Slope Entropy Characterisation: An Asymmetric Approach to Threshold Parameters Role Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
- The Bern–Barcelona EEG database [40]. This database comprises a collection of both focal and non-focal time series extracted from seizure-free recordings of patients afflicted with pharmacoresistant focal-onset epilepsy. For the purpose of our experiments, we employed 50 records, each possessing a length of 10,240 data points, sampled at a frequency of 512 Hz. This database has also been used in works such as [41], which includes a review of results achieved in other classification studies based on time series from this same database.
- The Fantasia RR database [42]: It presents a meticulously curated repository comprising a total of 40 individual time series, thoughtfully stratified into two cohorts of 20 records each (mature subjects and a counterpart assembly of youthful subjects). All subjects were, in principle, healthy, thereby obviating possible confounding health-related factors. They were monitored over an extended period of 120 min, with a sampling frequency of 250 Hz. This database has been used in many studies, such as [43,44].
- The Ford A dataset [45]: It is a repository of data gleaned from an automotive subsystem. The principal objective underpinning its creation was the empirical evaluation of the efficacy of classification schemes upon the acoustic characteristics of engine noise. Within the ambit of this experimental undertaking, a corpus of 40 discrete records was selected and subsequently employed for analysis from each distinct class.
- The House Twenty dataset [46,47]: It is a compendium of temporal sequences emanating from 40 distinct domestic entities. They are part of the Personalised Retrofit Decision Support Tools for UK Homes using Smart Home Technology (REFIT) project. This dataset includes data from 40 households, divided into two classes with 20 each: The first class represents the consumption of electricity in general, and the second class represents the specific electrical consumption of dryers and washing machines. We also used this dataset in our previous work [34].
- The PAF (Paroxysmal Atrial Fibrillation) prediction dataset [48]: This dataset comprises discrete 5-min temporal recordings corresponding to patients diagnosed with PAF. These temporal records were classified into two distinct categories: The first pertains to recordings that immediately precede the onset of a PAF episode, while the second encompasses instances temporally distant from any PAF manifestation. Each classification category comprises a total of 25 distinct files. This is a very well-known dataset used in a myriad of scientific works [49,50,51].
- The Worms two-class dataset [52,53]: It contains a time series intrinsically linked to the locomotive patterns exhibited by a distinct species of worm used in the realm of behavioural genetics research. We selected records from two classes: mutant and non-mutant worms. The first type contains 75 records of 900 samples, and the second type has 105 records with the same time series length. As with previous datasets, there are other works that used time series from this one [54,55].
- The Bonn EEG dataset [56,57]. This dataset encapsulates a corpus of 4097 electroencephalograms, each one with a duration of 23.6 s. These instances are distinctly categorised into five salient classes (A, B, C, D, and E), reflecting the underlying diversity of neural activity scenarios under consideration. Specifically, the classes include healthy subjects with eyes open (Class A) and those with eyes closed (Class B). Other instances pertain to epileptic subjects classified as Class C, D, and E (see further details in [57]). For the scope of the specific experiments in the present paper, the focus was directed solely towards classes D and E (seizure-free periods at the epileptogenic zone, and seizure activity from the hippocampal focus), with 100 records from each class. There are many examples available of works using this same dataset [58,59,60].
- The Synthetic database. As its name suggests, it is a collection of datasets that have been generated artificially by a computer. It is composed of three different sets: Synth1, Synth2, and Synth3. Each one of them is composed of two classes; the first one is generated by a normal (Gaussian) distribution, while the other is based on a uniform distribution. Each class contains 20 time series with a length of 3000 samples. Regarding the parameters used to generate the series, Synth1 uses mean = 0 and standard deviation equal to 5 (SD = 5) for its Gaussian class. Synth2’s Gaussian class uses mean = 0 and SD = 10. In the case of Synth3, mean = 0 and SD = 20. On the other hand, the uniform distribution used to generate the second class uses the same parameters for all three datasets, drawing samples uniformly from the range [−1, 1]. It has been included for reference purposes, but it is not used in all the experiments since it is not as illustrative as the real datasets.
- (True Positives) are instances correctly identified as belonging to a particular class.
- (True Negatives) are instances correctly identified as not belonging to that class.
- (False Positives) are instances incorrectly identified as belonging to that class.
- (False Negatives) are instances incorrectly identified as not belonging to that class.
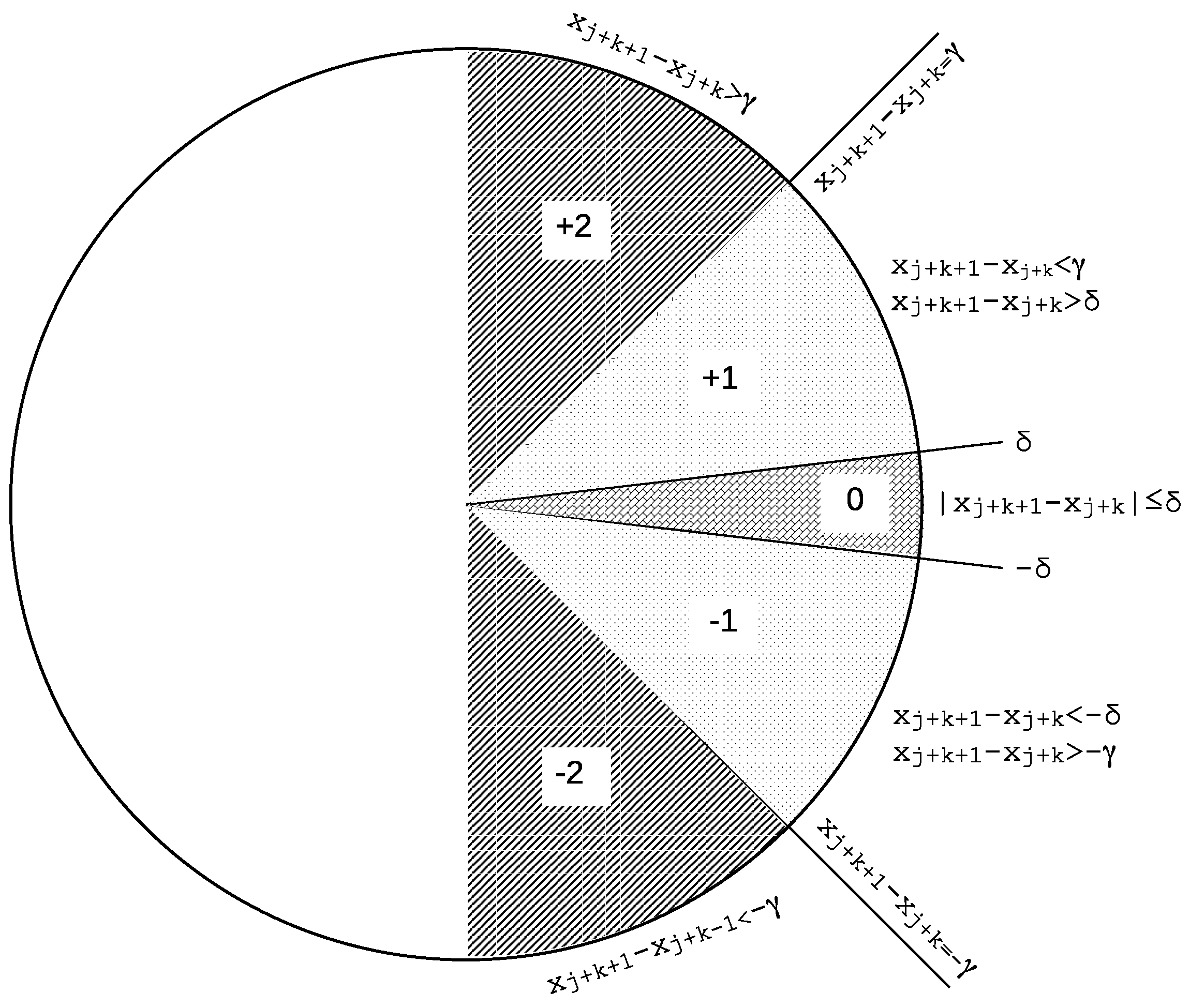
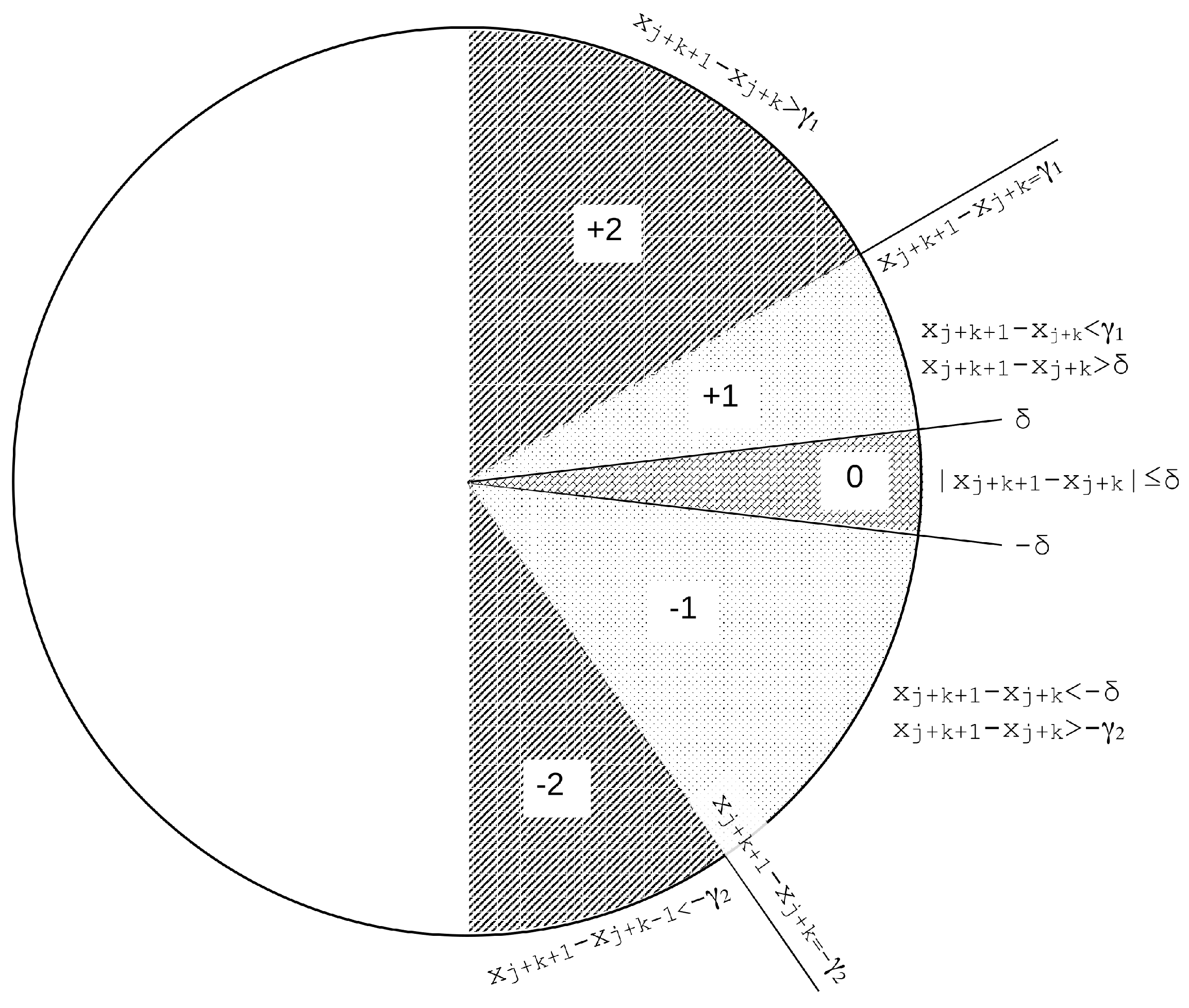
2.2. Slope Entropy
- If > , the symbol assigned to the current active symbolic pattern position is (or just 2), .
- Else, if > , the symbol assigned to the current active symbolic pattern position is (or just 1), .
- Else, if , the case when two consecutive values are very similar (depending on threshold ), which includes the case for ties when [61], the symbol assigned to the current active symbolic pattern position is 0, .
- Else, if but , the symbol assigned to the current active symbolic pattern position is , .
- Otherwise, the symbol assigned is , region where , .
- . In order to obtain the symbolic representation of this subsequence, we compute . Applying the thresholding method described above results in a symbolic pattern .
- . In order to obtain the symbolic representation of this subsequence, we compute . Applying the thresholding method described above results in a symbolic pattern .
- . In order to obtain the symbolic representation of this subsequence, we compute . Applying the thresholding method described above results in a symbolic pattern .
- . In order to obtain the symbolic representation of this subsequence, we compute . Applying the thresholding method described above results in a symbolic pattern .
3. Experiments and Results
3.1. Experiments
3.2. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Vargas, B.; Cuesta-Frau, D.; Ruiz-Esteban, R.; Cirugeda, E.; Varela, M. What Can Biosignal Entropy Tell Us About Health and Disease? Applications in Some Clinical Fields. Nonlinear Dyn. Psychol. Life Sci. 2015, 19, 419–436. [Google Scholar]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol.-Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, X.; Wang, D.; Wang, Z.; Hua, X. Analysis of Weak Fault in Hydraulic System Based on Multi-scale Permutation Entropy of Fault-Sensitive Intrinsic Mode Function and Deep Belief Network. Entropy 2019, 21, 425. [Google Scholar] [CrossRef] [PubMed]
- Danylchuk, H.; Chebanova, N.; Reznik, N.; Vitkovskyi, Y. Modeling of investment attractiveness of countries using entropy analysis of regional stock markets. Glob. J. Environ. Sci. Manag. 2019, 5, 227–235. [Google Scholar] [CrossRef]
- Garland, J.; Jones, T.R.; Neuder, M.; Morris, V.; White, J.W.C.; Bradley, E. Anomaly Detection in Paleoclimate Records Using Permutation Entropy. Entropy 2018, 20, 931. [Google Scholar] [CrossRef]
- Drzazga-Szczȩśniak, E.A.; Szczepanik, P.; Kaczmarek, A.Z.; Szczȩśniak, D. Entropy of Financial Time Series Due to the Shock of War. Entropy 2023, 25, 823. [Google Scholar] [CrossRef]
- Dastgoshadeh, M.; Rabiei, Z. Detection of epileptic seizures through EEG signals using entropy features and ensemble learning. Front. Hum. Neurosci. 2023, 16, 4061. [Google Scholar] [CrossRef]
- Aljalal, M.; Aldosari, S.A.; Molinas, M.; AlSharabi, K.; Alturki, F.A. Detection of Parkinson’s disease from EEG signals using discrete wavelet transform, different entropy measures, and machine learning techniques. Sci. Rep. 2022, 12, 22547. [Google Scholar] [CrossRef]
- Vargas, B.; Cuesta-Frau, D.; González-López, P.; Fernández-Cotarelo, M.J.; Vázquez-Gómez, O.; Colás, A.; Varela, M. Discriminating Bacterial Infection from Other Causes of Fever Using Body Temperature Entropy Analysis. Entropy 2022, 24, 510. [Google Scholar] [CrossRef]
- Chen, W.; Zhuang, J.; Yu, W.; Wang, Z. Measuring complexity using FuzzyEn, ApEn, and SampEn. Med. Eng. Phys. 2009, 31, 61–68. [Google Scholar] [CrossRef]
- Bandt, C.; Pompe, B. Permutation Entropy: A Natural Complexity Measure for Time Series. Phys. Rev. Lett. 2002, 88, 174102. [Google Scholar] [CrossRef] [PubMed]
- Parlitz, U.; Berg, S.; Luther, S.; Schirdewan, A.; Kurths, J.; Wessel, N. Classifying cardiac biosignals using ordinal pattern statistics and symbolic dynamics. Comput. Biol. Med. 2012, 42, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Amigó, J. Permutation Complexity in Dynamical Systems; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar] [CrossRef]
- Ravelo-Garcia, A.G.; Navarro-Mesa, J.L.; Casanova-Blancas, U.; González, S.; Quintana, P.; Guerra-Moreno, I.; Canino, J.; Hernandez-Pérez, E. Application of the Permutation Entropy over the Heart Rate Variability for the Improvement of Electrocardiogram-based Sleep Breathing Pause Detection. Entropy 2015, 17, 914–927. [Google Scholar] [CrossRef]
- Mateos, D.; Diaz, J.; Lamberti, P. Permutation Entropy Applied to the Characterization of the Clinical Evolution of Epileptic Patients under Pharmacological Treatment. Entropy 2014, 16, 5668–5676. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, M.; Niu, Y.; Li, C.; Cao, R.; Wang, B.; Yan, P.; Ma, Y.; Xiang, J. Epileptic Seizure Prediction Based on Permutation Entropy. Front. Comput. Neurosci. 2018, 12, 55. [Google Scholar] [CrossRef]
- Hight, D.; Obert, D.P.; Kratzer, S.; Schneider, G.; Sepulveda, P.; Sleigh, J.; García, P.S.; Kreuzer, M. Permutation entropy is not an age-independent parameter for EEG-based anesthesia monitoring. Front. Aging Neurosci. 2023, 15, 1173304. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, Y.; Liu, M.; Kong, L. Bearings fault diagnosis based on adaptive local iterative filtering–multiscale permutation entropy and multinomial logistic model with group–lasso. Adv. Mech. Eng. 2019, 11, 1687814019836311. [Google Scholar] [CrossRef]
- Gao, Y.; Villecco, F.; Li, M.; Song, W. Multi-Scale Permutation Entropy Based on Improved LMD and HMM for Rolling Bearing Diagnosis. Entropy 2017, 19, 176. [Google Scholar] [CrossRef]
- Li, Y.; Hu, C.; Shen, Y.; Han, B.; Yang, J.; Xu, G. A New Methodology for Early Detection of Thermoacoustic Combustion Oscillations Based on Permutation Entropy. J. Therm. Sci. 2023, 32, 2310–2320. [Google Scholar] [CrossRef]
- Henry, M.; Judge, G. Permutation Entropy and Information Recovery in Nonlinear Dynamic Economic Time Series. Econometrics 2019, 7, 10. [Google Scholar] [CrossRef]
- Zhang, Y.; Shang, P. Permutation entropy analysis of financial time series based on Hill’s diversity number. Commun. Nonlinear Sci. Numer. Simul. 2017, 53, 288–298. [Google Scholar] [CrossRef]
- Siokis, F. High short interest stocks performance during the Covid-19 crisis: An informational efficacy measure based on permutation-entropy approach. J. Econ. Stud. 2023, 50, 1570–1584. [Google Scholar] [CrossRef]
- Kilpua, E.K.J.; Good, S.; Ala-Lahti, M.; Osmane, A.; Koikkalainen, V. Permutation Entropy and Complexity Analysis of Large-scale Solar Wind Structures and Streams. EGUsphere 2023, 2023, 1–21. [Google Scholar] [CrossRef]
- Konstantinou, K.I.; Rahmalia, D.A.; Nurfitriana, I.; Ichihara, M. Permutation entropy variations in seismic noise before and after eruptive activity at Shinmoedake volcano, Kirishima complex, Japan. Earth Planets Space 2022, 74, 175. [Google Scholar] [CrossRef]
- Cheng, W.; Chen, H.; Tian, L.; Ma, Z.; Cui, X. Heart rate variability in different sleep stages is associated with metabolic function and glycemic control in type 2 diabetes mellitus. Front. Physiol. 2023, 14, 1157270. [Google Scholar] [CrossRef]
- Wang, Y.H.; Chen, I.Y.; Chiueh, H.; Liang, S.F. A Low-Cost Implementation of Sample Entropy in Wearable Embedded Systems: An Example of Online Analysis for Sleep EEG. IEEE Trans. Instrum. Meas. 2021, 70, 4002412. [Google Scholar] [CrossRef]
- Olbrys, J.; Majewska, E. Approximate entropy and sample entropy algorithms in financial time series analyses. Procedia Comput. Sci. 2022, 207, 255–264. [Google Scholar] [CrossRef]
- Zhou, X.; Lin, J.S.; Liang, X.; Xu, W. Rainfall Patterns From Multiscale Sample Entropy Analysis. Front. Water 2022, 4, 885456. [Google Scholar] [CrossRef]
- Cuesta-Frau, D. Slope Entropy: A New Time Series Complexity Estimator Based on Both Symbolic Patterns and Amplitude Information. Entropy 2019, 21, 1167. [Google Scholar] [CrossRef]
- Riedl, M.; Müller, A.; Wessel, N. Practical considerations of permutation entropy. Eur. Phys. J. Spec. Top. 2013, 222, 249–262. [Google Scholar] [CrossRef]
- Cuesta-Frau, D.; Schneider, J.; Bakštein, E.; Vostatek, P.; Spaniel, F.; Novák, D. Classification of Actigraphy Records from Bipolar Disorder Patients Using Slope Entropy: A Feasibility Study. Entropy 2020, 22, 1243. [Google Scholar] [CrossRef]
- Cuesta-Frau, D.; Dakappa, P.H.; Mahabala, C.; Gupta, A.R. Fever Time Series Analysis Using Slope Entropy. Application to Early Unobtrusive Differential Diagnosis. Entropy 2020, 22, 1034. [Google Scholar] [CrossRef] [PubMed]
- Kouka, M.; Cuesta-Frau, D. Slope Entropy Characterisation: The Role of the δ Parameter. Entropy 2022, 24, 1456. [Google Scholar] [PubMed]
- Li, D.; Liang, Z.; Wang, Y.; Hagihira, S.; Sleigh, J.W.; Li, X. Parameter selection in permutation entropy for an electroencephalographic measure of isoflurane anesthetic drug effect. J. Clin. Monit. Comput. 2013, 27, 113–123. [Google Scholar] [CrossRef]
- Zhan, J.; Gan, Z.; Chou, L.; Hu, L.; Zhou, Y.; Yang, H.; Chou, Y. A fast permutation entropy for pulse rate variability online analysis with one-sample recursion. Med. Eng. Phys. 2023, 120, 104050. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Liang, X.; Zhu, T.; Zhao, L.; Li, J.; Liu, C. A New Physically Meaningful Threshold of Sample Entropy for Detecting Cardiovascular Diseases. Entropy 2019, 21, 830. [Google Scholar] [CrossRef]
- Alcaraz, R.; Abásolo, D.; Hornero, R.; Rieta, J. Study of Sample Entropy ideal computational parameters in the estimation of atrial fibrillation organization from the ECG. In Proceedings of the 2010 Computing in Cardiology, Belfast, UK, 26–29 September 2010; pp. 1027–1030. [Google Scholar]
- Jiang, Y.; Mao, D.; Xu, Y. A fast algorithm for computing sample entropy. Adv. Adapt. Data Anal. 2011, 3, 167–186. [Google Scholar] [CrossRef]
- Andrzejak, R.G.; Schindler, K.; Rummel, C. Nonrandomness, nonlinear dependence, and nonstationarity of electroencephalographic recordings from epilepsy patients. Phys. Rev. E 2012, 86, 046206. [Google Scholar] [CrossRef]
- Acharya, U.R.; Hagiwara, Y.; Deshpande, S.N.; Suren, S.; Koh, J.E.W.; Oh, S.L.; Arunkumar, N.; Ciaccio, E.J.; Lim, C.M. Characterization of focal EEG signals: A review. Future Gener. Comput. Syst. 2018, 91, 290–299. [Google Scholar] [CrossRef]
- Iyengar, N.; Peng, C.; Morin, R.; Goldberger, A.L.; Lipsitz, L.A. Age-related alterations in the fractal scaling of cardiac interbeat interval dynamics. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 1996, 271, R1078–R1084. [Google Scholar] [CrossRef]
- Sharma, K.; Sunkaria, R.K. Cardiac arrhythmia detection using cross-sample entropy measure based on short and long RR interval series. J. Arrhythmia 2023, 39, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Mandic, D.P. Variational Embedding Multiscale Sample Entropy: A Tool for Complexity Analysis of Multichannel Systems. Entropy 2022, 24, 26. [Google Scholar] [CrossRef]
- FordA Description. Available online: http://www.timeseriesclassification.com/description.php?Dataset=FordA (accessed on 28 February 2022).
- HouseTwenty Description. Available online: http://www.timeseriesclassification.com/description.php?Dataset=HouseTwenty.htm (accessed on 28 February 2022).
- Murray, D.; Liao, J.; Stankovic, L.; Stankovic, V.; Hauxwell-Baldwin, R.; Wilson, C.; Coleman, M.; Kane, T.; Firth, S. A data management platform for personalised real-time energy feedback. In Proceedings of the 8th International Conference on Energy Efficiency in Domestic Appliances and Lighting, Lucerne, Switzerland, 26–28 August 2015. [Google Scholar]
- Moody, G.; Goldberger, A.; McClennen, S.; Swiryn, S. Predicting the onset of paroxysmal atrial fibrillation: The Computers in Cardiology Challenge 2001. In Proceedings of the Computers in Cardiology 2001. Vol. 28 (Cat. No. 01CH37287), Rotterdam, The Netherlands, 23–26 September 2001; IEEE: Piscataway, NJ, USA, 2001; pp. 113–116. [Google Scholar]
- Mendez, M.M.; Hsu, M.C.; Yuan, J.T.; Lynn, K.S. A Heart Rate Variability-Based Paroxysmal Atrial Fibrillation Prediction System. Appl. Sci. 2022, 12, 2387. [Google Scholar] [CrossRef]
- Wang, L.H.; Yan, Z.H.; Yang, Y.T.; Chen, J.Y.; Yang, T.; Kuo, I.C.; Abu, P.A.R.; Huang, P.C.; Chen, C.A.; Chen, S.L. A Classification and Prediction Hybrid Model Construction with the IQPSO-SVM Algorithm for Atrial Fibrillation Arrhythmia. Sensors 2021, 21, 5222. [Google Scholar] [CrossRef] [PubMed]
- Olier, I.; Ortega-Martorell, S.; Pieroni, M.; Lip, G.Y.H. How machine learning is impacting research in atrial fibrillation: Implications for risk prediction and future management. Cardiovasc. Res. 2021, 117, 1700–1717. [Google Scholar] [CrossRef] [PubMed]
- WormsTwoClass Description. Available online: http://www.timeseriesclassification.com/description.php?Dataset=WormsTwoClass.htm (accessed on 28 February 2022).
- Yemini, E.; Jucikas, T.; Grundy, L.J.; Brown, A.E.; Schafer, W.R. A database of Caenorhabditis elegans behavioral phenotypes. Nat. Methods 2013, 10, 877–879. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Park, C.M. Novel Features for Binary Time Series Based on Branch Length Similarity Entropy. Entropy 2021, 23, 480. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Bates, K.; Elchesen, A.; Hartsock, I.; Lu, H.; Bubenik, P. Topological Data Analysis of C. elegans Locomotion and Behavior. Front. Artif. Intell. 2021, 4, 668395. [Google Scholar] [CrossRef]
- Tsipouras, M.G. Spectral information of EEG signals with respect to epilepsy classification. EURASIP J. Adv. Signal Process. 2019, 2019, 10. [Google Scholar] [CrossRef]
- Andrzejak, R.G.; Lehnertz, K.; Mormann, F.; Rieke, C.; David, P.; Elger, C.E. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Phys. Rev. E 2001, 64, 061907. [Google Scholar]
- Zaid, Y.; Sah, M.; Direkoglu, C. Pre-processed and combined EEG data for epileptic seizure classification using deep learning. Biomed. Signal Process. Control 2023, 84, 104738. [Google Scholar] [CrossRef]
- Wong, S.; Simmons, A.; Rivera-Villicana, J.; Barnett, S.; Sivathamboo, S.; Perucca, P.; Ge, Z.; Kwan, P.; Kuhlmann, L.; Vasa, R.; et al. EEG datasets for seizure detection and prediction— A review. Epilepsia Open 2023, 8, 252–267. [Google Scholar] [CrossRef]
- Chen, W.; Wang, Y.; Ren, Y.; Jiang, H.; Du, G.; Zhang, J.; Li, J. An automated detection of epileptic seizures EEG using CNN classifier based on feature fusion with high accuracy. BMC Med. Inform. Decis. Mak. 2023, 23, 96. [Google Scholar] [CrossRef]
- Cuesta-Frau, D.; Varela-Entrecanales, M.; Molina-Picó, A.; Vargas, B. Patterns with Equal Values in Permutation Entropy: Do They Really Matter for Biosignal Classification? Complexity 2018, 2018, 1324696. [Google Scholar] [CrossRef]
- Li, Y.; Mu, L.; Gao, P. Particle Swarm Optimization Fractional Slope Entropy: A New Time Series Complexity Indicator for Bearing Fault Diagnosis. Fractal Fract. 2022, 6, 345. [Google Scholar] [CrossRef]
- Li, Y.; Gao, P.; Tang, B.; Yi, Y.; Zhang, J. Double Feature Extraction Method of Ship-Radiated Noise Signal Based on Slope Entropy and Permutation Entropy. Entropy 2022, 24, 22. [Google Scholar] [CrossRef]
- Cuesta–Frau, D. Permutation entropy: Influence of amplitude information on time series classification performance. Math. Biosci. Eng. 2019, 16, 6842. [Google Scholar] [CrossRef]
- Zunino, L.; Olivares, F.; Scholkmann, F.; Rosso, O.A. Permutation entropy based time series analysis: Equalities in the input signal can lead to false conclusions. Phys. Lett. A 2017, 381, 1883–1892. [Google Scholar] [CrossRef]
| Dataset | Baseline | SlpEn with | Parameters |
|---|---|---|---|
| Bern–Barcelona | 80% | 81% | , , , |
| Fantasia | 85% | 89% | , , , |
| Ford A | 83% | 94% | , , , |
| House Twenty | 95% | 97% | , , , |
| PAF prediction | 76% | 84% | , , , |
| Worms two-class | 70% | 92% | , , , |
| Bonn EEG | 94% | 95% | , , , |
| Synth1 | 92% | 95% | , , , |
| Synth2 | 87% | 89% | , , , |
| Synth3 | 95% | 95% | , , , |
| Dataset | Baseline | SlpEn without | Parameters |
|---|---|---|---|
| Bern–Barcelona | 76% | 77% | , , |
| Fantasia | 82% | 89% | , , |
| Ford A | 82% | 94% | , , |
| House Twenty | 95% | 95% | , , |
| PAF prediction | 76% | 80% | , , |
| Worms two-class | 69% | 72% | , , |
| Bonn EEG | 93% | 93% | , , |
| Synth1 | 89% | 89% | , , |
| Synth2 | 87% | 89% | , , |
| Synth3 | 86% | 89% | , , |
| Dataset | SlpEn with | SlpEn without | ||
|---|---|---|---|---|
| Mean | sd | Mea | sd | |
| Bern–Barcelona | 76.81% | 1.82 | 68.04% | 2.98 |
| Fantasia | 78.15% | 3.66 | 75.18% | 3.63 |
| Ford A | 93.87% | 2.30 | 93.98% | 2.00 |
| House Twenty | 97.36% | 0.00 | 75.25% | 1.44 |
| PAF prediction | 72.36% | 2.77 | 65.62% | 2.17 |
| Worms two-class | 92.02% | 1.18 | 69.49% | 1.63 |
| Bonn EEG | 85.41% | 4.42 | 81.63% | 7.01 |
| Synth1 | 75.70% | 3.21 | 69.54% | 5.26 |
| Synth2 | 71.02% | 2.11 | 68.34% | 4.80 |
| Synth3 | 72.06% | 3.31 | 70.63% | 3.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kouka, M.; Cuesta-Frau, D.; Moltó-Gallego, V. Slope Entropy Characterisation: An Asymmetric Approach to Threshold Parameters Role Analysis. Entropy 2024, 26, 82. https://doi.org/10.3390/e26010082
Kouka M, Cuesta-Frau D, Moltó-Gallego V. Slope Entropy Characterisation: An Asymmetric Approach to Threshold Parameters Role Analysis. Entropy. 2024; 26(1):82. https://doi.org/10.3390/e26010082
Chicago/Turabian StyleKouka, Mahdy, David Cuesta-Frau, and Vicent Moltó-Gallego. 2024. "Slope Entropy Characterisation: An Asymmetric Approach to Threshold Parameters Role Analysis" Entropy 26, no. 1: 82. https://doi.org/10.3390/e26010082
APA StyleKouka, M., Cuesta-Frau, D., & Moltó-Gallego, V. (2024). Slope Entropy Characterisation: An Asymmetric Approach to Threshold Parameters Role Analysis. Entropy, 26(1), 82. https://doi.org/10.3390/e26010082







