On Sequential Bayesian Inference for Continual Learning
Abstract
1. Introduction
2. Background
2.1. The Continual Learning Problem
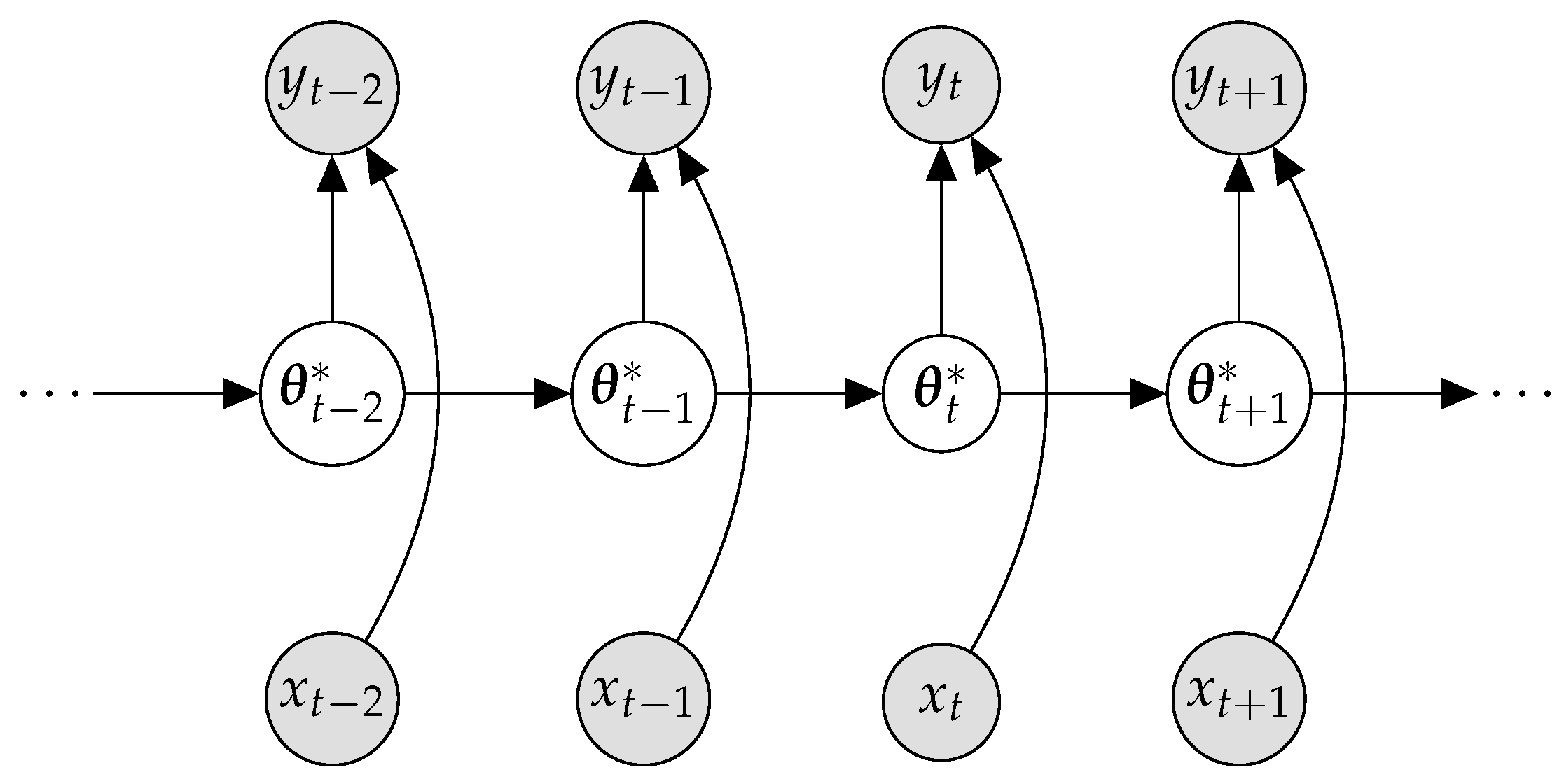
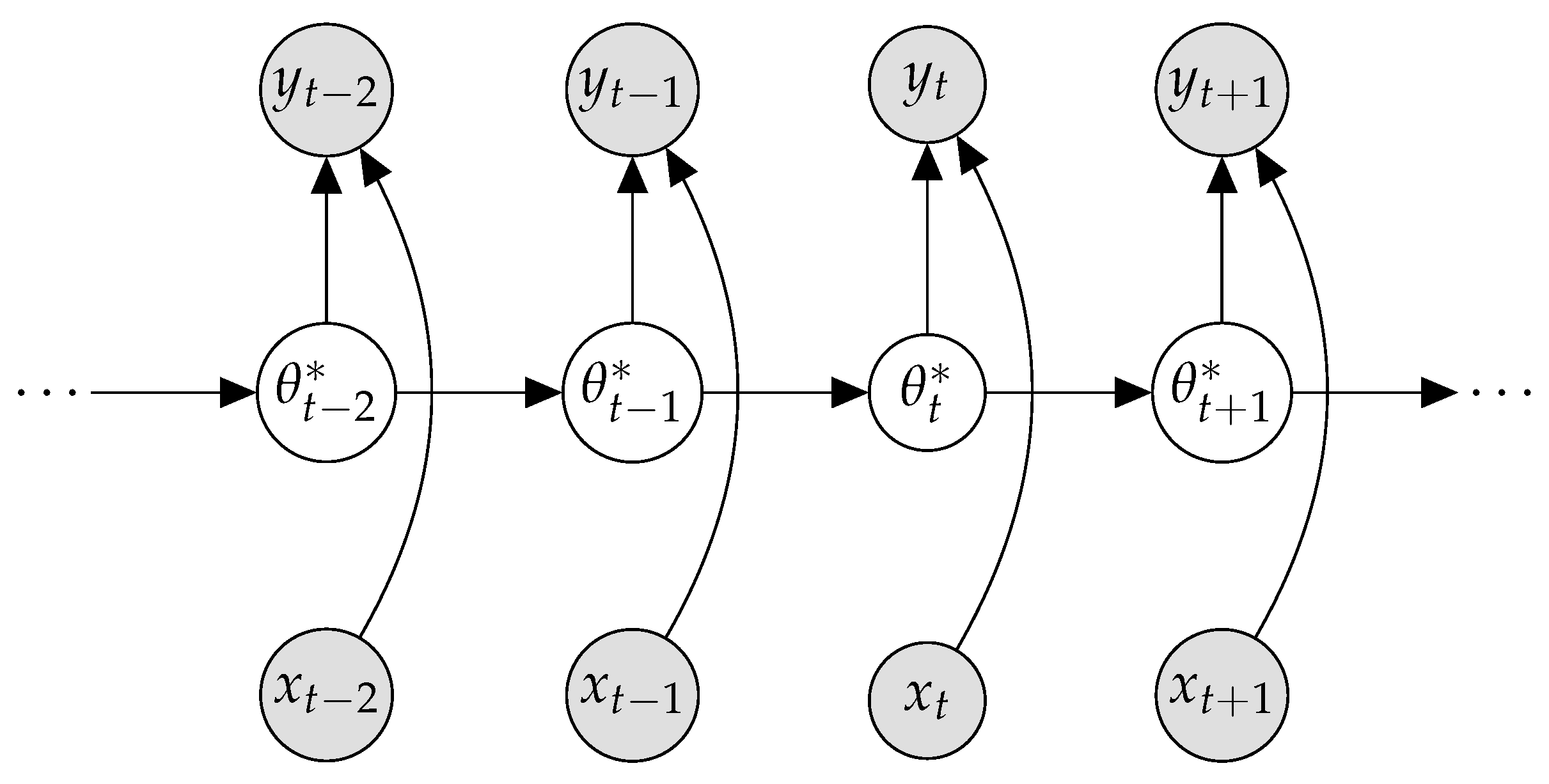
2.2. Bayesian Continual Learning
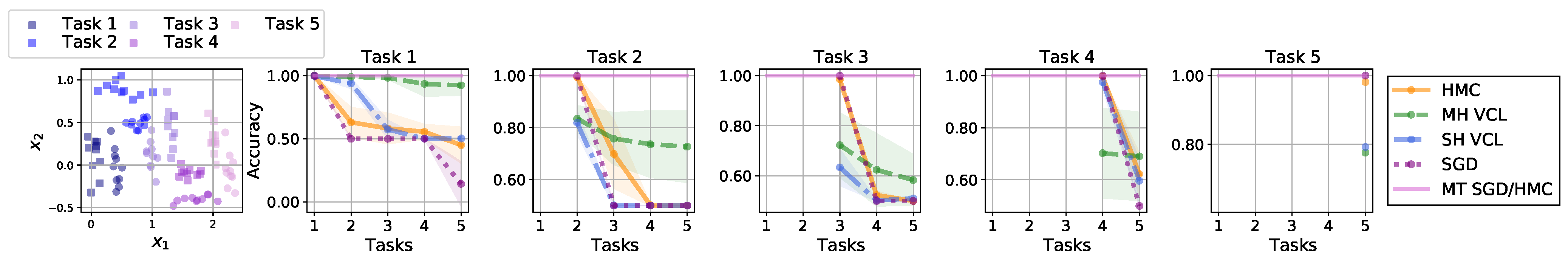
Continual Learning Example: Split-MNIST
2.3. Variational Continual Learning
3. Bayesian Continual Learning with Hamiltonian Monte Carlo
4. Bayesian Continual Learning and Model Misspecification
5. Sequential Bayesian Inference and Imbalanced Task Data
6. Related Work
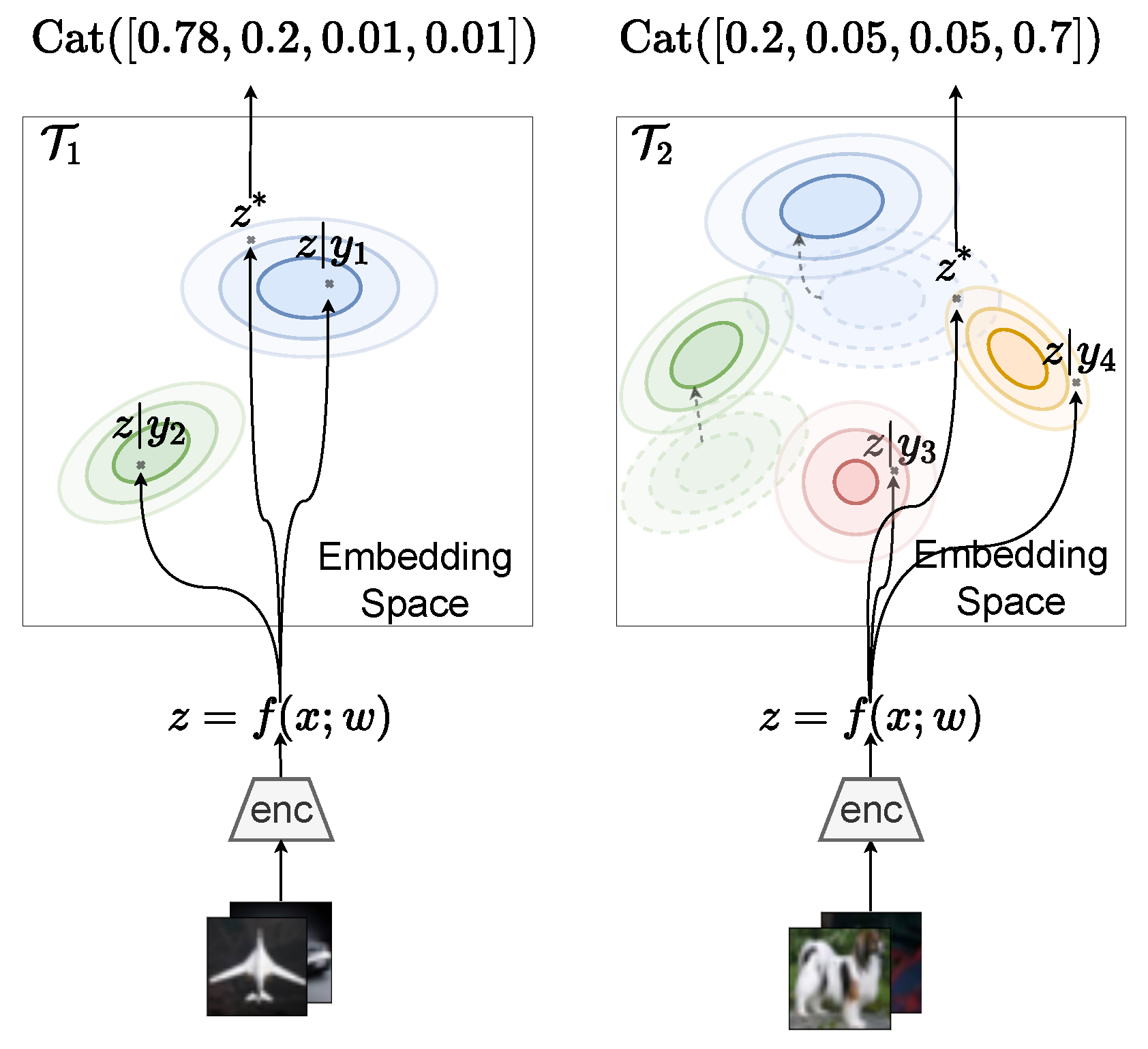
7. Prototypical Bayesian Continual Learning
| Algorithm 1 ProtoCL continual learning |
8. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CL | Continual Learning |
| NN | Neural Network |
| BNN | Bayesian Neural Network |
| HMC | Hamiltonian Monte Carlo |
| VCL | Variational Continual Learning |
| SGD | Stochastic Gradient Descent |
| SH | Single Head |
| MH | Multi-head |
| GMM | Gaussian Mixture Model |
| ProtoCL | Prototypical Bayesian Continual Learning |
Appendix A. The Toy Gaussians Dataset
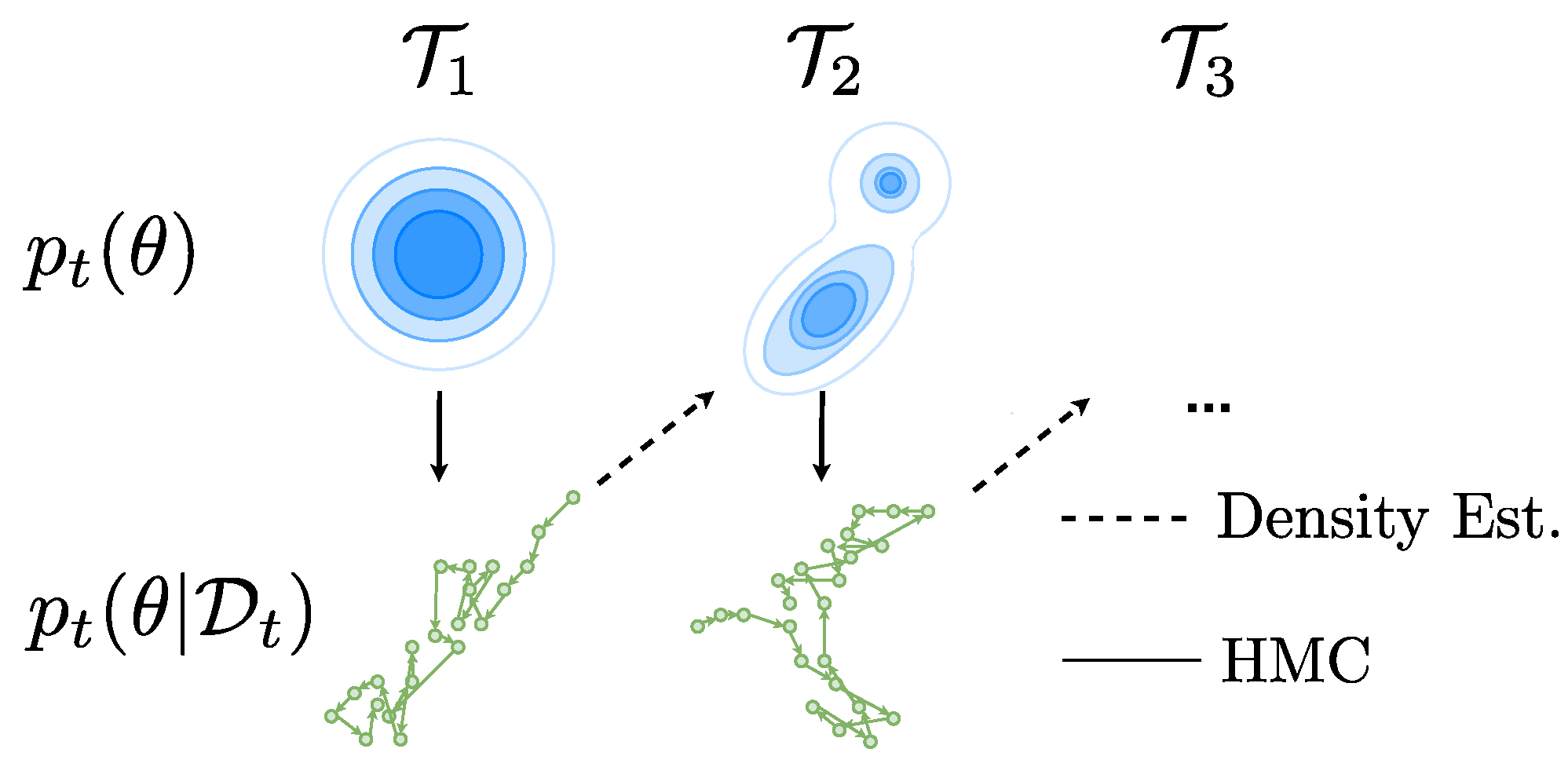
Appendix B. HMC Implementation Details
Appendix C. Density Estimation Diagnostics


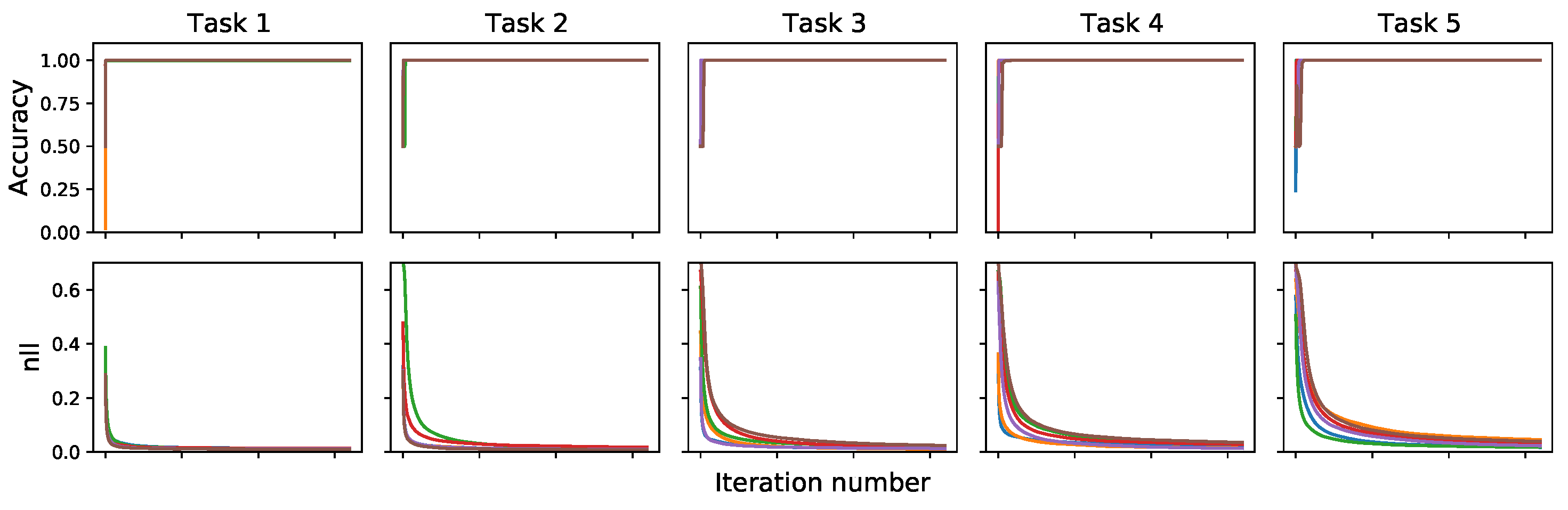
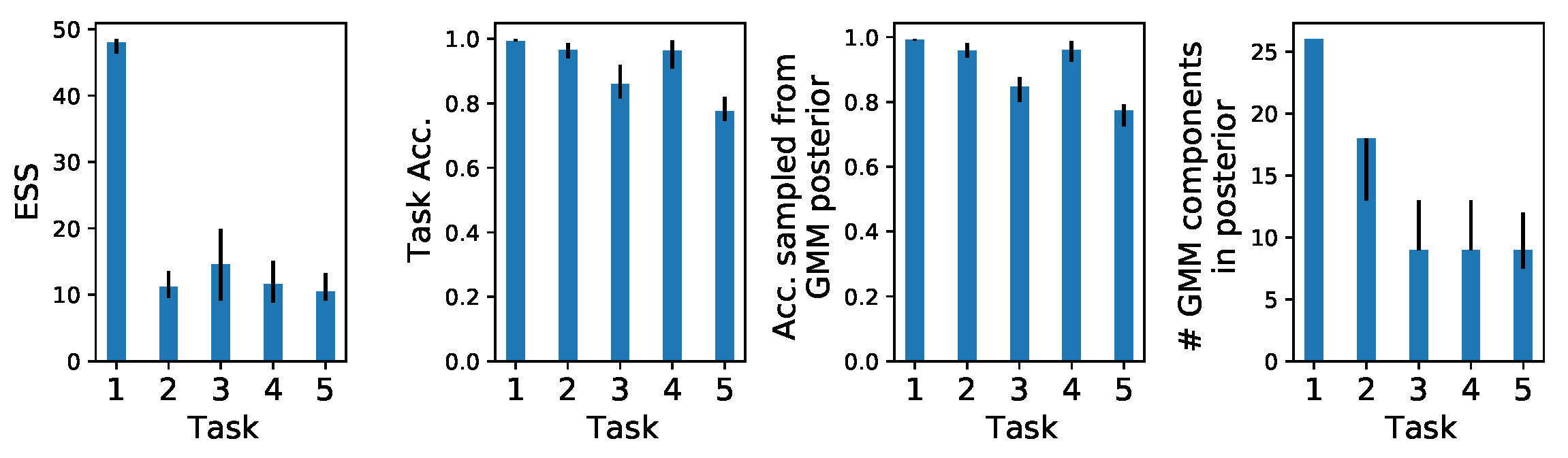
Appendix D. Prototypical Bayesian Continual Learning
Appendix D.1. Inference
Appendix D.2. Sequential Updates
Appendix D.3. ProtoCL Objective


Appendix D.4. Predictions
Appendix D.5. Experimental Setup
Appendix E. Sequential Bayesian Estimation as Bayesian Neural Network Optimization
References
- McCloskey, M.; Cohen, N.J. Catastrophic interference in connectionist networks: The sequential learning problem. In Psychology of Learning and Motivation; Elsevier: Amsterdam, The Netherlands, 1989; Volume 24, pp. 109–165. [Google Scholar]
- French, R.M. Catastrophic forgetting in connectionist networks. Trends Cogn. Sci. 1999, 3, 128–135. [Google Scholar] [CrossRef]
- Kirkpatrick, J.; Pascanu, R.; Rabinowitz, N.; Veness, J.; Desjardins, G.; Rusu, A.A.; Milan, K.; Quan, J.; Ramalho, T.; Grabska-Barwinska, A.; et al. Overcoming catastrophic forgetting in neural networks. Proc. Natl. Acad. Sci. USA 2017, 114, 3521–3526. [Google Scholar] [CrossRef]
- MacKay, D.J. A practical Bayesian framework for backpropagation networks. Neural Comput. 1992, 4, 448–472. [Google Scholar] [CrossRef]
- Graves, A. Practical variational inference for neural networks. Adv. Neural Inf. Process. Syst. 2011, 24, 1–9. [Google Scholar]
- Blundell, C.; Cornebise, J.; Kavukcuoglu, K.; Wierstra, D. Weight uncertainty in neural network. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 6–11 July 2015; pp. 1613–1622. [Google Scholar]
- Schwarz, J.; Czarnecki, W.; Luketina, J.; Grabska-Barwinska, A.; Teh, Y.W.; Pascanu, R.; Hadsell, R. Progress & compress A scalable framework for continual learning. In Proceedings of the International Conference on Machine Learning, PMLR, Stockholm, Sweden, 10–15 July 2018; pp. 4528–4537. [Google Scholar]
- Ritter, H.; Botev, A.; Barber, D. Online structured laplace approximations for overcoming catastrophic forgetting. Adv. Neural Inf. Process. Syst. 2018, 31, 1–11. [Google Scholar]
- Nguyen, C.V.; Li, Y.; Bui, T.D.; Turner, R.E. Variational Continual Learning. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Ebrahimi, S.; Elhoseiny, M.; Darrell, T.; Rohrbach, M. Uncertainty-Guided Continual Learning in Bayesian Neural Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Long Beach, CA, USA, 16–17 June 2019; pp. 75–78. [Google Scholar]
- Kessler, S.; Nguyen, V.; Zohren, S.; Roberts, S.J. Hierarchical indian buffet neural networks for bayesian continual learning. In Proceedings of the Uncertainty in Artificial Intelligence, PMLR, Online, 27–30 July 2021; pp. 749–759. [Google Scholar]
- Loo, N.; Swaroop, S.; Turner, R.E. Generalized Variational Continual Learning. In Proceedings of the International Conference on Learning Representations, Addis Ababa, Ethiopia, 26–30 April 2020. [Google Scholar]
- Neal, R.M. MCMC using Hamiltonian dynamics. Handbook of Markov Chain Monte Carlo; Chapman and Hall: New York, NY, USA, 2011; pp. 113–162. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Zenke, F.; Poole, B.; Ganguli, S. Continual learning through synaptic intelligence. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 3987–3995. [Google Scholar]
- Hsu, Y.C.; Liu, Y.C.; Ramasamy, A.; Kira, Z. Re-evaluating continual learning scenarios: A categorization and case for strong baselines. arXiv 2018, arXiv:1810.12488. [Google Scholar]
- Van de Ven, G.M.; Tolias, A.S. Three scenarios for continual learning. arXiv 2019, arXiv:1904.07734. [Google Scholar]
- van de Ven, G.M.; Tuytelaars, T.; Tolias, A.S. Three types of incremental learning. Nat. Mach. Intell. 2022, 4, 1185–1197. [Google Scholar] [CrossRef]
- Chopin, N.; Papaspiliopoulos, O. An Introduction to Sequential Monte Carlo; Springer: Cham, Switzerland, 2020; Volume 4. [Google Scholar]
- Cobb, A.D.; Jalaian, B. Scaling Hamiltonian Monte Carlo Inference for Bayesian Neural Networks with Symmetric Splitting. In Proceedings of the Thirty-Seventh Conference on Uncertainty in Artificial Intelligence, Online, 27–30 July 2021; pp. 675–685. [Google Scholar]
- Izmailov, P.; Vikram, S.; Hoffman, M.D.; Wilson, A.G.G. What are Bayesian neural network posteriors really like? In Proceedings of the International Conference on Machine Learning, PMLR, Online, 18–24 July 2021; pp. 4629–4640. [Google Scholar]
- Pan, P.; Swaroop, S.; Immer, A.; Eschenhagen, R.; Turner, R.; Khan, M.E.E. Continual deep learning by functional regularisation of memorable past. Adv. Neural Inf. Process. Syst. 2020, 33, 4453–4464. [Google Scholar]
- Dinh, L.; Sohl-Dickstein, J.; Bengio, S. Density estimation using real NVP. arXiv 2016, arXiv:1605.08803. [Google Scholar]
- Doucet, A.; De Freitas, N.; Gordon, N. An introduction to sequential Monte Carlo methods. In Sequential Monte Carlo Methods in Practice; Springer: Berlin/Heidelberg, Germany, 2001; pp. 3–14. [Google Scholar]
- Kalman, R.E. A new approach to linear filtering and prediction problems. J. Basic Eng. Mar. 1960, 82, 35–45. [Google Scholar] [CrossRef]
- Aljundi, R.; Lin, M.; Goujaud, B.; Bengio, Y. Gradient based sample selection for online continual learning. Adv. Neural Inf. Process. Syst. 2019, 32, 1–10. [Google Scholar]
- Aljundi, R.; Kelchtermans, K.; Tuytelaars, T. Task-free continual learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 11254–11263. [Google Scholar]
- De Lange, M.; Aljundi, R.; Masana, M.; Parisot, S.; Jia, X.; Leonardis, A.; Slabaugh, G.; Tuytelaars, T. A continual learning survey: Defying forgetting in classification tasks. arXiv 2019, arXiv:1909.08383. [Google Scholar]
- Wilson, A.G.; Izmailov, P. Bayesian deep learning and a probabilistic perspective of generalization. Adv. Neural Inf. Process. Syst. 2020, 33, 4697–4708. [Google Scholar]
- Ciftcioglu, Ö.; Türkcan, E. Adaptive Training of Feedforward Neural Networks by Kalman Filtering; Netherlands Energy Research Foundation ECN: Petten, The Netherlands, 1995. [Google Scholar]
- Aitchison, L. Bayesian filtering unifies adaptive and non-adaptive neural network optimization methods. Adv. Neural Inf. Process. Syst. 2020, 33, 18173–18182. [Google Scholar]
- Jacot, A.; Gabriel, F.; Hongler, C. Neural tangent kernel: Convergence and generalization in neural networks. Adv. Neural Inf. Process. Syst. 2018, 31, 1–10. [Google Scholar]
- Thrun, S.; Mitchell, T.M. Lifelong robot learning. Robot. Auton. Syst. 1995, 15, 25–46. [Google Scholar] [CrossRef]
- Zeno, C.; Golan, I.; Hoffer, E.; Soudry, D. Task agnostic continual learning using online variational bayes. arXiv 2018, arXiv:1803.10123. [Google Scholar]
- Ahn, H.; Cha, S.; Lee, D.; Moon, T. Uncertainty-based continual learning with adaptive regularization. Adv. Neural Inf. Process. Syst. 2019, 32, 1–11. [Google Scholar]
- Farquhar, S.; Osborne, M.A.; Gal, Y. Radial bayesian neural networks: Beyond discrete support in large-scale bayesian deep learning. In Proceedings of the International Conference on Artificial Intelligence and Statistics, PMLR, Online, 26–28 August 2020; pp. 1352–1362. [Google Scholar]
- Mehta, N.; Liang, K.; Verma, V.K.; Carin, L. Continual learning using a Bayesian nonparametric dictionary of weight factors. In Proceedings of the International Conference on Artificial Intelligence and Statistics, PMLR, Online, 13–15 April 2021; pp. 100–108. [Google Scholar]
- Kumar, A.; Chatterjee, S.; Rai, P. Bayesian structural adaptation for continual learning. In Proceedings of the International Conference on Machine Learning, PMLR, Online, 18–24 July 2021; pp. 5850–5860. [Google Scholar]
- Adel, T.; Zhao, H.; Turner, R.E. Continual learning with adaptive weights (claw). arXiv 2019, arXiv:1911.09514. [Google Scholar]
- Titsias, M.K.; Schwarz, J.; Matthews, A.G.d.G.; Pascanu, R.; Teh, Y.W. Functional Regularisation for Continual Learning with Gaussian Processes. In Proceedings of the ICLR, Addis Ababa, Ethiopia, 26–30 April 2020. [Google Scholar]
- Kapoor, S.; Karaletsos, T.; Bui, T.D. Variational auto-regressive Gaussian processes for continual learning. In Proceedings of the International Conference on Machine Learning, PMLR, Online, 18–24 July 2021; pp. 5290–5300. [Google Scholar]
- Buzzega, P.; Boschini, M.; Porrello, A.; Abati, D.; Calderara, S. Dark experience for general continual learning: A strong, simple baseline. Adv. Neural Inf. Process. Syst. 2020, 33, 15920–15930. [Google Scholar]
- Benjamin, A.; Rolnick, D.; Kording, K. Measuring and regularizing networks in function space. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Henning, C.; Cervera, M.; D’Angelo, F.; Von Oswald, J.; Traber, R.; Ehret, B.; Kobayashi, S.; Grewe, B.F.; Sacramento, J. Posterior meta-replay for continual learning. Adv. Neural Inf. Process. Syst. 2021, 34, 14135–14149. [Google Scholar]
- Swaroop, S.; Nguyen, C.V.; Bui, T.D.; Turner, R.E. Improving and understanding variational continual learning. arXiv 2019, arXiv:1905.02099. [Google Scholar]
- Rudner, T.G.J.; Chen, Z.; Teh, Y.W.; Gal, Y. Tractabe Function-Space Variational Inference in Bayesian Neural Networks. In Proceedings of the 36th Conference on Neural Information Processing Systems (NeurIPS 2022), New Orleans, LA, USA, 28 November–9 December 2022. [Google Scholar]
- Rudner, T.G.J.; Smith, F.B.; Feng, Q.; Teh, Y.W.; Gal, Y. Continual Learning via Sequential Function-Space Variational Inference. In Proceedings of the 38th International Conference on Machine Learning, PMLR, Virtual, 18–24 July 2022. [Google Scholar]
- Lavda, F.; Ramapuram, J.; Gregorova, M.; Kalousis, A. Continual classification learning using generative models. arXiv 2018, arXiv:1810.10612. [Google Scholar]
- van de Ven, G.M.; Li, Z.; Tolias, A.S. Class-incremental learning with generative classifiers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 3611–3620. [Google Scholar]
- Snell, J.; Swersky, K.; Zemel, R. Prototypical networks for few-shot learning. Adv. Neural Inf. Process. Syst. 2017, 30, 1–11. [Google Scholar]
- Rebuffi, S.A.; Kolesnikov, A.; Sperl, G.; Lampert, C.H. ICARL: Incremental classifier and representation learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2001–2010. [Google Scholar]
- Harrison, J.; Sharma, A.; Finn, C.; Pavone, M. Continuous meta-learning without tasks. Adv. Neural Inf. Process. Syst. 2020, 33, 17571–17581. [Google Scholar]
- Knoblauch, J.; Husain, H.; Diethe, T. Optimal continual learning has perfect memory and is NP-hard. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 13–18 July 2020; pp. 5327–5337. [Google Scholar]
- Petersen, K.B.; Pedersen, M.S. The Matrix Cookbook; Technical University of Denmark: Lyngby, Denmark, 2008; Volume 7, p. 510. [Google Scholar]


| Method | Coreset | Split-MNIST | Split-FMNIST |
|---|---|---|---|
| VCL [9] | ✗ | ||
| coreset | ✓ | ||
| HIBNN [11] | ✗ | ||
| FROMP [22] | ✓ | ||
| S-FSVI [47] | ✓ | ||
| ProtoCL (ours) | ✓ |
| Method | Training Time (s) | Split CIFAR-10 (Acc) |
|---|---|---|
| FROMP [22] | ||
| S-FSVI [47] | ||
| ProtoCL (ours) | ||
| Split CIFAR-100 (Acc) | ||
| S-FSVI [47] | ||
| ProtoCL (ours) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kessler, S.; Cobb, A.; Rudner, T.G.J.; Zohren, S.; Roberts, S.J. On Sequential Bayesian Inference for Continual Learning. Entropy 2023, 25, 884. https://doi.org/10.3390/e25060884
Kessler S, Cobb A, Rudner TGJ, Zohren S, Roberts SJ. On Sequential Bayesian Inference for Continual Learning. Entropy. 2023; 25(6):884. https://doi.org/10.3390/e25060884
Chicago/Turabian StyleKessler, Samuel, Adam Cobb, Tim G. J. Rudner, Stefan Zohren, and Stephen J. Roberts. 2023. "On Sequential Bayesian Inference for Continual Learning" Entropy 25, no. 6: 884. https://doi.org/10.3390/e25060884
APA StyleKessler, S., Cobb, A., Rudner, T. G. J., Zohren, S., & Roberts, S. J. (2023). On Sequential Bayesian Inference for Continual Learning. Entropy, 25(6), 884. https://doi.org/10.3390/e25060884







