1. Introduction
Gaussian processes are powerful and flexible statistical models that have gained significant popularity in different fields: signal processing, medical imaging, data science, machine learning, econometrics, shape analysis, etc. [
1,
2,
3,
4,
5]. They provide a nonparametric approach for modeling complex relationships and uncertainty estimation in data [
6]. The core idea of Gaussian processes is the assumption that any finite set of data points can be jointly modeled as a multivariate Gaussian distribution [
7]. Rather than explicit formulations, Gaussian processes allow for the incorporation of prior knowledge and inference of a nonparametric function
f that generates the Gaussian process for a set
with
;
and noisy measurements
. If
f is modeled with a Gaussian process prior, then it can be fully characterized by a mean
and a covariance function
k, satisfying
The mean function is usually assumed to be zero (
), whereas the covariance
provides the dependence between two inputs,
t and
s. Gaussian processes can be applied for various tasks, including regression [
8], classification [
9], and time series analysis [
10]. In regression, Gaussian processes can capture complex and non-linear patterns in data, while, in classification, they enable probabilistic predictions and can handle imbalanced datasets [
11]. Additionally, Gaussian processes have been successfully employed in optimization, experimental design, reinforcement learning, and more [
12].
One of the key advantages of Gaussian processes is their ability to provide a rich characterization of uncertainty. This makes them particularly suitable for applications where robust uncertainty quantification is crucial, such as in decision-making processes or when dealing with limited or noisy data. Significant efforts have been dedicated to the development of asymptotically efficient or approximate computational methods for modeling with Gaussian processes. However, Gaussian processes may also suffer from some computational challenges. When the number of observations
N increases, the computational complexity for inference and learning grows significantly and incurs an
computational cost, which is unfeasible for many modern problems [
13]. Another limitation of Gaussian processes is the memory scaling
in a direct implementation. To address these issues, various approximations and scalable algorithms, such as sparse Gaussian processes [
14,
15] and variational inference [
16], have been developed to make Gaussian processes applicable to larger datasets.
Certain approximations, as demonstrated in [
17,
18], involve reduced-rank Gaussian processes that rely on approximating the covariance function. For example, Ref. [
19] addressed the computational challenge of working with large-scale datasets by approximating the covariance matrix, which is often required for computations involving kernel methods. In addition, Ref. [
20] proposed an FFT-based method for stationary covariances as a technique that leverages the Fast Fourier Transform (FFT) to efficiently compute and manipulate covariance functions in the frequency domain. The link between state space models (SSMs) and Gaussian processes was explored in [
21]. This could avoid the cubic complexity in time using Kalman filtering inference methods [
22]. Recently, Ref. [
23] presented a novel method for approximating covariance functions as an eigenfunction expansion of the Laplace operator defined on a compact domain. More recently, Ref. [
24] introduced a reduced-rank algorithm for Gaussian process regression with a numerical scheme.
In this paper, we consider a specific Karhunen–Loève expansion of a Gaussian process with many advantages over other low-rank compression techniques [
25]. Initially, we express a Gaussian process as a series of basis functions and random coefficients. By selecting a subset of the most vital basis functions based on the dominant eigenvalues, the rank of the Gaussian process can be reduced. This proves especially advantageous when working with extensive datasets as it can alleviate computational and storage demands. By scrutinizing the eigenvalues linked to the eigenfunctions, one can gauge each eigenfunction’s contribution to noise. This insight can be used for noise modeling, estimation, and separation. The Karhunen–Loève expansion offers a natural framework for model selection and regularization in Gaussian process modeling. By truncating the decomposition to a subset of significant eigenfunctions, we can prevent overfitting. This regularization has the potential to enhance the Gaussian process generalization capability and mitigate the influence of noise or irrelevant features.
In contrast to conventional Karhunen–Loève expansions where eigenfunctions are derived directly from the covariance function or the integral operator, our approach involves differential operators in which the corresponding orthogonal polynomials serve as eigenfunctions. This choice holds significant importance because polynomials are tailored to offer numerical stability and good conditioning, resulting in more precise and stable computations, particularly when dealing with rounding errors. Additionally, orthogonal polynomials frequently possess advantageous properties for integration and differentiation. These properties streamline efficient calculations involving interpolated functions, making them exceptionally valuable for applications necessitating complex computations. On the whole, decomposing Gaussian processes using orthogonal polynomials provides benefits such as numerical stability, quicker convergence, and accurate approximations [
26]. However, their incorporation within the Gaussian process framework of the machine learning community has been virtually non-existent. The existing research predominantly revolves around the analysis of integral operators and numerical approximations to compute Karhunen–Loève expansions.
The paper is structured as follows. The first three sections review the necessary theoretical foundations:
Section 2 introduces operators in Hilbert spaces, the expansion theorem, and the general Karhunen–Loève theorem;
Section 3 focuses on the Gaussian process case; and
Section 4 highlights the challenges of canonical Gaussian process regression.
Section 5 explores low-complexity Gaussian processes and their computational advantages.
Section 6 presents the proposed solutions for various differential operators using orthogonal polynomial bases. Finally,
Section 7 discusses the experimental results, followed by a comprehensive discussion and conclusion in
Section 8.
2. Operators on Hilbert Spaces
In this section, we recall and prove some useful results about linear compact symmetric operators on the particular
Hilbert space. More general results of any Hilbert space
H are moved to
Appendix A.
Let be a probability space, and let X and Y be second-order real-valued random variables, meaning and . By the Cauchy–Schwarz inequality, , allowing us to center the variables, assuming without loss of generality that they have zero mean. Thus, is the variance of X and is the covariance of X and Y. The set of these random variables forms a Hilbert space with inner product and norm , leading to mean square convergence: converges in norm to X if ; equivalently, as . X and Y are orthogonal, written , if . If , then . For mutually orthogonal variables , we have .
Lemma 1. Let , be a real-valued second-order random process with zero mean and covariance function . Then, the covariance k is a symmetric non-negative definite function.
Proof. First, by definition of
, its symmetry is trivial. Next, it holds that, for all possible choices of
,
and all possible functions
, we have
Thus,
k is non-negative definite. □
In this section, we are interested in the Hilbert space
, the space of all the real-valued Borel-measurable functions
on the interval
such that
with a positive weight function
, which inherits all proprieties from
Appendix A. Consider the Hilbert–Schmidt integral operator
, expressed as
Theorem 1 (Mercer’s Theorem)
. Let be continuous symmetric non-negative definite and let be the corresponding Hilbert–Schmidt operator. Let be an orthonormal basis for the space spanned by the eigenvectors corresponding to the non-zero eigenvalues of . If is the eigenvector corresponding to the eigenvalue , thenwhere - (i)
the series converges absolutely in both variables jointly.
- (ii)
the series converges to uniformly in both variables jointly.
- (iii)
the series converges to in .
Theorem 2 (Karhunen–Loève Theorem)
. For a real-valued second-order random process with zero mean and continuous covariance function on , we can decompose each aswhere - (i)
are eigenfunctions of , which form an orthonormal basis, i.e., , where is the Kronecker’s delta.
- (ii)
is the coefficient given by the projection of onto the j-th deterministic element of the Karhunen–Loève basis in .
- (iii)
are pairwise orthogonal random variables with zero mean and variance , corresponding to the eigenvalue of the eigenfunction .
Moreover, the series converges to in mean square uniformly for all .
Proof. We have
and
due to orthonormality of
. Thus,
are pairwise orthogonal in
and
. To show the mean square convergence, let
and
. Then,
uniformly in
by Mercer’s theorem. □
Definition 1. The mean square error is defined as . The mean integrated square error (MISE), denoted by , is then given by , representing the mean square error integrated over the basis onto which is projected for every .
Proposition 1. The MISE tends to 0 as .
Proof. The MISE of
satisfies
which tends to 0 as
since
are absolutely summable. □
Now, we highlight the crucial role of the Karhunen–Loève basis in minimizing the error incurred by truncating the expansion of . By aligning the basis functions with the dominant modes of variation captured by the process, we achieve an optimal representation in terms of mean integrated square error.
Proposition 2. The MISE is minimized if and only if constitutes an orthonormalization of the eigenfunctions of the Fredholm equationwith arranged to correspond to the eigenvalues in decreasing magnitude: . 3. Expansion and Convergence of Gaussian Processes
A stochastic process is said to be a Gaussian process (GP) if for all positive integers and all choices of the random variables form a Gaussian random vector, which means that they are jointly Gaussian. One of the main advantages of a GP is that it can be represented as a series expansion involving a complete set of deterministic basis functions with corresponding random Gaussian coefficients.
Theorem 3. If , is a zero-mean GP of covariance denoted , then the Karhunen–Loève expansion projections are independent Gaussian random variables: .
Now, we provide a result that establishes important convergence properties, connecting mean square convergence, convergence in probability, and convergence almost surely for sequences of random variables.
Lemma 2. 1. The mean square convergence of any sequence of real-valued random variables implies its convergence in probability.
- 2.
If is a sequence of independent real-valued random variables, then the convergence of the series in probability implies its convergence almost surely.
Corollary 1. For each , converges to almost surely.
Proof. Since are independent, are also independent because the eigenfunctions are deterministic. Thus, Lemma 2 can be applied to the series of independent random variables . This means that its convergence in probability implies almost sure convergence. Therefore, we only need to prove convergence in probability. This argument is straightforward because, by Lemma 2, convergence in mean square implies convergence in probability. The mean square convergence of the partial sums is a result of the Karhunen–Loève theorem. □
Example 1. 1. The Karhunen–Loève expansion of the Brownian motion on as a centered GP with covariance is given bywhere and . - 2.
The Karhunen–Loève expansion of the Brownian bridge on as a centered GP with covariance is given by where and .
4. Ill-Conditioned Canonical Gaussian Process Regression
In a regression task, a nonparametric function
f is assumed to be a realization of a stochastic GP prior, whereas the likelihood term holds from observations corrupted by a noise term according to the canonical form
where
is a Gaussian noise. Given a training dataset
, the posterior distribution over
is also Gaussian:
. From Bayes’ rule, we state that the mean and the covariance posterior are expressed as
where
is the prior covariance matrix and
is the
identity matrix. The predictive distribution at any test input
can be computed in closed form as
with
where
.
The covariance function
k usually depends on a set of hyperparameters denoted
that need to be estimated from the training dataset. The log marginal likelihood for GP regression serves as an indicator of the degree to which the selected model accurately captures the observed patterns. The log marginal likelihood is typically used for model selection and optimization. Let
denote the set of all model parameters, and then the log marginal likelihood
is given by
Here,
denotes the determinant. The goal is to estimate
that maximizes the log marginal likelihood. This can be achieved using different methods, such as gradient-based algorithm [
27]. The weakness of inferring the posterior mean or the mean prediction or even learning the hyperparameters from the log marginal likelihood is the need to invert the
Gram matrix
. This operation costs
, which limits the applicability of standard GPs when the sample size
N increases significantly. Furthermore, the memory requirements for GP regression scale with a computational complexity of
.
A covariance function
is said to be stationary (isotropic) if it is invariant to translation, i.e., a function of
only. Two commonly used stationary covariance functions for GP regression are the Squared Exponential (SE) and Matérn-
kernels defined by
respectively, where
is the variance parameter controlling the amplitude of the covariance,
is the shape parameter, and
;
is the half integer smoothness parameter controlling its differentiability. Here,
is the gamma function and
is the modified Bessel function of the second kind. Both the SE and Matérn covariance functions have hyperparameters that needs to be estimated from the data during the model training process. A GP with a Matérn-
covariance is
times differentiable in the mean-square sense. The SE covariance is the limit of Matérn-
as the smoothness parameter
approaches infinity. When choosing between the SE and Matérn covariance functions, it is often a matter of balancing the trade-off between modeling flexibility and computational complexity. The SE covariance function is simpler and more computationally efficient but may not capture complex patterns in data as well as the Matérn covariance function with an appropriate choice of smoothness parameter.
In order to compute (
10), we also need to invert the
Gram matrix
. This task is impractical when the size sample
N is large because inverting the matrix leads to
memory and
time complexities [
28]. There are several methods to overcome this difficulty. For instance, the variational inference proceeds by introducing
n inducing points and corresponding
n inducing variables. The variational parameters of inducing variables are learned by minimizing the Kullback–Leibler divergence [
16]. Picking
, the complexity reduces to
in prediction and
in minimizing the Kullback–Leibler divergence. The computational complexity of conventional sparse Gaussian process (SGP) approximations typically scales as
in time for each step of evaluating the marginal likelihood [
14]. The storage demand scales as
. This arises from the unavoidable cost of re-evaluating all results involving the basis functions at each step and the need to store the matrices required for these calculations.
5. Low-Complexity Gaussian Process Regression
In order to avoid the inversion of the
Gram matrix
, we use the approximation scheme presented in
Section 3 and rewrite the GP with a truncated set of
m basis functions. Hence, the truncated
f at an arbitrary order
is given by
with an approximation error
. The canonical GP regression model (
7) becomes
with a covariance function approximated by
. The convergence degree of the Mercer series in (
4), that is,
with
depends heavily on the eigenvalues and the differentiability of the covariance function. Ref. [
29] showed that the speed of the uniform convergence varies in terms of the decay rate of eigenvalues and demonstrated that, for a
times differentiable covariance
k, the truncated covariance
approximates
k as
. For infinitely differentiable covariances, the latter is
for any
. To summarize, smoother covariance functions tend to exhibit faster convergence, while less smooth or non-differentiable covariance functions may exhibit slower or no convergence.
The resulting covariance falls into the class of reduced-rank approximations based on approximating the covariance matrix
with a matrix
, where
is an
diagonal matrix with eigenvalues such that
and
is an
matrix with eigenfunctions such that
. Note that the approximate covariance matrix
becomes ill-conditioned when the ratio
is large. This ill-conditioning occurs particularly when the observation points
are too close to each other [
30]. In practice, this can lead to significant numerical errors when inverting
, resulting in unstable solutions, amplified errors in parameter estimation, and unreliable model predictions.
Now, we show how the approximated regression models make use of GP decomposition to achieve low complexity. We write down the expressions needed for inference and discuss the computational requirements. Applying the matrix inversion lemma [
31], we rewrite the predictive distribution (
10) and (
11) as
where
is an
m-dimensional vector with the
j-th entry being
. When the number of observations is much higher than the number of required basis functions (
), the use of this approximation is advantageous. Thus, any prediction mean evaluation is dominated by the cost of constructing
, which means that the method has an overall asymptotic computational complexity of
.
The approximate log marginal likelihood satisfies
Consequently, evaluating the approximate log marginal likelihood has a complexity of
needed to inverse the
matrix
. In practice, if the sample size
N is large, it is preferable to store the result of
in memory, leading to a memory requirement that scales as
. For efficient implementation, matrix-to-matrix multiplications can often be avoided, and the inversion of
can be performed using Cholesky factorization for numerical stability. This factorization (
) can also be used to compute the term
. Algorithm 1 outlines the main steps for estimating the hyperparameters of the low-complexity GP.
| Algorithm 1 Gradient Ascent for Hyperparameter Learning. |
- Require:
Data , initial hyperparameters , learning rate , tolerance , maximum iterations T. - Ensure:
Optimized hyperparameters . - 1:
Initialize . - 2:
for to T do - 3:
Construct and depending on . - 4:
Compute . - 5:
Evaluate . - 6:
for each hyperparameter do - 7:
Compute gradient . - 8:
Update hyperparameter: . - 9:
end for - 10:
if then - 11:
break - 12:
end if - 13:
end for - 14:
return optimized hyperparameters .
|
6. Closed Solutions from Differential Operators
In this section, we highlight the connection between Hilbert–Schmidt integral operators in (
3) and differential operators. Thus, we describe explicit solutions for the low complexity GP (LCGP) with covariances derived from differential operators. Unlike previous works where the eigen-decomposition is determined from the Mercer series or the integral operator directly, this paper focuses on constructing covariance functions that incorporate orthogonal polynomials as eigenfunctions. It is worth noting that polynomials can approximate a wide range of functions with various degrees of complexity. They can be adjusted to predict different data patterns and capture both linear and non-linear relationships [
32].
The connection between a differential operator denoted
and the integral operator
has been largely used. We follow the same idea in [
33,
34,
35] and define Green’s function
G of the differential operator
as its “right inverse”, i.e.,
where
denotes the Dirac delta function.
Theorem 4. Let be continuous symmetric non-negative definite and let be the corresponding Hilbert–Schmidt operator. Let be an orthonormal basis for the space spanned by the eigenfunctions corresponding to the non-zero eigenvalues of . If is the eigenfunction associated with the eigenvalue and the covariance function acts as a Green’s function of a differential operator , then the eigenvalues of correspond to reciprocal eigenvalues of , while the corresponding eigenfunctions are still the same.
Proof. We have
Finally, we obtain
which completes the proof. □
At this stage, we compute eigenvalues and eigenfunctions of
, from which we deduce the Mercer decomposition of
given in (
4) replacing
by
. We select a list of some interesting and useful differential operators that act on
, for example: Matérn, Legendre, Laguerre, Hermite, Chebyshev, and Jacobi, from which we explicitly find the corresponding eigen-decompositions.
Table 1 summarizes each class of
, the index set
I, the weight function
, the eigenvalues
, the eigenfunctions as polynomials
, and the resulting MISE. Note that, for Laguerre, Hermite, and Chebyshev polynomials, the eigenvalues
of
were initially negatives. Therefore, we consider the iterated operator
with squared eigenvalues and unchanged eigenfunctions. Further, for Legendre, Hermite, and Chebyshev, we state that
, which means that
should be normalized to form an orthonormal basis. For Jacobi, the differential operator is
with parameters
greater than
. More details about the corresponding eigenfunctions
and the
-norm are provided in [
36].
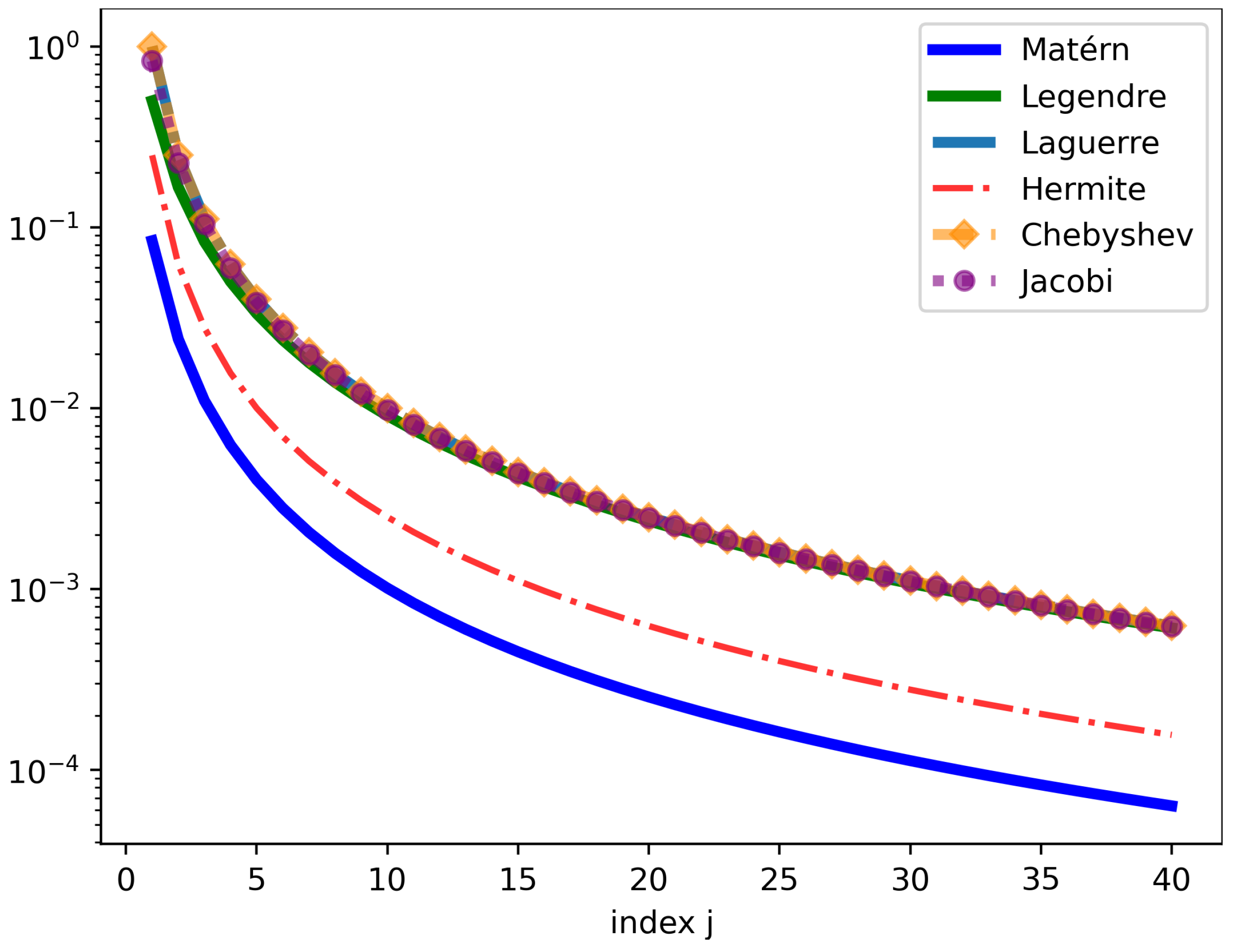
Figure 1 illustrates the behavior of the eigenvalues
of the integral operator
as the index
j varies from 1 to 40. This is attributed to the smoothness of the true covariance as
m is growing.
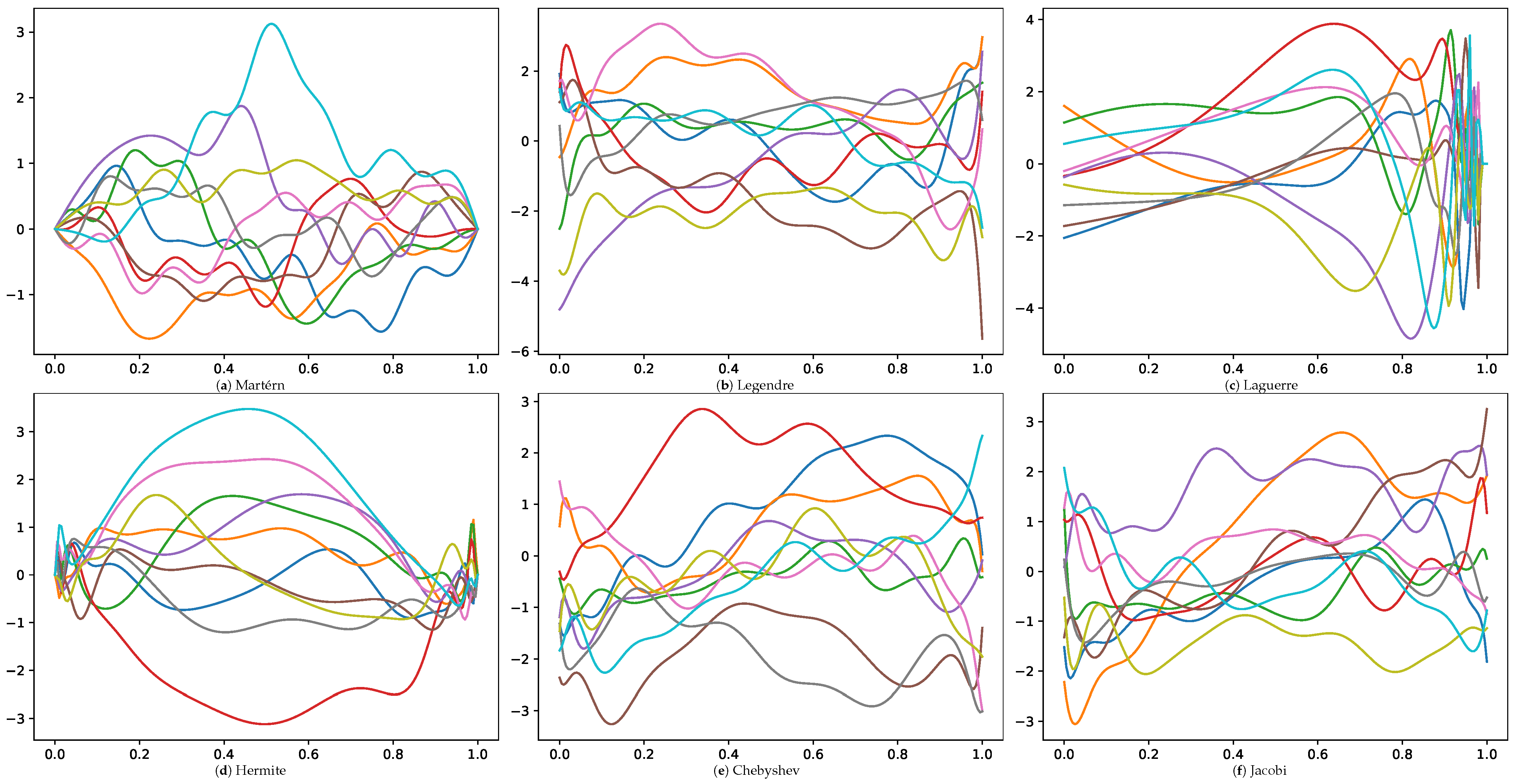
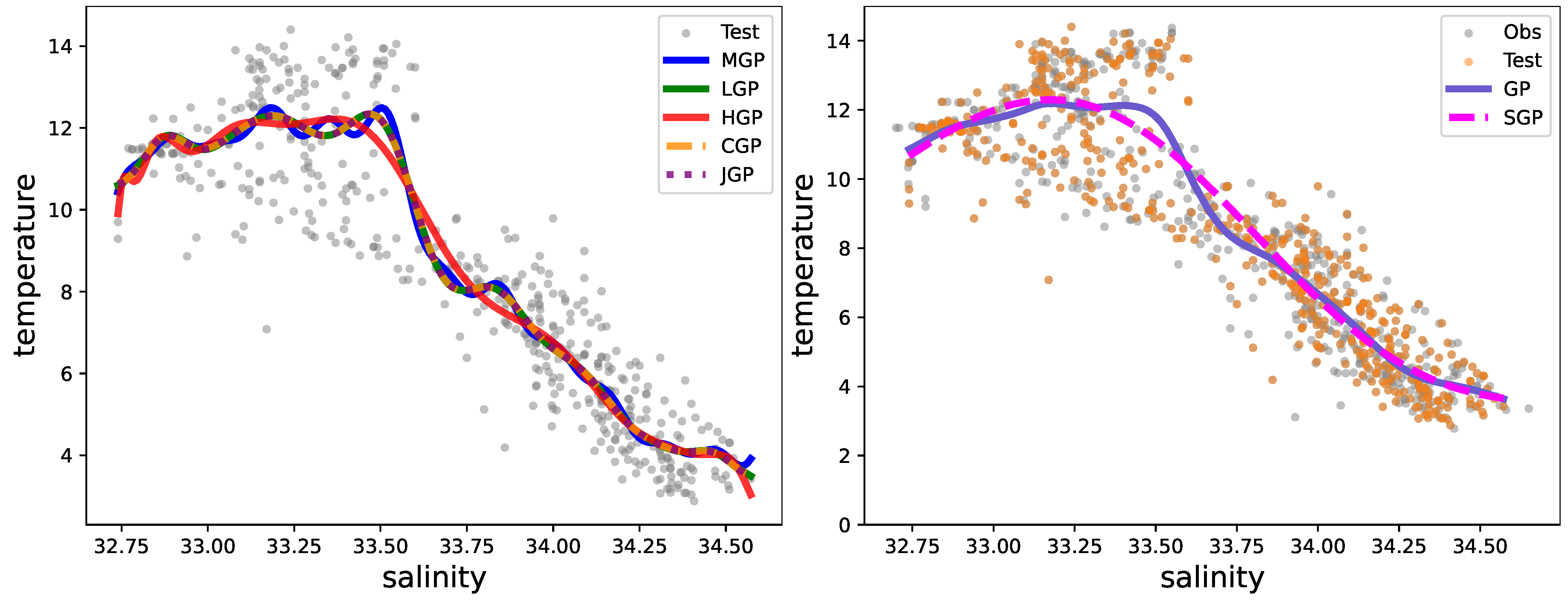
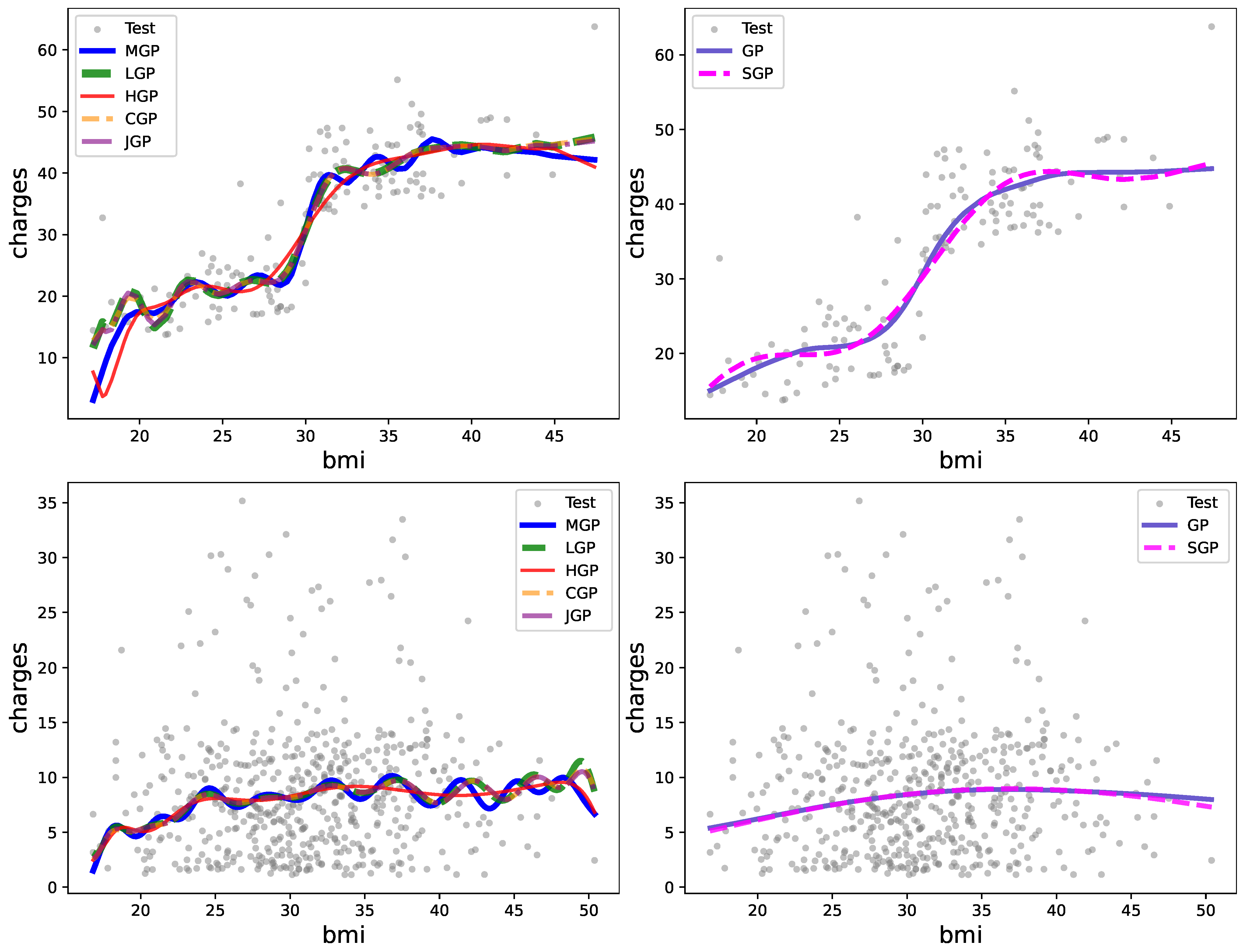
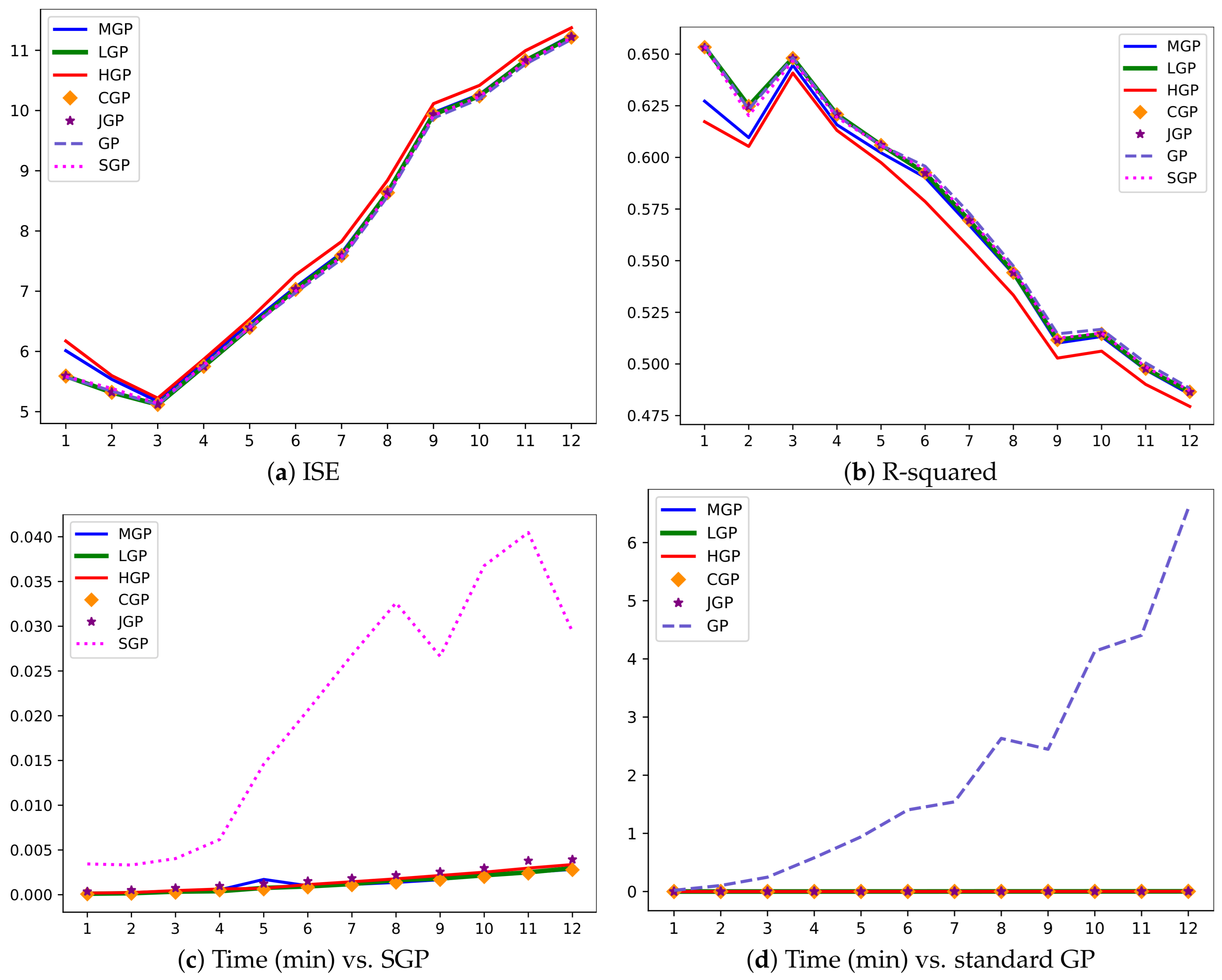
Figure 2 visually depicts several GP realizations across various differential operator settings.