Using Nearest-Neighbor Distributions to Quantify Machine Learning of Materials’ Microstructures
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental System and Imaging
2.2. U-Net Implementation
2.3. Methodology for Nearest-Neighbor Analysis
2.4. Learning Functions
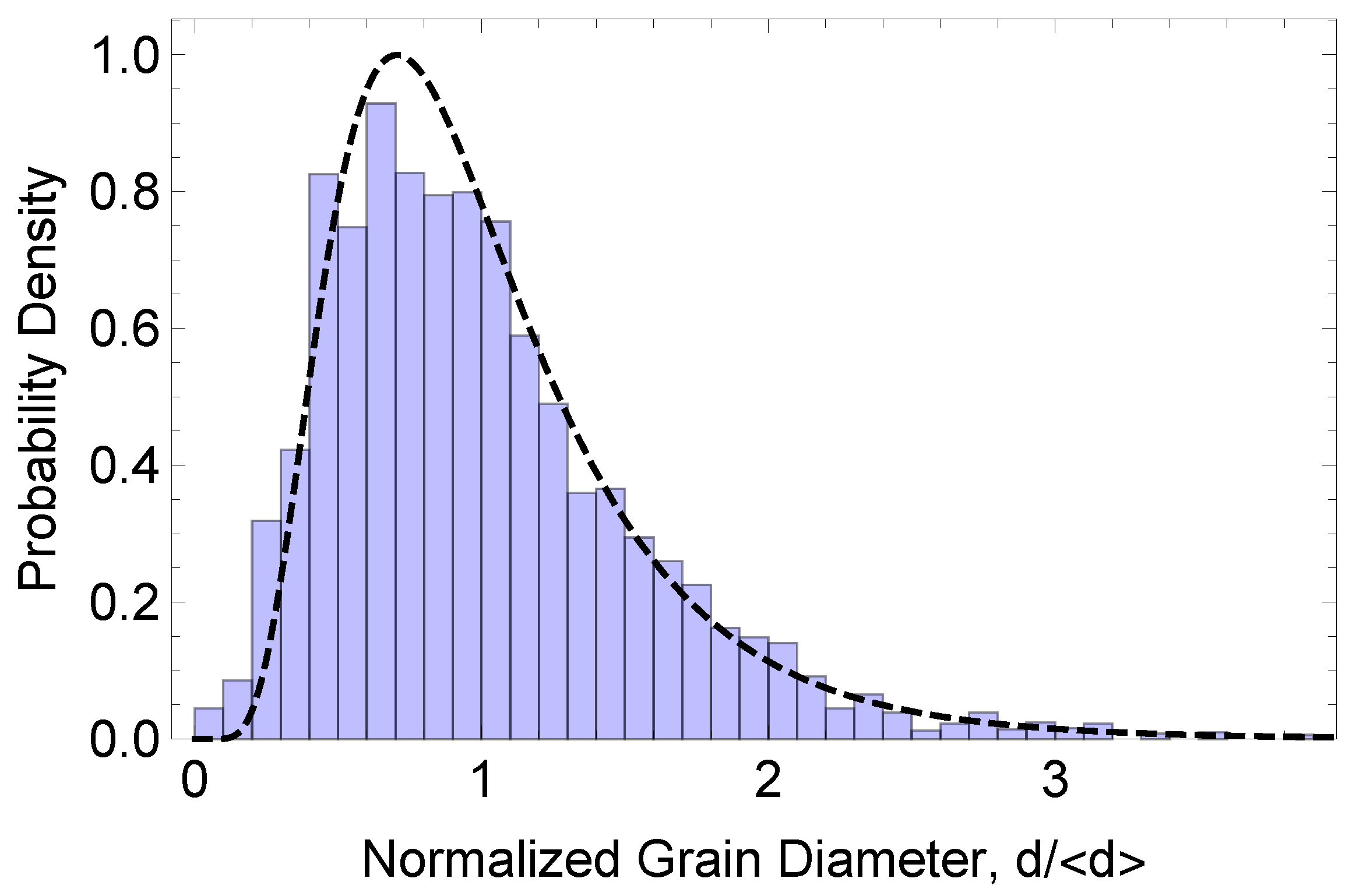
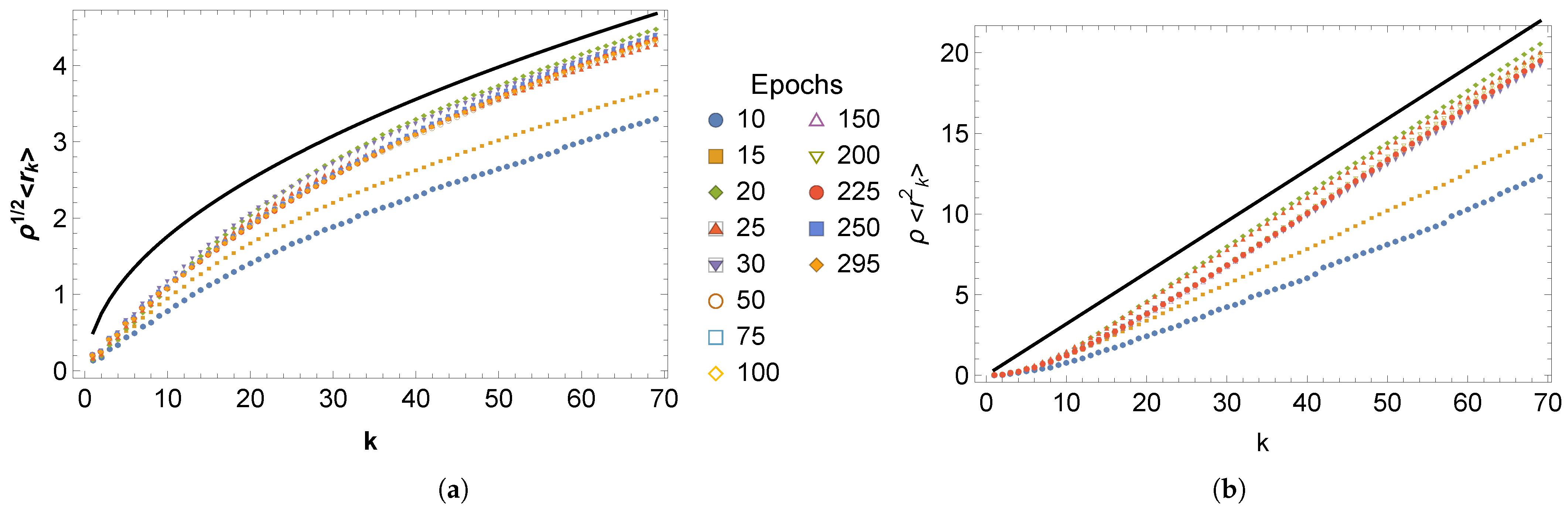
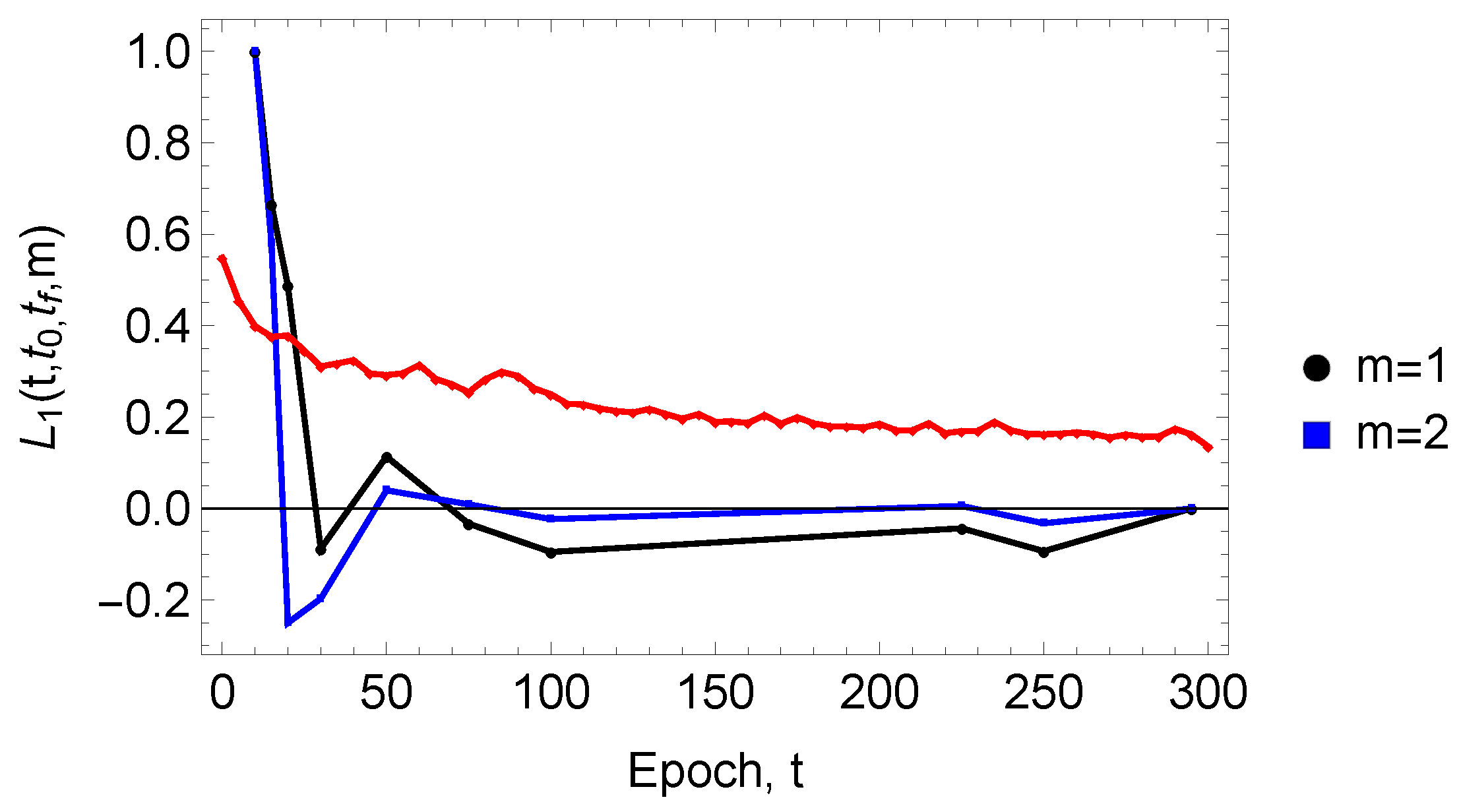
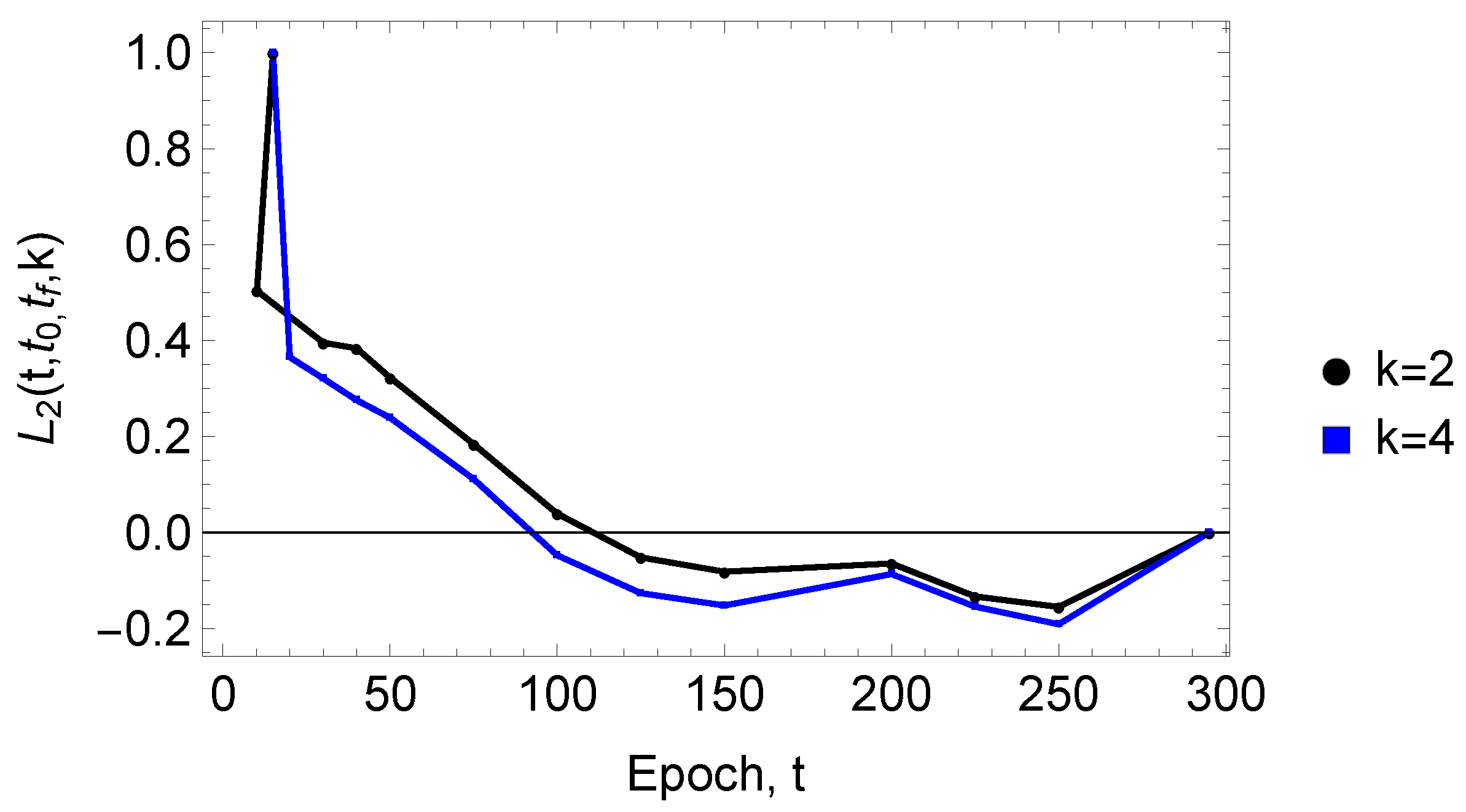
3. Results
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hansen, N. Hall-Petch relation and boundary strengthening. Scr. Mater. 2004, 51, 801–806. [Google Scholar] [CrossRef]
- Hanaor, D.A.H.; Xu, W.; Ferry, M.; Sorrell, C.C. Abnormal grain growth of rutile TiO2 induced by ZrSiO4. J. Cryst. Growth 2012, 359, 83–91. [Google Scholar] [CrossRef]
- Zhu, M.-L.; Xuan, F.-Z. Correlation between microstructure, hardness and strength in HAZ of dissimilar welds of rotor steels. Mat. Sci. Eng. A 2010, 527, 4035–4042. [Google Scholar] [CrossRef]
- Yoo, E.; Moon, J.H.; Sang, Y.; Kim, Y.; Ahn, J.-P.; Kim, Y.K. Electrical resistivity and microstructural evolution of electrodeposited Co and Co-W nanowires. Mater. Charact. 2020, 166, 110451. [Google Scholar] [CrossRef]
- Barmak, K.; Lu, X.; Darbal, A.; Nuhfer, N.T.; Choi, D.; Sun, T.; Warren, A.P.; Coffey, K.R.; Toney, M.F. On twin density and resistivity of nanometric Cu thin films. J. Appl. Phys. 2016, 120, 065106. [Google Scholar] [CrossRef]
- Bedu-Amissah, K.; Rickman, J.M.; Chan, H.M.; Harmer, M.P. Grain-boundary diffusion of Cr in pure and Y-doped alumina. J. Am. Ceram. Soc. 2007, 90, 1551–1555. [Google Scholar] [CrossRef]
- Wang, C.-M.; Cho, J.; Chan, H.M.; Harmer, M.P.; Rickman, J.M. Influence of dopant concentration on creep properties of Nd2O3-doped alumina. J. Am. Ceram. Soc. 2001, 84, 1010–1016. [Google Scholar] [CrossRef]
- Cho, J.; Wang, C.M.; Chan, H.M.; Rickman, J.M.; Harmer, M.P. Improved tensile creep properties of yttrium- and lanthanum-doped alumina: A solid solution effect. J. Mater. Res. 2001, 16, 425–429. [Google Scholar] [CrossRef]
- Carpenter, D.T.; Rickman, J.M.; Barmak, K. A methodology for automated quantitative microstructural analysis of transmission electron micrographs. J. Appl. Phys. 1998, 84, 5843. [Google Scholar] [CrossRef]
- Patrick, M.J.; Eckstein, J.K.; Lopez, J.R.; Toderas, S.; Levine, S.; Rickman, J.M.; Barmak, K. Automated grain boundary detection for bright-field transmission electron microscopy images via U-Net. Microsc. Microanal. 2023, 29, 1968–1979. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.J.; Eckstein, J.K.; Lopez, J.R.; Toderas, S.; Levine, S.; Barmak, K. U-Net implementation for high throughput grain boundary detection in bright field TEM micrographs: Toward in situ grain growth studies. Microsc. Microanal. 2023, 29 (Suppl. S1), 1581–1582. [Google Scholar] [CrossRef]
- Williams, D.B.; Carter, C.B. Transmission Electron Microscopy: A Textbook for Materials Science, 2nd ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- Jocher, G.; Chaurasia, A.; Qiu, J. Ultralytics YOLOv8, version 8.0.0; Ultralytics: San Francisco, CA, USA, 2023. Available online: https://github.com/ultralytics/ultralytics (accessed on 9 May 2025).
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Via del Mar, Chile, 27–29 October 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Ciampiconi, L.; Elwood, A.; Leonardi, M.; Mohamed, A.; Rozza, A. A survey and taxonomy of loss functions in machine learning. arXiv 2023, arXiv:2301.05579. [Google Scholar]
- Barmak, K.; Rickman, J.M.; Patrick, M.J. Advances in experimental studies of grain growth in thin films. JOM 2024, 76, 3622–3636. [Google Scholar] [CrossRef]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-net and its variants for medical image segmentation: A review of theory and applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention (MICCAI); Springer: Berlin/Heidelberg, Germany, 2015; Volume 9351, pp. 234–241. [Google Scholar]
- Gao, B.; Pavel, L. On the properties of the softmax function with application in game theory and reinforcement learning. arXiv 2017, arXiv:1704.00805. Available online: https://arxiv.org/pdf/1704.00805 (accessed on 14 May 2025).
- Diggle, P.J. Statistical Analysis of Spatial and Spatio-Temporal Point Patterns; CRC Press: New York, NY, USA, 2014. [Google Scholar]
- van Kampen, N.G. Stochastic Processes in Physics and Chemistry, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Banerjee, A.; Abel, T. Nearest neighbour distributions: New statistical measures for cosmological clustering. Mon. Not. R. Astron. Soc. 2021, 500, 5479–5499. [Google Scholar] [CrossRef]
- Arfken, G.B.; Weber, H.J. Mathematical Methods for Physicists, 6th ed.; Academic Press: New York, NY, USA, 2005. [Google Scholar]
- Luscombe, J.H. Thermodynamics; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Schilling, M.F. Multivariate two-sample tests based on nearest neighbors. J. Am. Stat. Assoc. 1986, 81, 799–806. [Google Scholar] [CrossRef]
- Friedman, J.H.; Steppel, S. A Nonparametric Procedure for Comparing Multivariate Point Sets; SLAC Computation Group (Internal) Technical Memo 153 [U.S. Atomic Energy Contract AT(043)515]; Stanford University: Stanford, CA, USA, 1974. [Google Scholar]
- Fogolari, F.; Borelli, R.; Dovier, A.; Esposito, G. The kth nearest neighbor method for estimation of entropy changes from molecular ensembles. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2024, 14, e1691. [Google Scholar] [CrossRef]
- Li, S.; Mnatsakanov, R.M.; Andrew, M.E. k-nearest neighbor based consistent entropy estimation for hyperspherical distributions. Entropy 2011, 13, 650–667. [Google Scholar] [CrossRef]
- Cover, T.M.; Thomas, J.A. Elements of Information Theory; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Zhao, P.; Lai, L. Analysis of k nearest neighbor KL divergence estimation for continuous distributions. In Proceedings of the 2020 IEEE International Symposium on Information Theory (ISIT), Los Angeles, CA, USA, 21–26 June 2020; pp. 2562–2567. [Google Scholar]
- Miyagawa, M. An approximation for the kth nearest distance and its application to locational analysis. J. Oper. Res. Soc. Jpn. 2012, 55, 146–157. [Google Scholar] [CrossRef][Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rickman, J.M.; Barmak, K.; Patrick, M.J.; Mensah, G.A. Using Nearest-Neighbor Distributions to Quantify Machine Learning of Materials’ Microstructures. Entropy 2025, 27, 536. https://doi.org/10.3390/e27050536
Rickman JM, Barmak K, Patrick MJ, Mensah GA. Using Nearest-Neighbor Distributions to Quantify Machine Learning of Materials’ Microstructures. Entropy. 2025; 27(5):536. https://doi.org/10.3390/e27050536
Chicago/Turabian StyleRickman, Jeffrey M., Katayun Barmak, Matthew J. Patrick, and Godfred Adomako Mensah. 2025. "Using Nearest-Neighbor Distributions to Quantify Machine Learning of Materials’ Microstructures" Entropy 27, no. 5: 536. https://doi.org/10.3390/e27050536
APA StyleRickman, J. M., Barmak, K., Patrick, M. J., & Mensah, G. A. (2025). Using Nearest-Neighbor Distributions to Quantify Machine Learning of Materials’ Microstructures. Entropy, 27(5), 536. https://doi.org/10.3390/e27050536





