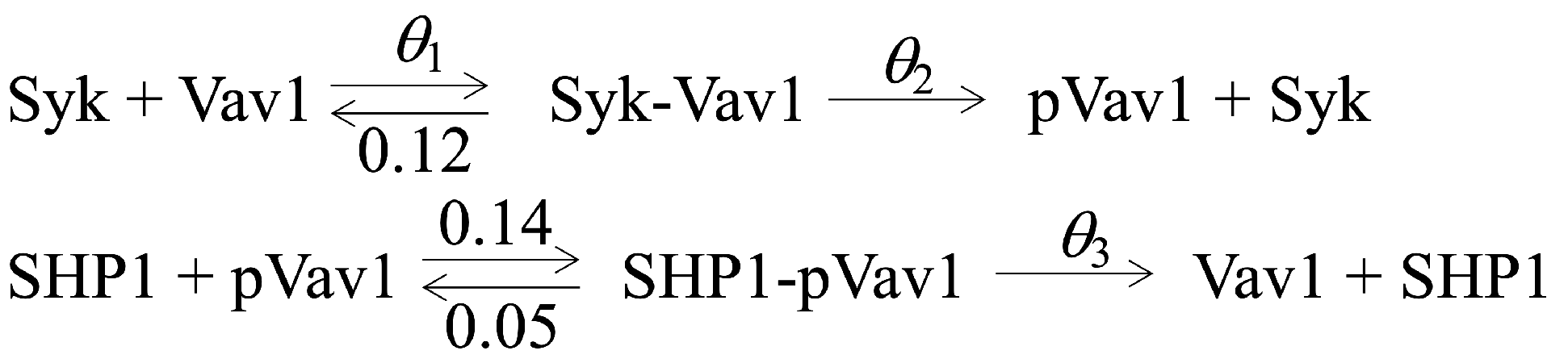1. Introduction
Ordinary differential equations (ODEs) are commonly used to model the sub-cellular dynamics of proteins and mRNA [
1,
2,
3]. Usually, ODEs describe the deterministic dynamics of average concentrations, which facilitates the estimation of reaction rates (
also known as model parameters) from experimental data. This is often an important step towards building mechanistic biological models, but the task of estimating model parameters from experimental data is challenging for a variety of reasons [
4,
5,
6], especially when the number of distinct proteins measured in experiments is smaller than the number of model parameters. Recent developments in single-cell experimental techniques for the longitudinal measurements of transcripts and proteins in an individual cell, such as single-cell RNA-seq [
7] or single-cell mass cytometry by time-of-flight (CyTOF) [
8,
9], which can simultaneously measure over a thousand different RNA sequences or more than thirty different protein species in a single cell, appear to alleviate this problem. Since individual cells are not tracked across time in these experiments, the measurements generate a large collection of time-stamped snapshot (TSS) data. Another challenge stems from the cell-to-cell differences in the copy number of a protein (or abundance), which contains variation present at the pre-stimulus state (
also known as extrinsic noise), as well as variation arising from the inherent stochasticity of biochemical reactions over time (
also known as intrinsic noise) [
10,
11]. When the observed protein abundances are large, intrinsic noise can be ignored, extrinsic noise is known to play a significant role [
12], and single-cell protein signaling kinetics can be well approximated by ODEs. By comparing the distribution of protein abundances in single cells observed at time
t to the predictions obtained from an ODE or a stochastic model that evolves single-cell protein abundances seen at an earlier time (e.g.,
,
also known as initial conditions), we can estimate the parameters of the candidate model using a generalized method of moments (GMM) approach [
13,
14]. GMM, which contrasts sample moments and their corresponding expectations, is widely used in econometrics [
15]. In this paper, we propose an entropy-based approach to address the larger question of model selection for systems that can be described by deterministic dynamical models (e.g., sets of ODEs) with randomness arising from initial conditions. Specifically, for the models considered here, we show that our cross-entropy [
16] approach can find the
best ODE model from a set of competing candidate models, where the best model neither over-fits nor under-fits the available data.
The primary goal of model selection is to find the best model relative to some defensible criterion, and two attractive criteria are cross-entropy and Kullback–Leibler (KL) divergence [
17]. The latter is a non-negative number that, for a pair of random variables can provide a useful measure of dependence known as mutual information [
18]. But more generally, KL divergence measures the “distance” between two probability distributions [
13] where the KL divergence vanishes for a pair of identical distributions. Usually, the distribution that gave rise to the protein abundances observed at time
t (denoted
f) is considered to be the “ground truth”, while the other distribution is most often a candidate distribution (or model) that is presumed to be “close” to
f. A defining feature of KL divergence is that it is zero if and only if the candidate model and
f are the same. Typically, the best candidate model will strike a balance between under-fitting (i.e., over-estimating the random error) and over-fitting (i.e., under-estimating the random error). Since KL divergence is a difference in expectations taken with respect to
f (i.e., cross-entropy minus entropy), and since entropy depends only on
f, it suffices (for the purpose of model selection) to find the model with the smallest cross-entropy. Given a finite set of candidate models, the model with the smallest cross-entropy is also the model with the smallest KL divergence, which makes it the best approximating model to
f.
There are however two very important challenges when performing model selection from TSS data. First, the multivariate probability density of the observed protein abundances at time
t (denoted symbolically as
f, and in words as the “ground truth”) is rarely known. Second, while we can estimate the parameters of each candidate model, we cannot evaluate the likelihood of any model. Fortunately, we can deal with the first challenge by minimizing cross-entropy (instead of KL divergence), as cross-entropy only requires
realizations from
f, not complete knowledge of
f. As for the second challenge, which cannot be avoided, we tackle it head-on by estimating the likelihood for each candidate model (see
Section 2 and
Appendix B for more details) from the time evolution of initial conditions (see
Section 3 and
Appendix A for more details).
The remaining sections of this paper are organized as follows. In the first subsection of Methods, we give mathematical descriptions of KL divergence and cross-entropy, and we briefly explain how parameters of a candidate model are estimated. Then, in the second subsection of Methods, we outline our model selection approach, leaving technical details (such as the estimation of the Gaussian copula, marginal densities, and marginal cumulative distribution functions) to
Appendix A and
Appendix B. Furthermore, in the second subsection of Methods, we briefly describe a complementary model selection approach that applies Akaike Information Criterion [
19] corrected for small samples (AICc) [
20] to differences between the mean protein abundances observed at time
t and the mean protein abundances predicted at time
t. In
Section 3, we describe how synthetic TSS data are generated, and we describe the time-evolution of initial conditions using different candidate models. Finally, we demonstrate the utility of our model selection approach in
Section 4, and we give some interesting insights and suggestions for further improvements in
Section 5.
3. Data Description
In most real-world applications that model the time-dependent kinetics of protein abundances, the “ground truth” (denoted
f) is rarely known. However, because we are primarily concerned with improving model selection (i.e., increasing accuracy, increasing usability, and developing additional measures of relative support), we chose to mimic observed TSS data
y by simulating (without intrinsic noise) protein abundances at time
t from three scenarios of interest: SMALL, MEDIUM, and LARGE (see
Table A1 for more details). For the SMALL, MEDIUM, and LARGE scenarios, we create ground truth models where we vary one, two, and three parameters of
. For instance, in the ground truth MEDIUM scenario, candidate model
is expected to explain the TSS data better than
, which is too simplistic, and better than
, which over-fits to noise in the data (see
Table 1).
For each “ground truth” scenario, we always know which candidate model should yield the best fit. For example, consider candidate model
which has two freely varying parameters,
and
, because we set
. Relative to
and
, candidate model
should be the
closest to the “ground truth” MEDIUM scenario, provided that under-fitting and over-fitting are accounted for appropriately (see
Table 2). Similarly, candidate models
and
are expected to to be
closest to “ground truth” scenarios SMALL and LARGE, respectively, since in
we set
and
with only
varying freely; and since
has all three parameters varying freely. Of course, when implementing ACE (and AICc), we pretend that the “ground truth” is unknown.
Table 1.
Under-fitting and over-fitting. The cost (shown in brackets) decreases as the complexity of the candidate model increases, so
has the lowest cost. Yet ACE selects
, the correct candidate model, 95% of the time (see
Table 3), which implies that ACE appropriately balances the under-fitting of
and the over-fitting of
. The parameter estimates
are shown for each candidate model; and as a point of reference, the “ground truth” MEDIUM scenario is shown in bold.
Table 1.
Under-fitting and over-fitting. The cost (shown in brackets) decreases as the complexity of the candidate model increases, so
has the lowest cost. Yet ACE selects
, the correct candidate model, 95% of the time (see
Table 3), which implies that ACE appropriately balances the under-fitting of
and the over-fitting of
. The parameter estimates
are shown for each candidate model; and as a point of reference, the “ground truth” MEDIUM scenario is shown in bold.
| Model: | | | | Cost |
|---|
| MEDIUM: | 0.10 | 0.90 | 0.18 | [–NA–] |
| : | 0.093 | (9 × 0.093) | (2 × 0.093) | [0.0370] |
| : | 0.100 | (9 × 0.100) | 0.182 | [0.0074] |
| : | 0.097 | 0.896 | 0.182 | [0.0071] |
To generate TSS data, we begin by simulating uncorrelated initial conditions from a multivariate log-normal distribution with parameters = (5.25, 7.60, 5.25, 7.60, 5.25, 5.25), and = (0.15, 0.06, 0.15, 0.06, 0.15, 0.15). Now, let us consider simulating data for the “ground truth” LARGE scenario. To accomplish this, we take half of the initial conditions (discussed immediately above) and we evolve them to time t using coupled ODEs and parameters (0.10, 0.95, and 0.18). Computing ACE and model selection probabilities for all three candidate models takes about 2 h on a 2.5 GHz computer, but apart from computational time, there is no limit on the number of candidate models one can consider.
5. Discussion
5.1. New Tools for Model Selection with Single-Cell Data
Mechanistic models based on ODEs describing subcellular kinetics of proteins are widely used in computational biology for gleaning mechanisms and generating predictions. On the other hand, to model random gene expression data, stochastic mechanistic models such as the telegraph model have been used [
25,
26]. It is common to have multiple candidate mechanistic models that can be set up to probe different hypotheses describing the same biological phenomena, and an important task in model development is to rank order the candidate models according to their ability to describe the measured data. The availability of large, high-dimensional, single-cell datasets allows for estimation of model parameters using mean values and higher-order moments of the measured data; however, rank ordering candidate models from such data may not be straightforward when using standard approaches in model selection (e.g., AIC, AICc, and Kullback–Leibler). Here we propose a model selection approach based on cross-entropy for ODE-based models that are calibrated against means and higher-order moments of the measured data. We show as “proof-of-concept” that our proposed approach successfully rank orders a set of ODE models against synthetic single-cell datasets.
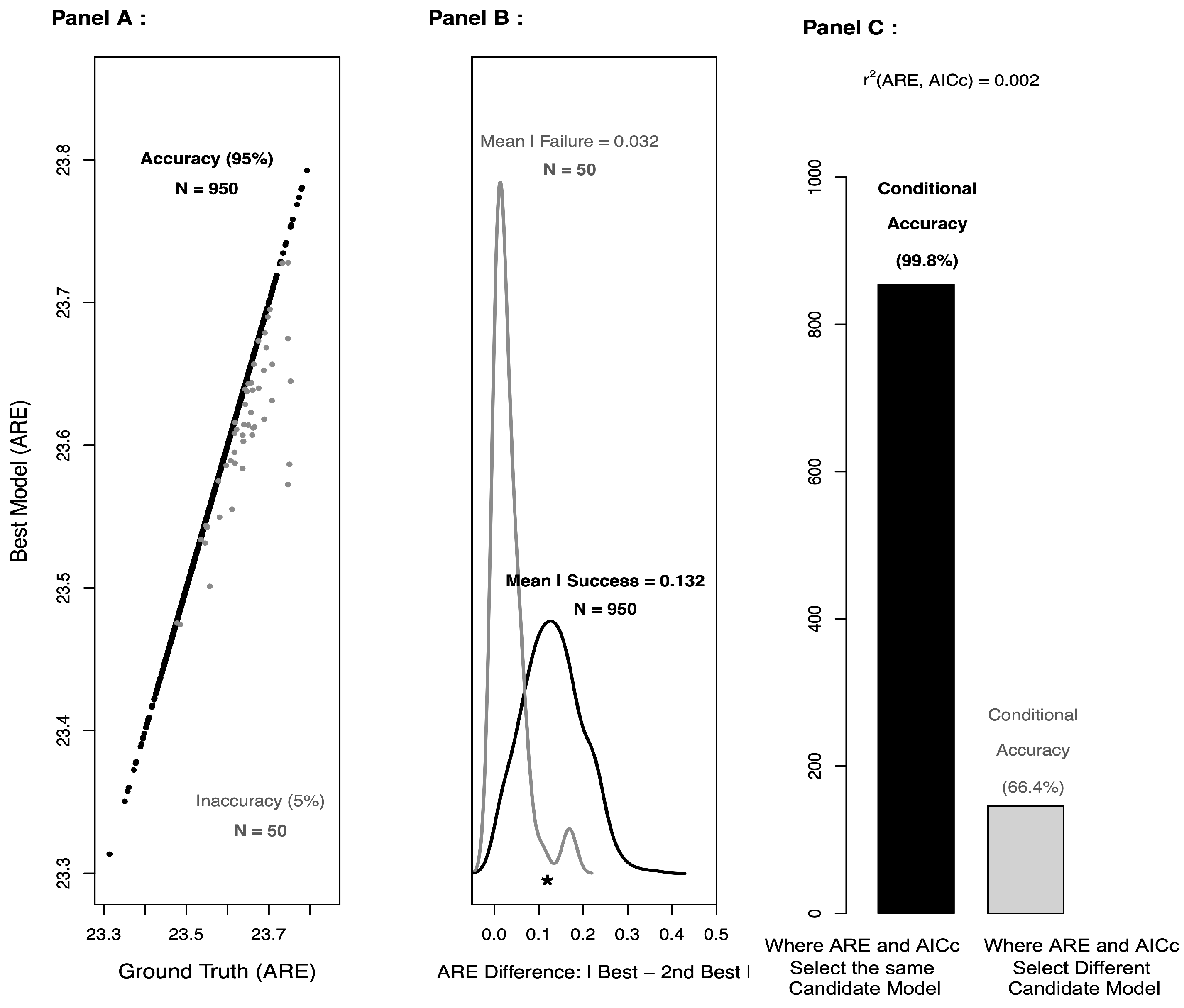
5.2. ACE Complements Approximate AICc
As shown in
Figure 2 (Panel C), approximate AICc is virtually independent of ACE, and concordance between ACE and approximate AICc appears to provide additional support for the candidate model selected by ACE. So, any improvement in approximate AICc would likely only benefit ACE. One potential area for improvement might be a recalibration of the penalties used in approximate AICc. In particular, it’s not immediately clear what the penalty should be for approximate AICc, as the parameter estimation is based on thousands of cells, whereas the model selection is based on differences in only six means. Moreover, when one combines a split-sample approach with model selection (as we have done here), the penalty terms in AIC and AICc may no longer be needed [
27]. Indeed, the so-called
penalties are actually
corrections for the bias that arises when parameter estimation and model selection are performed on the same dataset.
By design, the additional penalty term in AICc depends on the corresponding sample size (here, AICc uses only six means). As such, the number of distinct proteins must be larger than the number of freely varying parameters; otherwise, the denominator of the additional penalty term will be zero or negative. Also, because the AICc likelihood is base on the differences in means, AICc may have more difficulty than ACE accurately rank ordering two or more candidate models with similar means. Note that ACE (as implemented here, with a split-sample approach) does not have either design limitation: (1) ACE penalizes indirectly for complexity and does not require a bias correction, and (2) ACE makes use of the entire multivariate density, not just the means.
For the MEDIUM and LARGE “ground truth” scenarios, ACE outperforms approximate AICc, but for the SMALL “ground truth” scenario, approximate AICc does better than ACE. This suggests that the means tend to carry the bulk of the information in the “ground truth” SMALL scenario, so there’s very little benefit to estimating the other candidate models (which contain information about higher-order moments and cross-moments). However, when the higher-order moments and cross-moments begin to matter, as is likely the case with the more complex “ground truth” scenarios, the benefit of estimating the candidate models with 2 and 3 freely varying parameters is likely greater.
5.3. Limitations and Future Directions
Presently, our ACE approach to model selection based on TSS data has three main limitations: (1) it is not designed to handle intrinsic noise, (2) there is considerable latitude in terms of multivariate density estimation that we have only scratched the surface of here, and (3) there may be more efficient ways to incorporate split-sample techniques. Extending our ACE approach to include
both extrinsic and intrinsic noise may be possible for relatively short evolution times and/or for networks with a relatively small number of interacting proteins. Further, for other applications, users may want to include higher-order moments and/or different copulas or kernel density estimators [
28,
29].
When researchers are unable to specify the full candidate model, the likelihood is not known and model selection is often challenging. However, when the sample size is large and consistent estimators of the model parameters and candidate models exist, we propose a split-sample entropy-based approach that allows users to find the best approximating model to the “ground truth”. Furthermore, our approach is quite flexible with respect to (1) parameter estimation (e.g., choosing which moments to use—first moments only, first and second moments, etc.), and (2) multivariate density estimation (e.g., choosing a “good” kernel density estimator and/or copula for each candidate model).