1. Introduction
Decision tree algorithms are classic machine learning methods used for handling classification and regression problems. Despite being around for nearly half a century, they remain active in various fields of machine learning, thanks to their strong performance characteristics, such as high accuracy, simple algorithmic processes, and excellent model interpretability [
1].
The principle of decision tree algorithms can be summarized as a recursive process that repeatedly selects features to partition the dataset, with the goal of reducing the disorder or complexity of the variable to be classified or regressed as much as possible. For new data points, the algorithm returns the value of the variable to be classified or regressed based on the selected features, thus completing the classification or regression task [
2]. Therefore, the questions of how to measure the disorder or uncertainty of variables, how to find features to divide the dataset, and what conditions should trigger the termination of recursion become key issues when building a tree. To address these challenges in tree construction, many well-known decision tree algorithms have been developed. For example, in the ID3 algorithm proposed by Quinlan, the concept of Shannon entropy from information theory is used to describe the disorder of classification variables. Subsequently, he also introduced an improved version of the C4.5 algorithm, which not only incorporated the Gini index as a new measure of classification variable uncertainty but also introduced the concept of information gain ratio [
2,
3]. Breiman’s CART decision tree algorithm combines both classification and regression problems and introduces stopping criteria for dividing trees, allowing for control over the complexity of the trees and enabling them to handle regression problems [
3]. As decision tree algorithms continue to mature, they are increasingly applied to disciplines like management science, medicine, biology, etc. In combination with other statistical methods, decision trees can also achieve more outstanding performance [
4,
5,
6,
7]. Kumar et al. proposed a hybrid classification model combining support vector machines with decision trees that significantly improves computational efficiency without sacrificing accuracy [
8]. Bibal et al. designed a method called DT-SNE for visualizing high-dimensional discrete data, which combines the visualization capabilities of t-SNE with the interpretability of decision trees [
9].
However, decision tree algorithms themselves also have some drawbacks. Rokach et al. point out that decision trees often perform poorly when faced with attributes involving complex interactions, and duplication problems are an example of this deficiency [
10]. When faced with imbalanced datasets, decision trees may also produce fragmentation problems, which result in low predictive credibility due to too few samples before and after splitting, or even overfitting [
11]. The Random Forest algorithm, introduced by Breiman, is an ensemble algorithm based on decision trees. Compared to decision trees, it has stronger identification capabilities for complex attributes and better accuracy and robustness to noise [
12]. However, decision trees as base learners still have some drawbacks. When constructing single decision trees, the algorithm only considers the best decision at each step, exhibiting myopia due to its greedy nature. Both decision trees and random forests typically construct trees based on specified criteria for category complexity reduction, such as entropy or Gini Index, limiting decision flexibility. Moreover, numerous practical applications show that different criteria lead to varying levels of performance among decision tree algorithms, indicating that there is no one-size-fits-all segmentation standard for building trees suitable for all datasets and performing well.
To achieve higher flexibility in decision trees, we aim to introduce some generalized entropy measures for complexity that allow the segmentation criteria based on these entropies to adjust according to different datasets. Many generalized entropies have been proposed, such as Rényi entropy, Tsallis entropy, and
r-type entropy [
13]. These are single-parameter generalized entropies, with Shannon entropy being a special case for specific parameter values. Similarly to Shannon entropy, the Gini index is a value calculated based on a probability distribution, used to measure sample category impurity [
14]. Wang et al. pointed out that Tsallis entropy possesses better properties [
15]. Under different parameter values, Tsallis entropy can converge to both Shannon entropy and the Gini index. Therefore, Tsallis entropy not only unifies Shannon entropy and the Gini index within a single framework but also allows for searching other potentially more suitable parameter values for the current dataset. Experiments have shown that this improvement significantly enhances the accuracy of decision trees when handling classification tasks. Optimal construction of binary decision trees has been proven an NP-hard problem, meaning that it is difficult to build an optimal decision tree in polynomial time [
16]. Therefore, the construction process of most decision trees is greedy, where at each step, the seemingly optimal option is chosen to generate the tree structure. Wang et al. proposed the two-term information measures based on Tsallis entropy and design a less-greedy tree construction algorithm TEIM. Experiments on various datasets demonstrate that this algorithm outperforms traditional decision trees constructed using a single information measure in terms of accuracy and robustness while partially overcoming the short-sightedness issue of decision trees. It is also worth noting that this approach of optimizing decision trees by modifying the splitting criterion does not alter the tree generation structure. Consequently, neither the time complexity nor the search complexity of tree construction undergoes any change [
17].
This paper presents an improvement to this less-greedy tree construction method. Inspired by generalized entropy and two-term information methods, we seek a more general unified framework through considering a broader range of entropy functions. At the same time, we re-examine the two-term information method, treating it as a split criterion with penalties to further improve decision tree algorithm performance. In summary, our innovations are as follows:
We introduce a two-parameter generalized entropy framework—the
entropy, which unifies Rényi entropy, Tsallis entropy, and the
r-type entropy under a common framework—to enhance decision tree generalization ability further [
18].
Utilizing the second term in the two-term information method as a penalty term in the split criteria and introducing a penalty coefficient to the said term, leading to increased interpretability and adaptability to diverse datasets.
Incorporation of Genetic Algorithm (GA) for rapid determination of parameter values within our improved decision tree construction algorithm. This optimization approach ensures effective parameter tuning for fast and accurate tree generation.
In terms of evaluating model performance, we conducted time and space complexity analyses in comparison with traditional algorithms. Meanwhile, we combined the performance of each model on the test set with non-parametric tests to assess the significance of differences in model superiority.
The remainder of this paper will focus on designing our improved decision tree algorithm. In
Section 2, we discuss relevant concepts of information entropy serving as the theoretical foundation for our algorithm.
Section 3 delves into details regarding improvements made to existing algorithms along with the corresponding steps involved. Lastly, in
Section 4, we evaluate our enhanced model through comparison with conventional methods, highlighting its superiority.
3. Classification Algorithm Based on RS-Entropy
In this section, we describe the classification tasks that decision tree algorithms address. We then elucidate the application of information entropy in the algorithmic process and present the steps of our designed classification algorithm based on RS-entropy.
3.1. Problem Description
For a given dataset D, which contains samples, each data sample is represented by . X represents the attribute vector for each data sample, where each serves as a random variable taking values from their respective finite sets . Y represents the class of the data sample, serving as a random variable, taking values from the class set .
The task is to construct a decision tree for classification based on the dataset. In this tree, each node represents a subset of the dataset, and the branching of the tree signifies a division of the dataset associated with that node. Nodes between different levels are connected by branches, indicating the splitting criteria used from one level to the next. To stop the growth of the tree, a criterion is set to determine when no further branching should occur. Nodes that do not branch further are called leaves, and they return the class with the most samples within that leaf as the predicted class.
For a new data example , we can start from the root node of the constructed tree and, based on the feature values of the sample, follow the branches sequentially until you reach a leaf node in the decision tree. The class associated with the leaf node is the model’s predicted classification result , thus completing the classification task.
3.2. Split Criterion
Different splitting and stopping criteria can lead to variations in the constructed tree, thereby influencing the predicted categories. Stopping criteria are typically used to limit the complexity of the tree, control computational resources, and prevent overfitting. These criteria often involve restrictions related to the overall depth of the tree, the number of samples in leaf nodes, or the complexity of samples in leaf nodes [
20].
This article primarily focuses on splitting criteria. The objective of splitting can be summarized as finding the most suitable attribute and attribute value to partition the dataset in such a way that the complexity of categories in the resulting partitions is minimized as much as possible. Various methods are available to characterize complexity, such as Shannon entropy and Gini index. The entropy of degree r exhibits favorable properties and achieves a unification of Shannon entropy and the Gini index.
3.2.1. The Two-Term Information Method
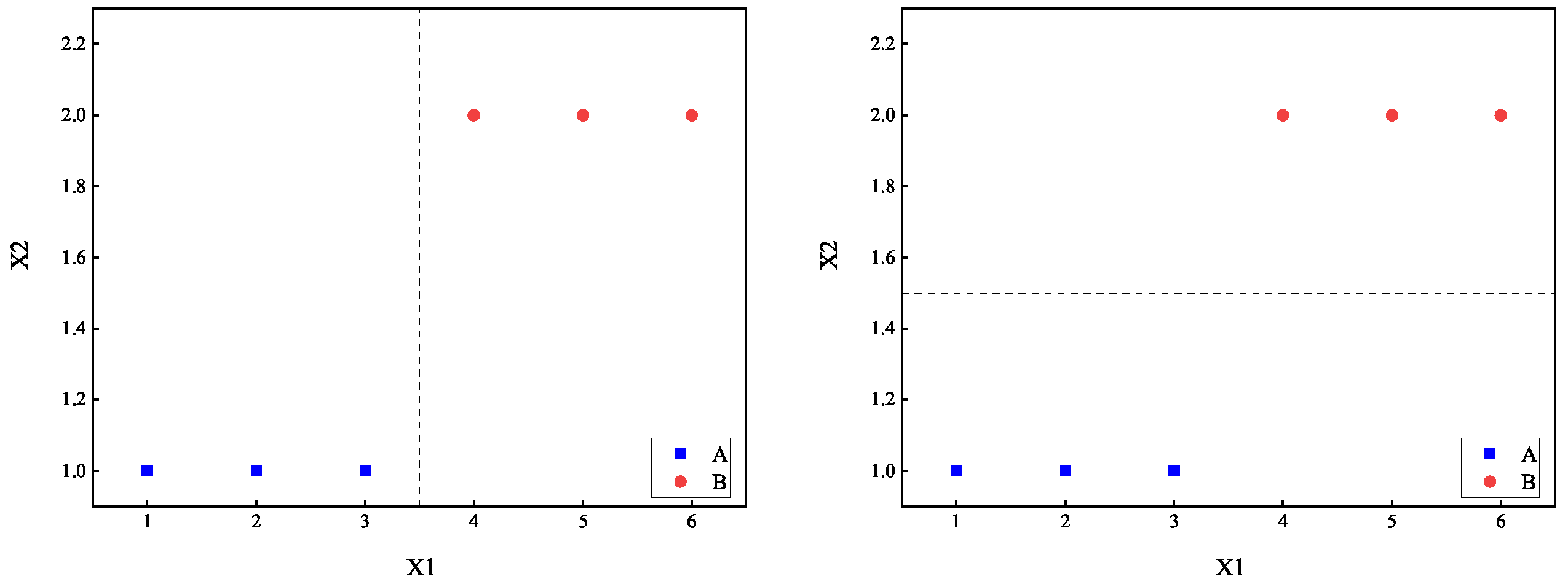
The two-term information method in decision trees was introduced by Wang. This measure represents a novel splitting criterion for decision trees. It not only computes the complexity of categories within the subsets generated by splitting on a particular attribute but also takes into account the complexity of attributes within the same category within those subsets. The sum of these two complexities is used as the splitting criterion. Compared to traditional methods, this construction approach is less greedy. While it may not guarantee that each individual split is optimal according to traditional standards, it achieves an overall better outcome. We will use the dataset from
Table 1 as an example to illustrate this approach.
To facilitate the description, we establish the following notations:
The splitting point is denoted as , and it is used to partition the dataset D into two subdatasets, represented as and .
The complexity of category Y in dataset D is denoted as , which calculates the empirical entropy of category Y in dataset D. For ease of calculation in the example, we use empirical Shannon entropy to quantify the complexity.
The complexity of the attribute
with respect to the category
Y in the dataset
D is denoted as
. Its calculation formula is
where
.
The variance of the attribute
in dataset
D is denoted as
, and its calculation formula is
The complexity of splitting point is denoted as . It represents the weighted sum of the complexities of subsets and after partitioning dataset D with respect to the sample proportions.
It is straightforward to calculate that when using the splitting point
to divide dataset
D into two subsets
and
, where
contains data examples with index 1, 2, and 3. The complexity with respect to category
Y for the subsets is
. Similarly, we have
. By weighting the complexities of the two subsets according to the sample proportions, we can calculate the category complexity under the splitting point
as follows:
Similarly, we can calculate the category complexity under splitting point
:
We should seek the splitting point that minimizes the complexity of categories after splitting. The results from Equations (
16) and (
17) indicate that both splitting points
and
have the same effect, resulting in a category complexity of 0 after splitting. However, it is evident that splitting point
is superior to
because attribute
appears to have a stronger correlation with
Y, indicating that the values of
may directly influence category
Y. In essence, this method of determining splitting points based on the complexity of categories within the resulting subsets has limitations as it overlooks the relationship between attributes and categories during the splitting process.
The two-term information methods have improved the way splitting point complexity is calculated, enhancing the identification capability of splitting points. In characterizing the complexity of subset
, this method calculates the sum of the set’s category complexity
and attribute complexity
, differently from the traditional splitting approach that solely considers category complexity while ignoring attribute complexity. Then we can calculate the category complexity under splitting point
:
Similarly, we have the category complexity under splitting point
:
Comparing (
18) and (
19), since
, this supports our choice to split the dataset using splitting point
. The splitting effect of
and
are illustrated in the left and right sub-figures of
Figure 1.
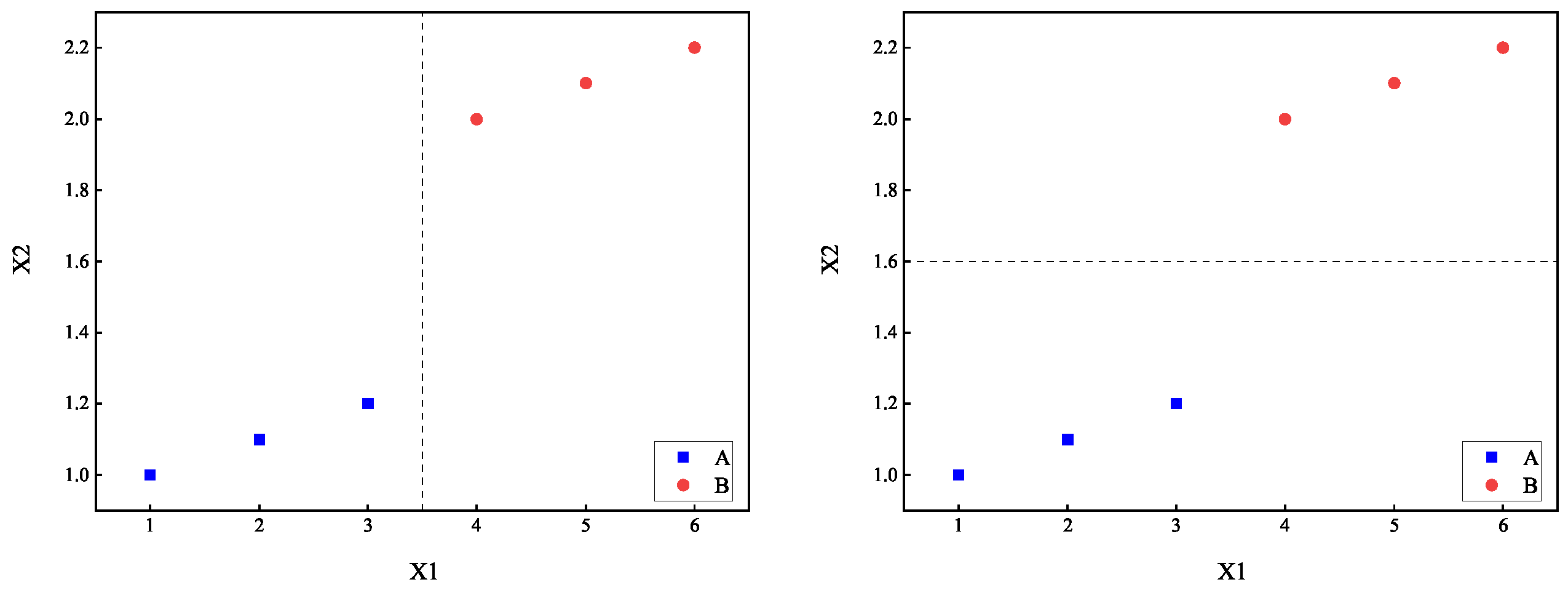
The two-term information method evidently performs better in attribute selection compared to traditional methods. The concept of this method can be understood as adding a penalty term on top of the traditional information measures. A good splitting point should not only effectively differentiate categories but also have a strong association between the split attribute and categories. This type of classification method is ideal as it combines both classification accuracy and interpretability. In other words, after splitting based on attribute , if the values of in the resulting subset are too dispersed, it implies that the relationship between this attribute and categories is not strong enough. Clearly, such a splitting point is not optimal, which will be reflected in the relatively larger value of the penalty term . This leads to a higher overall , thereby avoiding the selection of that splitting point as much as possible.
This penalty term has practical significance, as it means that the splitting criterion takes into account the degree of feature confusion. Especially in the random forest algorithm, we need to evaluate the importance of features based on the splitting method of each decision tree. Compared with the traditional feature importance calculated only based on the category Y, this penalty-term-included approach is bound to make the feature ranking more scientific. However, this paper does not elaborate further on this aspect.
3.2.2. Improved Two-Term Information Method
The two-term information method mentioned above still appears to have potential areas for improvement, which can be summarized in the following two aspects:
The types of attribute have not been taken into consideration. In reality, can be either a numerical or a categorical attribute, and the computed penalty term should differ accordingly.
The penalty term should be more adaptable to better suit different datasets. Introducing a penalty coefficient before , controlling the weight of by adjusting , can enhance its flexibility.
For example, consider the dataset in
Table 2.
Using the two-term information method from
Section 3.2.1, we calculate
, indicating that the splitting points
and
are equally optimal. However, it is noticeable that
should be more suitable as a splitting point. This suggests that the calculation of
may not be suitable for numerical attributes
. For such attributes, we define the conditional variance
to characterize their complexity. Similar to (
14), it is defined as
Therefore, the improved computation of the two-term information method can be provided by Equations (
21)–(
23).
where
and
denote the sets of numerical and categorical attributes in the dataset and
and
are defined as follows:
For example, considering the dataset in
Table 2, let us assume that both attributes,
and
, are continuous. Given
, the calculated improved
is as follows:
Similarly,
can be computed as well:
Therefore,
, indicating that selecting
as the splitting point results in subsets with both attributes and categories having smaller complexities, making it superior to
as a splitting point.
Figure 2 illustrates the comparison of the dataset partitioning effects between
and
.
3.3. Evaluation of Feature Importance
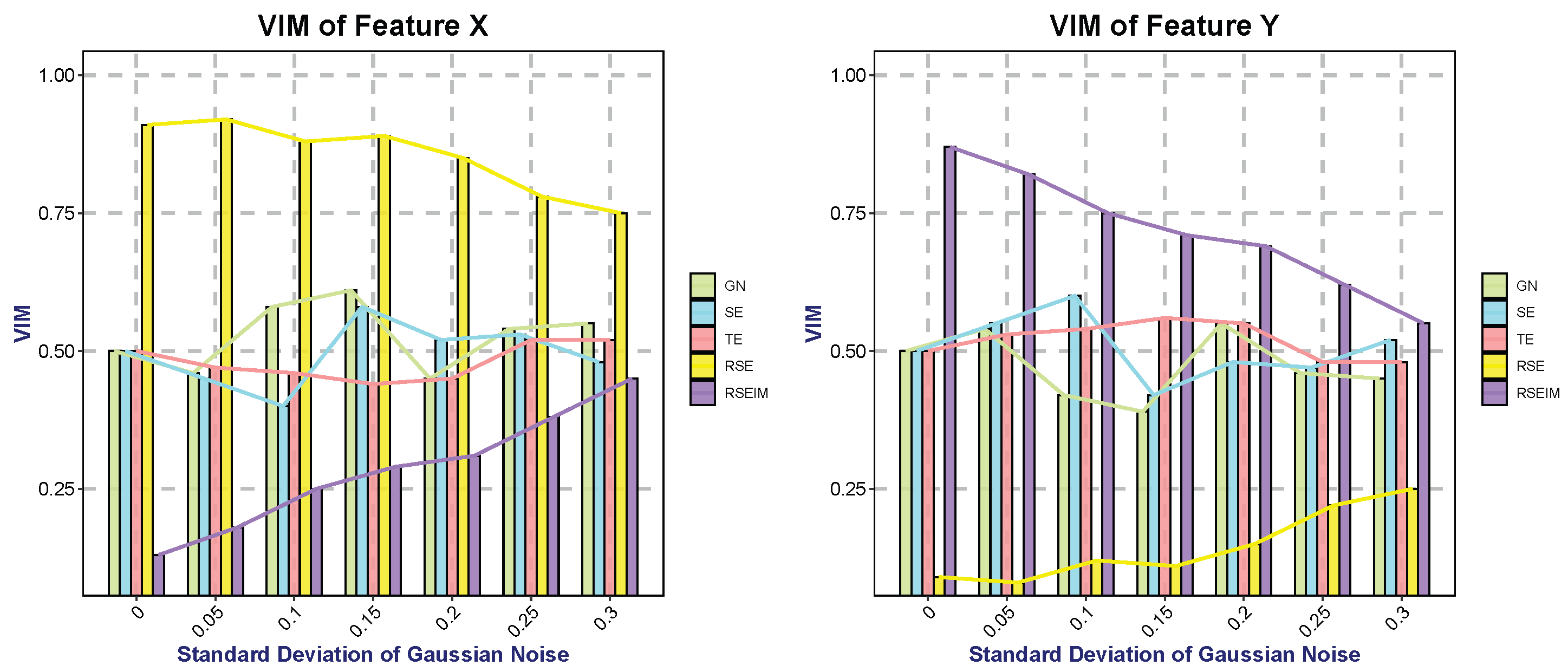
The two-term information method differs from traditional measurement approaches based on entropy or Gini index in that it incorporates the degree of feature dispersion under the constraint of sample categories. Subsets partitioned using this measurement method not only maximize the distinction between sample categories but also prioritize features with the minimum possible variance as the target for node splitting. Consequently, this method naturally revises the conventional evaluation criteria for feature importance—a modification of great significance in the random forest algorithm.
For two features equally capable of fully distinguishing sample categories, their importance scores are identical under traditional metrics. However, after integrating the measurement of feature variance, the feature with a smaller variance will yield a higher importance score. More specifically, we denote the feature importance measurement as and the Gini index as . Assuming that there are c features , the importance score of feature is denoted as , which represents the average reduction in node splitting impurity attributed to feature across all decision trees within the random forest.
The calculation formula for the Gini index of node
m is as follows:
where
denotes the number of categories and
represents the proportion of category
k in node
m. Similarly to Equation (
23), assuming that
is a continuous feature, the two-term Gini index for
is defined as follows:
The importance of feature
at node
m is defined as the difference between the Gini index
of node
m and the variance-corrected Gini indices
and
of the two child nodes (denoted as
l and
r) formed after splitting [
21]. Specifically, it is expressed as follows:
Assuming there are
n trees in the random forest, the normalized importance score of feature
is given by
When
, this calculation method for
degenerates into the feature importance score of a random forest constructed using the traditional Gini index as the splitting criterion. The
calculated with the improved splitting criterion includes
and
; specifically, the greater the variance of feature
in the split child subsets, the lower the resulting importance score. Considering the inconsistent units among different features, it is necessary to normalize the features in advance and attach a penalty coefficient
to the variance. In this way, features with high calculated scores will simultaneously possess the ability to distinguish category
Y and a low variance of their own.
3.4. RSEIM Algorithm
In
Section 2.2, we introduced the concept of
entropy, and in
Section 3.2.2, we proposed an improved two-term information method. Now, we can integrate these two by replacing Shannon entropy with
entropy in
Section 3.2. This amalgamation constitutes the RSEIM algorithm proposed in this paper. Initially, we present the symbols and equations employed in this algorithm:
The specific steps are outlined by the pseudocode in Algorithm 1.
| Algorithm 1: RSEIM algorithm |
- Input:
Data D, Attribute X, Class Y - Output:
A decision tree - 1:
while not satisfying the stop condition do - 2:
Initialize Inf - 3:
for each attribute do - 4:
for each cutting point do - 5:
if then - 6:
- 7:
- 8:
else - 9:
- 10:
- 11:
end if - 12:
Compute according to ( 30) - 13:
end for - 14:
if < then - 15:
- 16:
- 17:
- 18:
end if - 19:
end for - 20:
Grow the tree using X and a, partitioning the data via binary split. - 21:
Go to the beginning for and - 22:
// Recursively repeat the procedure to grow a tree - 23:
end while - 24:
Return: A decision tree - 25:
// Tree is built by nodes from the root to the leaf
|
It is evident that besides the parameters included in the stopping criteria, the algorithm also comprises four free parameters: r and s from the entropy , along with penalty coefficients and from the improved two-term information method. Particularly, when and , the algorithm degenerates into a decision tree algorithm based on entropy without penalty terms. In this paper, we refer to this algorithm as RSE. Additionally, by setting the condition on the RSE algorithm, it degrades to a decision tree algorithm based on Tsallis entropy, termed TE. Setting on the TE algorithm leads to its degradation into a decision tree algorithm based on Shannon entropy, denoted as SE in this paper. If the condition is altered to , it degrades into a decision tree algorithm based on Gini coefficient, labeled as GN in this paper.
The time complexity of a single split in RSEIM depends on the number of features
m and the number of samples
n. For continuous features, the time complexity of model training is
, while for discrete features, it is
, which is comparable to that of traditional decision tree models without hyperparameters [
17]. In fact, the construction process of RSEIM can be simply regarded as building several decision trees with different parameters and selecting the one with the best performance. The number of such trees depends on the density of parameter pairs selected in the space. Compared with a single tree, the difference in complexity only lies in a larger constant factor, and the increase in the number of features and samples will not lead to higher time complexity. However, the storage of multiple parameters will result in higher space complexity, which depends on the spatial density of the selected parameters.
5. Conclusions
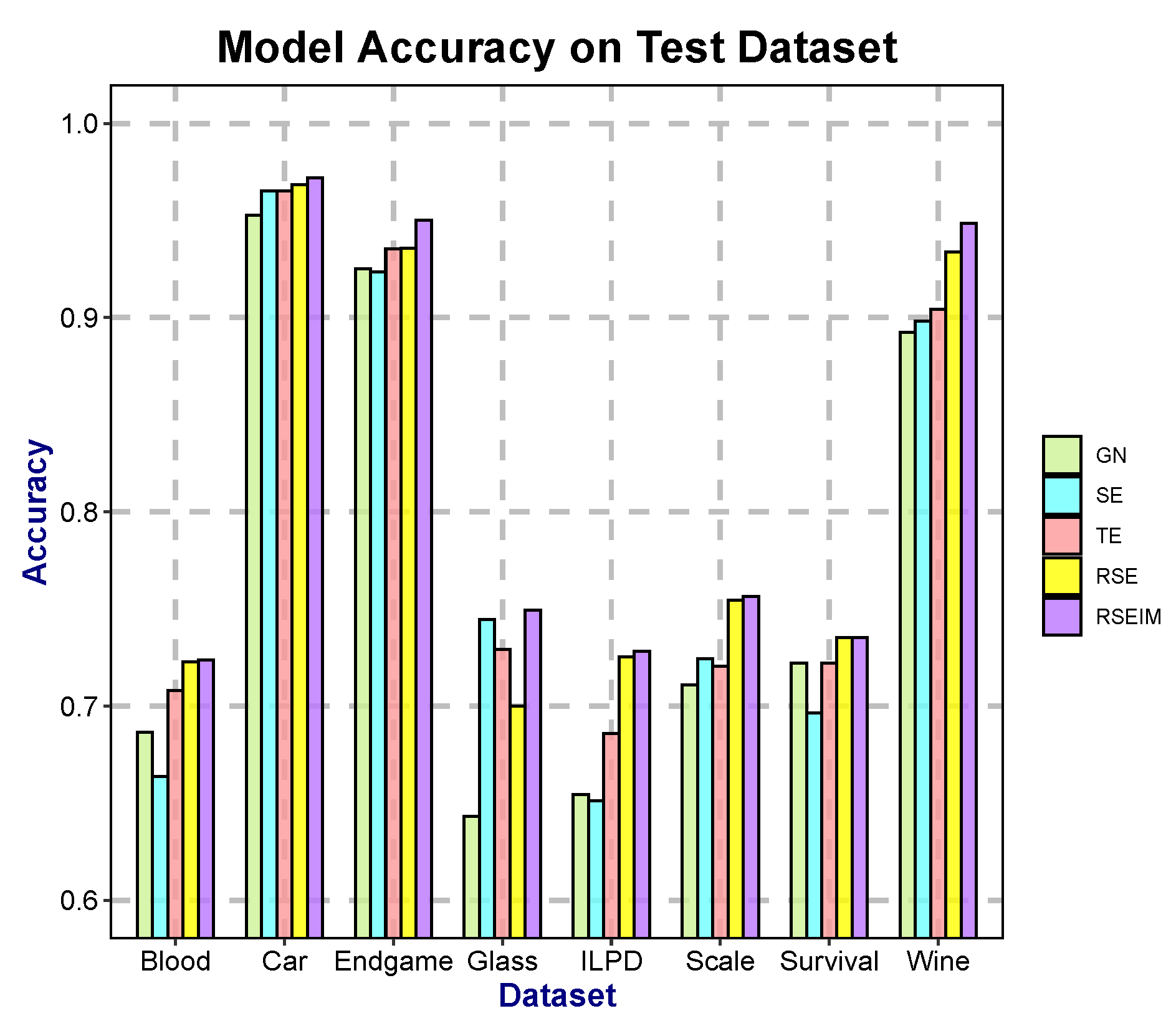
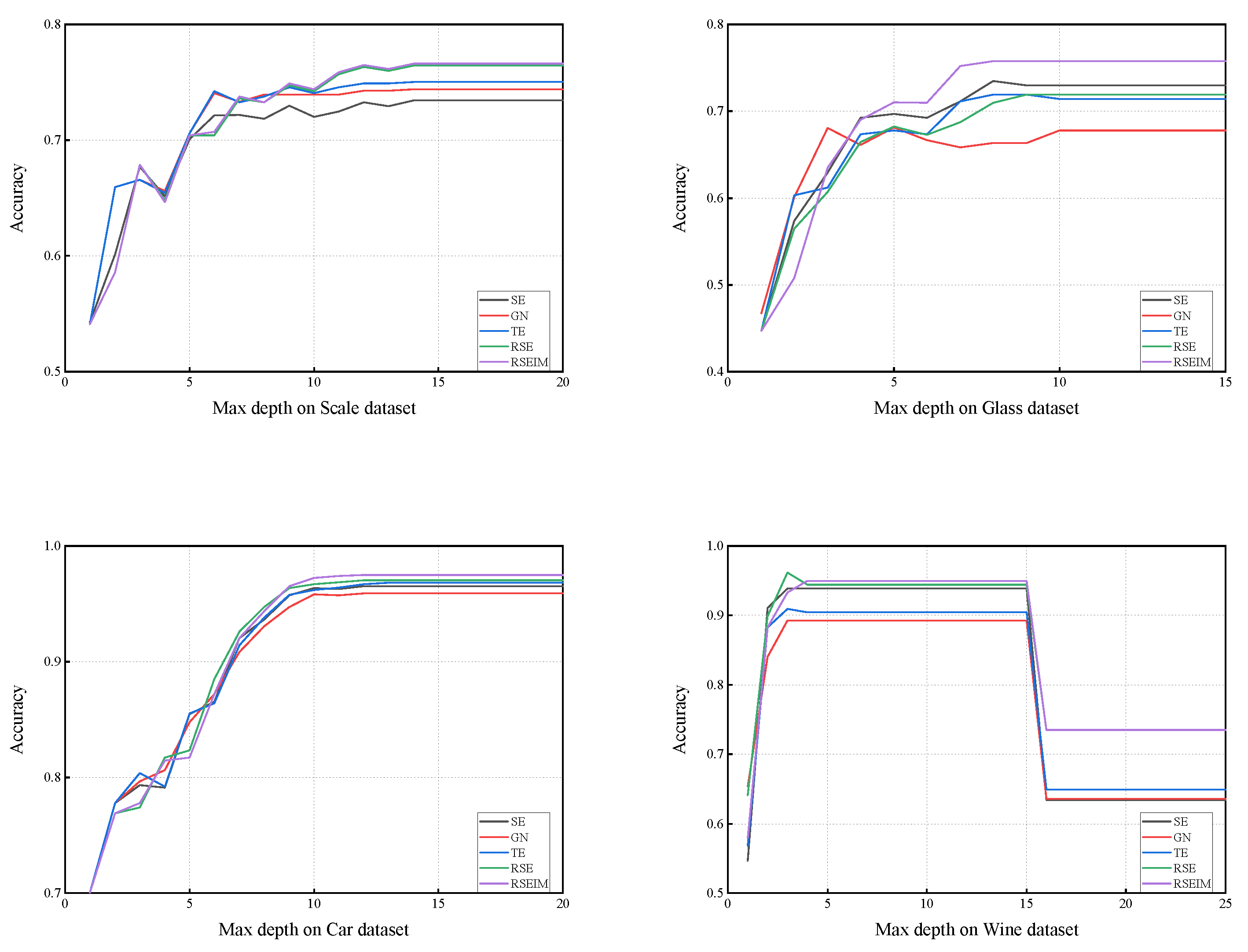
In this paper, we focused on improving the partitioning criteria of decision trees by introducing generalized entropy and decision penalty terms, proposing new decision tree algorithms called RSE and RSEIM. The generalized entropy serves as an extension of traditional partitioning criteria, inheriting their strengths while enhancing flexibility to accommodate a wider range of datasets. The decision penalty term is an enhancement based on two information-measuring methods, considering attribute types and introducing adjustable penalty coefficients. Theoretically, RSE and RSEIM are expected to outperform the algorithms before the enhancement. These enhanced algorithms contain multiple free parameters, and we utilized a genetic algorithm to optimize these parameters in pursuit of higher classification accuracy. Experiments conducted across various datasets demonstrate a significant improvement in accuracy with RSE and RSEIM algorithms compared to traditional decision tree algorithms. Notably, the constructed trees did not exhibit increased complexity despite the improvements achieved by the enhanced algorithms.
In fact, compared with decision tree algorithms, random forests, which are ensembles of decision trees, are now more widely used in machine learning. Random forests do have stronger robustness and stability than individual trees, and they can evaluate the importance of features. In research on optimizing random forest algorithms, there are optimizations in sampling methods, optimizations targeting errors, etc., but there are relatively few studies from the perspective of optimizing the individual trees that constitute the forest. The individual decision trees optimized from the perspective of splitting criteria introduced in this paper may become a new direction for optimizing random forests. Moreover, considering the entropy-based splitting of features can provide more reference information for the final feature importance ranking.