, 1823, 818–825. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alam, T.; An, M.R.; Papaconstantinou, J. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem. 1992, 267, 5021–5024. [Google Scholar] [CrossRef]
- Akira, S.; Isshiki, H.; Sugita, T.; Tanabe, O.; Kinoshita, S.; Nishio, Y.; Nakajima, T.; Hirano, T.; Kishimoto, T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990, 9, 1897–1906. [Google Scholar] [CrossRef] [PubMed]
- Nerlov, C. C/EBPs: Recipients of extracellular signals through proteome modulation. Curr. Opin. Cell Biol. 2008, 20, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Kowenz-Leutz, E.; Xu, H.; Leutz, A. Ras induces mediator complex exchange on C/EBP beta. Mol. Cell 2004, 13, 241–250. [Google Scholar] [CrossRef]
- Li, X.; Kim, J.W.; Grønborg, M.; Urlaub, H.; Lane, M.D.; Tang, Q.Q. Role of cdk2 in the sequential phosphorylation/activation of C/EBPbeta during adipocyte differentiation. Proc. Natl. Acad. Sci. USA 2007, 104, 11597–11602. [Google Scholar] [CrossRef]
- Mahoney, C.W.; Shuman, J.; McKnight, S.L.; Chen, H.C.; Huang, K.P. Phosphorylation of CCAAT-enhancer binding protein by protein kinase C attenuates site-selective DNA binding. J. Biol. Chem. 1992, 267, 19396–19403. [Google Scholar] [CrossRef]
- Nakajima, T.; Kinoshita, S.; Sasagawa, T.; Sasaki, K.; Naruto, M.; Kishimoto, T.; Akira, S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 1993, 90, 2207–2211. [Google Scholar] [CrossRef]
- Wegner, M.; Cao, Z. Rosenfeld MG. Calcium-regulated phosphorylation within the leucine zipper of C/EBP beta. Science 1992, 256, 370–373. [Google Scholar] [CrossRef]
- Tang, Q.Q. Grønborg, M.; Huang, H.; Kim, J.W.; Otto, T.C.; Pandey, A.; Lane, M.D. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 9766–9771. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.L.; Zuo, X.; Yang, F. Ubeda, O.J.; Gant, D.J.; Alaverdyan, M.; Kiosea, N.C.; Nazari, S.; Chen, P.P.; Nothias, F.; et al. Loss of MAP function leads to hippocampal synapse loss and deficits in the Morris Water Maze with aging. J. Neurosci. 2014, 34, 7124–7136. [Google Scholar] [CrossRef] [PubMed]
- Scemes, E.; Spray, D.C. The astrocytic syncytium. Adv. Mol. Cell Biol. 2003, 31, 165–179. [Google Scholar]
- Giaume, C.; McCarthy, K.D. Control of gap-junctional communication in astrocytic networks. Trends Neurosci. 1996, 19, 319–325. [Google Scholar] [CrossRef]
- Baum, J.R.; Dolmatova, E.; Tan, A.; Duffy, H.S. Omega 3 fatty acid inhibition of inflammatory cytokine-mediated Connexin43 regulation in the heart. Front. Physiol. 2012, 3, 272. [Google Scholar] [CrossRef]
- Champeil-Potokar, G.; Hennebelle, M.; Latour, A.; Vancassel, S.; Denis, I. Docosahexaenoic acid (DHA) prevents corticosterone-induced changes in astrocyte morphology and function. J. Neurochem. 2016, 136, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Thévenin, A.F.; Kowal, T.J.; Fong, J.T.; Kells, R.M.; Fisher, C.G.; Falk, M.M. Proteins and mechanisms regulating gap-junction assembly, internalization, and degradation. Physiology 2013, 28, 93–116. [Google Scholar] [CrossRef] [PubMed]
- Solan, J.L.; Lampe, P.D. Specific Cx43 phosphorylation events regulate gap junction turnover in vivo. FEBS Lett. 2014, 588, 1423–1429. [Google Scholar] [CrossRef]
- Mirnikjoo, B.; Brown, S.E.; Kim, H.F.; Marangell, L.B.; Sweatt, J.D.; Weeber, E.J. Protein kinase inhibition by omega-3 fatty acids. J. Biol. Chem. 2001, 276, 10888–10896. [Google Scholar] [CrossRef]
- Seung Kim, H.F.; Weeber, E.J.; Sweatt, J.D.; Stoll, A.L.; Marangell, L.B. Inhibitory effects of omega-3 fatty acids on protein kinase C activity in vitro. Mol. Psychiatry 2001, 6, 246–248. [Google Scholar] [CrossRef][Green Version]
- Lampe, P.D.; TenBroek, E.M.; Burt, J.M.; Kurata, W.E.; Johnson, R.G.; Lau, A.F. Phosphorylation of connexin43 on serine368 by protein kinase C regulates gap junctional communication. J. Cell Biol. 2000, 149, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Cone, A.C.; Cavin, G.; Ambrosi, C.; Hakozaki, H.; Wu-Zhang, A.X.; Kunkel, M.T.; Newton, A.C.; Sosinsky, G.E. Protein kinase Cδ-mediated phosphorylation of Connexin43 gap junction channels causes movement within gap junctions followed by vesicle internalization and protein degradation. J. Biol. Chem. 2014, 289, 8781–8798. [Google Scholar] [CrossRef] [PubMed]
- Laing, J.G.; Beyer, E.C. The gap junction protein connexin43 is degraded via the ubiquitin proteasome pathway. J. Biol. Chem. 1995, 270, 26399–26403. [Google Scholar] [CrossRef]
- Liao, C.K.; Jeng, C.J.; Wang, H.S.; Wang, S.H.; Wu, J.C. Lipopolysaccharide induces degradation of connexin43 in rat astrocytes via the ubiquitin-proteasome proteolytic pathway. PLoS ONE 2013, 8, e79350. [Google Scholar] [CrossRef] [PubMed]
- Claud, E.C.; McDonald, J.A.; He, S.M.; Yu, Y.; Duong, L.; Sun, J.; Petrof, E.O. Differential expression of 26S proteasome subunits and functional activity during neonatal development. Biomolecules 2014, 4, 812–826. [Google Scholar] [CrossRef]
- Shanley, K.L.; Hu, C.L.; Bizzozero, O.A. Proteasome Composition in Cytokine-Treated Neurons and Astrocytes is Determined Mainly by Subunit Displacement. Neurochem. Res. 2020, 45, 860–871. [Google Scholar] [CrossRef]

| Subunit | Sequence |
|---|---|
| β1 | Forward 5′-CTTATGCCTTCAACGGAGGT-3′ |
| Reverse 5′-GTGTCTGAAGCAACGATGGA-3′ | |
| β1i | Forward 5′-GACGGGAGAAGTCCACACC-3′ |
| Reverse 5′-ATCAGAGCCCACCACGAC-3′ | |
| β2 | Forward 5′-GAGGGCAGTGGAGCTTCTTA-3′ |
| Reverse 5′-AGGTGGGCAGATTCAAGATG-3′ | |
| β2i | Forward 5′-TTCCAGCCAAACATGACG-3′ |
| Reverse 5′-AGTGATCACACAGGCATCCA-3′ | |
| β5 | Forward 5′-CTCCAAACTGCTTGCCAAC-3′ |
| Reverse 5′-CCTGTTCCCCTCACTGTCTA-3′ | |
| β5i | Forward 5′-CGCAGGAAGTTACATTGCTAC-3′ |
| Reverse 5′-CCATTCCGCAGATAGTATAGCC-3′ | |
| GAPDH | Forward 5′-TGACAACTTTGGCATCGTGG-3′ |
| Reverse 5′-TACTCCTTGGAGGCCATGT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zgorzynska, E.; Dziedzic, B.; Markiewicz, M.; Walczewska, A. Omega-3 PUFAs Suppress IL-1β-Induced Hyperactivity of Immunoproteasomes in Astrocytes. Int. J. Mol. Sci. 2021, 22, 5410. https://doi.org/10.3390/ijms22115410
Zgorzynska E, Dziedzic B, Markiewicz M, Walczewska A. Omega-3 PUFAs Suppress IL-1β-Induced Hyperactivity of Immunoproteasomes in Astrocytes. International Journal of Molecular Sciences. 2021; 22(11):5410. https://doi.org/10.3390/ijms22115410
Chicago/Turabian StyleZgorzynska, Emilia, Barbara Dziedzic, Monika Markiewicz, and Anna Walczewska. 2021. "Omega-3 PUFAs Suppress IL-1β-Induced Hyperactivity of Immunoproteasomes in Astrocytes" International Journal of Molecular Sciences 22, no. 11: 5410. https://doi.org/10.3390/ijms22115410
APA StyleZgorzynska, E., Dziedzic, B., Markiewicz, M., & Walczewska, A. (2021). Omega-3 PUFAs Suppress IL-1β-Induced Hyperactivity of Immunoproteasomes in Astrocytes. International Journal of Molecular Sciences, 22(11), 5410. https://doi.org/10.3390/ijms22115410








