Evidence of an Absence of Inbreeding Depression in a Wild Population of Weddell Seals (Leptonychotes weddellii)
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Organism
2.2. Sample Selection and Genotyping
2.3. Two-Step Maximum Likelihood Analysis
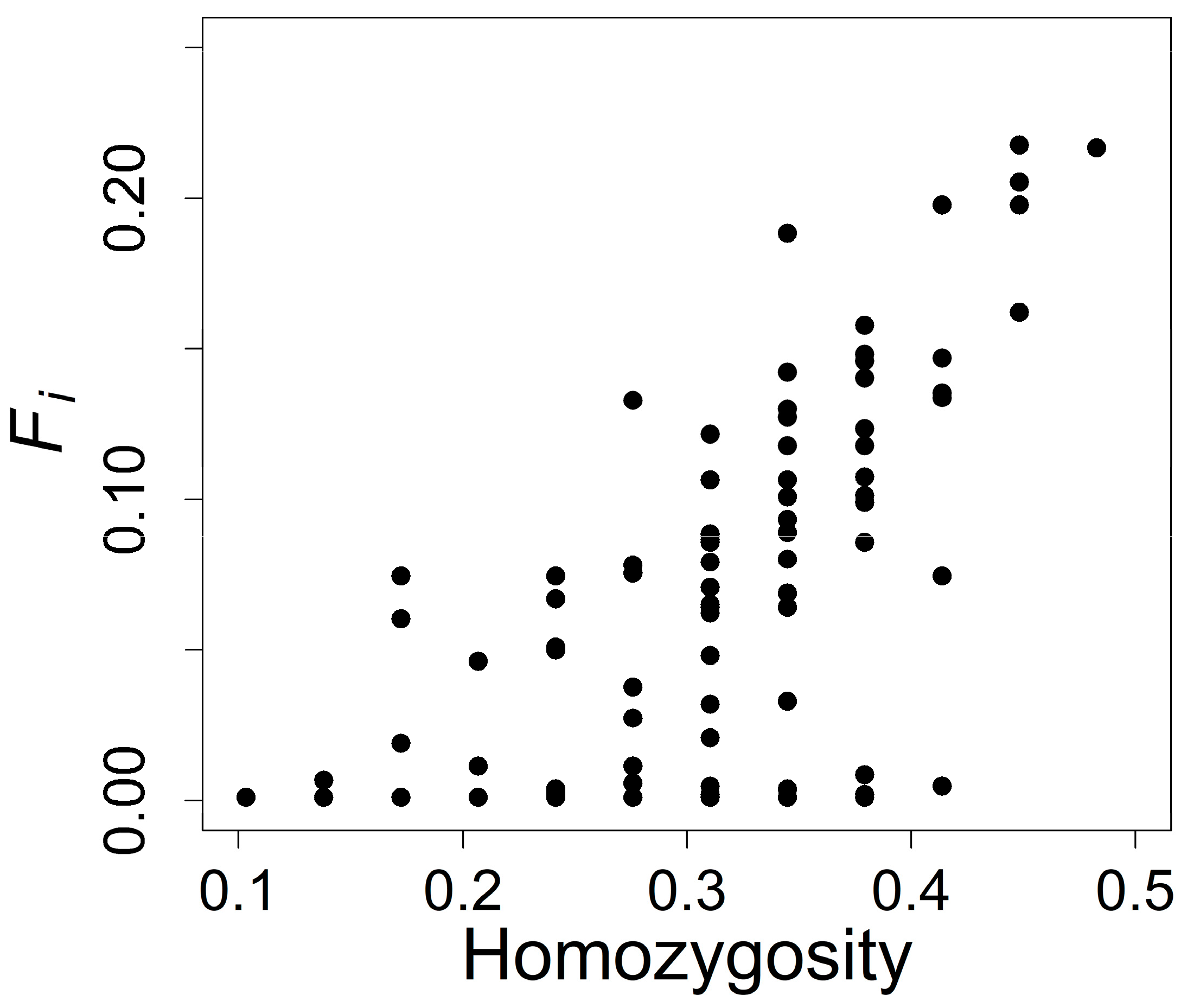
2.4. Estimating Individual Inbreeding Coefficients
2.5. Modeling the Association between Inbreeding and Fitness
2.5.1. Lifetime Reproductive Success
2.5.2. Survival across the Study
2.5.3. Age at First Reproduction
2.5.4. Frequency of Reproduction
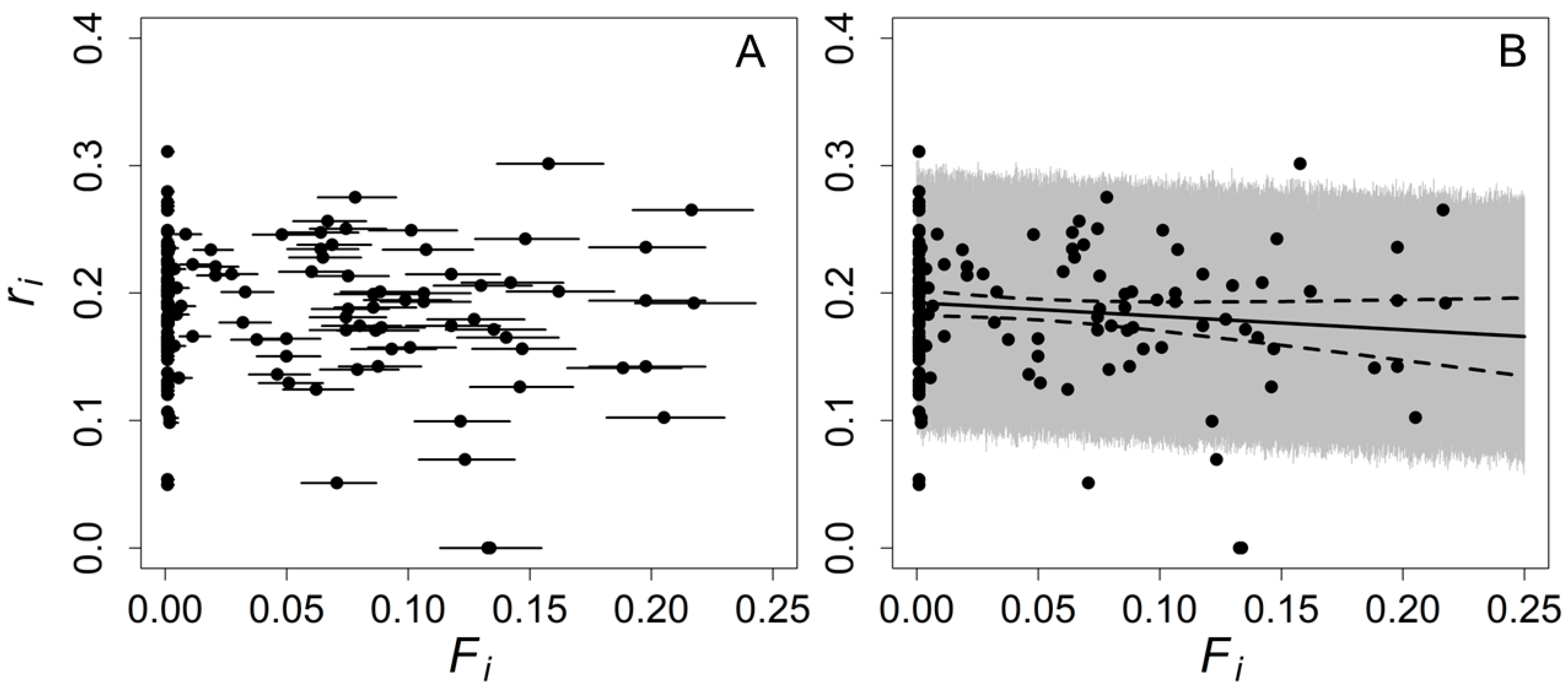
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Popper, K. The Logic of Scientific Discovery; Routledge: Oxfordshire, UK, 1959. [Google Scholar]
- Dennis, B.; Ponciano, J.M.; Taper, M.L.; Lele, S.R. Errors in statistical inference under model misspecification: Evidence, hypothesis testing, and AIC. Front. Ecol. Evol. 2019, 7, 372. [Google Scholar] [CrossRef]
- Sterling, T.D. Publication decisions and their possible effects on inferences drawn from tests of significance—Or vice versa. J. Am. Statl. Assoc. 1959, 54, 30–34. [Google Scholar]
- Rosenthal, R. The “file drawer problem” and tolerance for null results. Psychol. Bull. 1979, 86, 638–641. [Google Scholar] [CrossRef]
- Taper, M.L.; Ponciano, J.M.; Toquenaga, Y. Editorial: Evidential statistics, model identification, and science. Front. Ecol. Evol. 2022, 10, 883456. [Google Scholar] [CrossRef]
- Ives, A.R.; Whitlock, M.C. Inbreeding and metapopulations. Science 2002, 295, 454–455. [Google Scholar] [CrossRef]
- Darwin, C.R. Variation of Animals and Plants under Domestication; John Murry: London, UK, 1868. [Google Scholar]
- Ralls, K.; Brugger, K.; Ballou, J. Inbreeding and juvenile mortality in small populations. Science 1987, 206, 1101–1103. [Google Scholar] [CrossRef]
- Ralls, K.; Ballou, J.D.; Templeton, A. Estimates of lethal equivalents and the cost of inbreeding in mammals. Conserv. Boil. 1988, 2, 185–193. [Google Scholar] [CrossRef]
- Crnokrak, P.; Roff, D.A. Inbreeding depression in the wild. Heredity 1999, 83, 260–270. [Google Scholar] [CrossRef]
- Hedrick, P.W.; Kalinowski, S.T. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 2000, 31, 139–162. [Google Scholar] [CrossRef]
- Keller, L.F.; Waller, D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002, 17, 230–241. [Google Scholar] [CrossRef]
- Gilpin, M.E.; Soulé, M.E. Minimum viable populations: The process of species extinctions. In Conservation Biology the Science of Scarcity and Diversity; Soulé, M.E., Ed.; Sinauer Associates: Sunderland, MA, USA, 1986; pp. 13–34. [Google Scholar]
- Goodman, S.N. A dirty dozen: Twelve p-value misconceptions. Semin. Hematol. 2008, 45, 135–140. [Google Scholar] [CrossRef]
- Royall, R.M. Statistical Evidence: A Likelihood Paradigm; Chapman & Hall: London, UK, 1997. [Google Scholar]
- Ponciano, J.M.; Taper, M.L. Model Projections in Model Space: A geometric interpretation of the AIC allows estimating the distance between truth and approximating models. Front. Ecol. Evol. 2019, 10, 413. [Google Scholar] [CrossRef]
- Locke, J. An Essay Concerning Humane Understanding (Book IV, Chapter XVII); Thomas Basset: London, UK, 1690. [Google Scholar]
- Akaike, H. Information Theory as an Extension of the Maximum Likelihood Principle. In Second International Symposium on Information Theory; Petrov, B.N., Csaki, F., Eds.; Akademiai Kiado: Budapest, Hungary, 1973. [Google Scholar]
- Taper, M.L.; Lele, S.R.; Ponciano, J.M.; Dennis, B.; Jerde, C.L. Assessing the Global and Local Uncertainty of Scientific Evidence in the Presence of Model Misspecification. Front. Ecol. Evol. 2021, 10, 668. [Google Scholar] [CrossRef]
- Morton, N.E.; Crow, J.F.; Muller, H.J. An estimate of the mutational damage in man from data on consanguineous marriages. Proc. Nat. Acad. Sci. USA 1956, 42, 855–863. [Google Scholar] [CrossRef]
- Pemberton, J. Measuring inbreeding depression in the wild: The old ways are the best. Trends Ecol. Evol. 2004, 19, 613–615. [Google Scholar] [CrossRef]
- Slate, J.; David, P.; Dodds, K.G.; Veenvliet, B.A.; Glass, B.C.; Broad, T.E.; McEwan, J.C. Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: Theoretical expectations and empirical data. Heredity 2004, 93, 255–265. [Google Scholar] [CrossRef]
- Balloux, F.; Amos, W.; Coulson, T. Does heterozygosity estimate inbreeding in real populations? Mol. Ecol. 2004, 13, 3021–3031. [Google Scholar] [CrossRef]
- Slate, J.; Pemberton, J.M. Comparing molecular measures for detecting inbreeding depression. J. Evol. Biol. 2002, 15, 20–31. [Google Scholar] [CrossRef]
- Miller, J.M.; Malenfant, R.M.; David, P.; Davis, C.S.; Poissant, J.; Hogg, J.T.; Festa-Bianchet, M.; Coltman, D.W. Estimating genome-wide heterozygosity: Effects of demographic history and marker type. Heredity 2014, 112, 240–247. [Google Scholar] [CrossRef]
- Miller, J.M.; Coltman, D.W. Assessment of identity disequilibrium and its relation to empirical heterozygosity fitness correlations: A meta-analysis. Mol. Ecol. 2014, 23, 1899–1909. [Google Scholar] [CrossRef]
- Andrews, K.R.; Good, J.M.; Miller, M.R.; Luikart, G.; Hohenlohe, P.A. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 2016, 17, 81–92. [Google Scholar] [CrossRef]
- Kardos, M.; Luikart, G.; Allendorf, F.W. Measuring individual inbreeding in the age of genomics: Marker-based measures are better than pedigrees. Heredity 2015, 115, 63–72. [Google Scholar] [CrossRef]
- Coltman, D.; Slate, J. Microsatellite measures of inbreeding: A meta-analysis. Evolution 2003, 57, 971–983. [Google Scholar]
- Chapman, J.R.; Nakagawa, S.; Coltman, D.W.; Slate, J.; Sheldon, B.C. A quantitative review of heterozygosity-fitness correlations in animal populations. Mol. Ecol. 2009, 18, 2746–2765. [Google Scholar] [CrossRef]
- Powell, J.H. Investigating the Role of Dispersal on the Genetic Structure of Wild Populations. Ph.D. Thesis, Montana State University, Bozeman, MT, USA, 2014. [Google Scholar]
- Lele, S.R.; Dennis, B.; Lutscher, F. Data cloning: Easy maximum likelihood estimation for complex ecological models using Bayesian Markov chain Monte Carlo methods. Ecol. Lett. 2007, 10, 551–563. [Google Scholar] [CrossRef]
- Proffitt, K.M.; Garrott, R.A.; Rotella, J.J.; Wheatley, K.E. Environmental and senescent related variations in Weddell seal body mass: Implications for age-specific reproductive performance. Oikos 2007, 116, 1683–1690. [Google Scholar]
- Rotella, J.J.; Link, W.A.; Nichols, J.D.; Hadley, G.L.; Garrott, R.A.; Proffitt, K.M. An evaluation of density-dependent and density-independent influences on population growth rates in Weddell seals. Ecology 2009, 90, 975–984. [Google Scholar] [CrossRef]
- Cameron, M.F.; Siniff, D.B. Age-specific survival, abundance, and immigration rates of a Weddell seal (Leptonychotes weddellii) population in McMurdo Sound, Antarctica. Can. J. Zool. 2004, 82, 601–615. [Google Scholar] [CrossRef]
- Hadley, G.L.; Rotella, J.J.; Garrott, R.A.; Nichols, J.D. Variation in probability of first reproduction of Weddell seals. J. Anim. Ecol. 2006, 75, 1058–1070. [Google Scholar] [CrossRef]
- Hadley, G.L.; Rotella, J.J.; Garrott, R.A. Spatial variation in age-specific probabilities of first reproduction for Weddell seals. Oikos 2008, 117, 1165–1174. [Google Scholar] [CrossRef]
- Gelatt, T.S.; Davis, C.S.; Siniff, D.B.; Strobeck, C. Molecular evidence for twinning in Weddell seals (Leptonychotes weddellii). J. Mammal. 2001, 82, 491–499. [Google Scholar] [CrossRef]
- Hadley, G.L.; Rotella, J.J.; Garrott, R.A. Evaluation of reproductive costs for Weddell seals in Erebus Bay, Antarctica. J. Anim. Ecol. 2007, 76, 448–458. [Google Scholar] [CrossRef]
- Hastings, K.K.; Testa, J.W. Maternal and birth colony effects on survival of Weddell seal offspring from McMurdo Sound, Antarctica. J. Anim. Ecol. 1998, 67, 722–740. [Google Scholar] [CrossRef]
- Garrott, R.A.; Rotella, J.J.; Siniff, D.B.; Parkinson, C.L.; Stauffer, G.E. Environmental variation and cohort effects in an Antarctic predator. Okios 2011, 120, 1027–1040. [Google Scholar] [CrossRef]
- Gelatt, T.S.; Davis, C.S.; Stirling, I.; Siniff, D.B.; Strobeck, C.; Delisle, I. History and fate of a small isolated population of Weddell seals at White Island, Antarctica. Conserv. Genet. 2010, 11, 721–735. [Google Scholar] [CrossRef]
- Proffitt, K.M.; Garrott, R.A.; Rotella, J.J. Variation in offspring sex ratio among individual Weddell seal (Leptonychotes weddellii) females of different quality. Behav. Ecol. Sociobiol. 2008, 62, 1679–1687. [Google Scholar] [CrossRef]
- Abdelkrim, J.; Robertson, B.C.; Stanton, J.A.L.; Gimmel, N.J. Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. Biotechniques 2009, 46, 185–191. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Davis, C.S.; Gelatt, T.S.; Siniff, D.; Strobeck, A. Dinucleotide microsatellite markers from the Antarctic seals and their use in other Pinnipeds. Mol. Ecol. Notes 2002, 2, 203–208. [Google Scholar]
- Paetkau, D.; Strobeck, C. The molecular basis and evolutionary history of a microsatellite null allele in bears. Mol. Ecol. 1995, 4, 519–520. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6, genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5, genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Guo, S.W.; Thompson, E.A. Performing the exact test for Hardy-Weinberg proportion for multiple alleles. Biometrics 1992, 48, 361–371. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Rousset, F. Genepop ‘007, a complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Notes 2008, 4, 137–138. [Google Scholar]
- Lele, S.R. Consequences of lack of parameterization invariance of non-informative Bayesian analysis for wildlife management: Survival of San Joaquin kit fox and declines in amphibian populations. Front. Ecol. Evol. 2020, 7, 501. [Google Scholar] [CrossRef]
- Lele, S.R.; Nadeem, K.; Schmuland, B. Estimability and likelihood inference for generalized linear mixed models using data cloning. J. Am. Stat. Assoc. 2010, 105, 1617–1625. [Google Scholar] [CrossRef]
- Ponciano, J.M.; Burleigh, J.G.; Braun, E.L.; Taper, M.L. Assessing Parameter Identifiability in Phylogenetic Models Using Data Cloning. Syst. Biol. 2012, 61, 955–972. [Google Scholar] [CrossRef]
- Ponciano, J.M.; Taper, M.L.; Dennis, B.; Lele, S.R. Hierarchical models in ecology: Confidence intervals, hypothesis testing, and model selection using data cloning. Ecology 2009, 90, 356–362. [Google Scholar] [CrossRef]
- Sólymos, P. dclone: Data cloning in R. R J. 2010, 2, 29–37. [Google Scholar] [CrossRef]
- Walker, A.M. On asymptotic behaviour of posterior distributions. J. R. Stat. Soc. Ser. B Stat. Methodol. 1969, 31, 80–88. [Google Scholar] [CrossRef]
- Vogl, C.; Karhu, A.; Moran, G.; Savolainen, O. High resolution analysis of mating systems: Inbreeding in natural populations of Pinus radiata. J. Evol. Biol. 2002, 15, 433–439. [Google Scholar] [CrossRef]
- Lunn, D.J.; Thomas, A.; Best, N.; Spiegelhalter, D. WinBUGS—A Bayesian modeling framework: Concepts, structure, and extensibility. Stat. Comput. 2000, 10, 325–337. [Google Scholar] [CrossRef]
- Lesaffre, E.; Lawson, A. Bayesian Biostatistics; John Wiles & Sons: West Sussex, UK, 2012. [Google Scholar]
- Tuyl, F.; Gerlach, R.; Mengersen, K. A comparison of Bayes-Laplace, Jeffreys, and other priors: The case of zero events. Am. Stat. 2008, 62, 40–44. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- R Core Development Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012; Available online: http://www.R-project.org (accessed on 17 February 2023).
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Rubin, D.B. Bayesian Data Analysis, 2nd ed.; Chapman & Hall: Boca Raton, FL, USA, 2004. [Google Scholar]
- Plummer, M.; Best, N.; Cowles, K.; Vines, K. CODA: Convergence diagnostics and output analysis for MCMC. R News 2006, 6, 7–11. [Google Scholar]
- David, P.; Pujol, P.; Viard, F.; Castella, V.; Goudet, J. Reliable selfing rate estimates from imperfect population genetic data. Mol. Ecol. 2007, 16, 2474–2487. [Google Scholar] [CrossRef]
- Stoffel, M.A.; Esser, M.; Kardos, M.; Humble, E.; Nichols, H.; David, P.; Hoffman, J.I. inbreedR: An R package for the analysis of inbreeding based on genetic markers. Methods Ecol. Evol. 2016, 7, 1331–1339. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Ramsey, F.L.; Schafer, D.W. The Statistical Sleuth a Course in Methods of Data Analysis, 2nd ed.; Duxbury: Pacific Grove, CA, USA, 2002. [Google Scholar]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Taper, M.L. Model identification from many candidates. In The Nature of Scientific Evidence: Statistical, Philosophical and Empirical Considerations; Taper, M.L., Lele, S.R., Eds.; The University of Chicago Press: Chicago, IL, USA, 2004; pp. 448–524. [Google Scholar]
- Jerde, C.L.; Kraskura, K.; Eliason, E.J.; Csik, S.R.; Stier, A.C.; Taper, M.L. Strong evidence for an intraspecific metabolic scaling coefficient near 0.89 in fish. Front. Physiol. 2019, 10, 1166. [Google Scholar] [CrossRef]
- Bengtsson, T.; Cavanaugh, J.E. An improved Akaike information criterion for state-space model selection. Comput. Stat. Data Anal. 2006, 50, 2635–2654. [Google Scholar] [CrossRef]
- Harris, R.B. Reliability of trend lines obtained from variable counts. J Wildl. Manag. 1986, 50, 165–171. [Google Scholar] [CrossRef]
- Hone, J. On the rate of increase (r): Patterns of variation in Australian mammals and the implications for wildlife management. J. Appl. Ecol. 1999, 36, 709–718. [Google Scholar] [CrossRef]
- Lunn, D. WBDevDJLTruncatedNormal Documentation. 2003. Available online: http://www.winbugs-development.org.uk/ (accessed on 29 December 2013).
- Gelman, A.; Hill, J. Data Analysis Using Regression and Multilevel/Hierarchical Models; Cambridge University Press: New York, NY, USA, 2007. [Google Scholar]
- Gelman, A. Prior distributions for variance parameters in hierarchical models. Bayesian Anal. 2006, 1, 515–533. [Google Scholar] [CrossRef]
- Paterson, J.T.; Rotella, J.J.; Link, W.A.; Garrott, R. Variation in the vital rates of an Antarctic marine predator: The role of individual heterogeneity. Ecology 2018, 99, 2385–2396. [Google Scholar] [CrossRef]
- Garner, A.; Rachlow, J.L.; Hicks, J.F. Patterns of genetic diversity and its loss in mammalian populations. Conserv. Biol. 2005, 19, 1215–1221. [Google Scholar] [CrossRef]
- Stirling, I. Population aspects of Weddell Seal harvesting at McMurdo Sound, Antarctica. Polar Rec. 1971, 15, 653–667. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Brookfield, J.F.Y. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol. Ecol. 1996, 5, 453–455. [Google Scholar] [CrossRef]
- Lande, R.; Schemske, D.W. The evolution of self fertilization and inbreeding depression in plants. I. Genetic models. Evolution 1985, 39, 24–40. [Google Scholar]
- Frankel, O.H.; Soulé, M.E. Conservation and Evolution; Cambridge University Press: Cambridge, UK, 1981. [Google Scholar]
- Cole, L.C. The population consequences of life history phenomena. Q. Rev. Biol. 1954, 29, 103–137. [Google Scholar] [CrossRef]
- Hamilton, W.D. The moulding of senescence by natural selection. J. Theor. Biol. 1966, 12, 12–45. [Google Scholar] [CrossRef]
- Kardos, M.; Allendorf, F.W.; Luikart, G. Evaluating the role of inbreeding depression in heterozygosity-fitness correlations: How useful are tests for identity disequilibrium. Mol. Ecol. Resour. 2014, 14, 519–530. [Google Scholar] [CrossRef]
| Locus | Primer Sequences 5′-3′ | Repeat Motif | Size (bp) | Number of Alleles | HE | |
|---|---|---|---|---|---|---|
| Lew-001339 | F | GCACTCCAGTTTCCTTGGAC | (AC)14 | 106–135 | 10 | 0.631 |
| R | AGGAGCTTAGTAGGCAATCC | |||||
| Lew-001677 | F | ACAAGGGATTCTTAGGGAACTG | (AC)15 | 214–231 | 15 | 0.799 |
| R | TCCAGTGGTAATAACTTGCAAAC | |||||
| Lew-001845 | F | TGTAACCTCAAGGGTCCCAC | (TG)12 | 147–153 | 4 | 0.519 |
| R | GCGTCTGGAGTGTGGAATTG | |||||
| Lew-001859 | F | TCTCCCTGTTCATTAGATCCTG | (AC)13 | 98–114 | 8 | 0.622 |
| R | GAGCCAACTTGCATTGTGTTC | |||||
| Lew-001873 | F | TGTTTCCGGTTGGGCTATTC | (TG)14 | 133–155 | 8 | 0.624 |
| R | ACGATAGATTGGGCCTTGTC | |||||
| Lew-002658 | F | ATTCTCAGACCTCAGGGAGC | (GT)16 | 122–146 | 8 | 0.709 |
| R | CATCCTGAGTTTGGCCTTGG | |||||
| Lew-002762 | F | TGTTCCATCTCCTGCCACTC | (AC)13 | 164–185 | 9 | 0.759 |
| R | ATCTGGGGAAAGGTGGGTTC | |||||
| Lew-004467 | F | TGCACAGTATAAAACAGGATAGAGG | (AC)14 | 90–111 | 11 | 0.788 |
| R | CCAGAGAGAGCCTGTGTACG | |||||
| Lew-005761 | F | AGAGAGGGTCATTAGAGACAGC | (AC)13 | 158–174 | 8 | 0.782 |
| R | ATGACTCTTCATGGGCGTGC | |||||
| Lew-006174 | F | TGGTGAACTCAACAAGGGAAAG | (AC)15 | 101–112 | 7 | 0.738 |
| R | TGTATTGCTCAGCCCAACTC | |||||
| Lew-006657 | F | GCATGCTGGGTCATGAGTG | (GT)13 | 82–103 | 11 | 0.785 |
| R | GCCCCACGATGTTACTAAGTTG | |||||
| Lew-007425 | F | AAGTTTTATGTGGGCATCCG | (GT)12 | 96–106 | 5 | 0.560 |
| R | GCCGTTCACATTTCTGCCTC | |||||
| Model Structure | K | ΔBIC | ΔAICc | |
|---|---|---|---|---|
| ri | ||||
| 3 | 0.00 | 2.63 | ||
| 4 | 0.32 | 0.00 | ||
| 5 | 3.34 | 0.09 | ||
| 2 | 8.39 | 2.24 | ||
| Minimum age | ||||
| 1 | 0.00 | 0.00 | ||
| 2 | 4.53 | 1.55 | ||
| 3 | 9.44 | 3.5 | ||
| 4 | 14.34 | 5.47 | ||
| Age at maturity | ||||
| 1 | 0.00 | 0.00 | ||
| 2 | 4.55 | 1.57 | ||
| 3 | 9.51 | 3.57 | ||
| 4 | 14.55 | 5.68 | ||
| Pupping interval | ||||
| + | 1 | 0.00 | 0.00 | |
| 2 | 3.99 | 1.04 | ||
| 3 | 7.96 | 2.07 | ||
| 4 | 11.61 | 2.83 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Powell, J.H.; Kalinowski, S.T.; Taper, M.L.; Rotella, J.J.; Davis, C.S.; Garrott, R.A. Evidence of an Absence of Inbreeding Depression in a Wild Population of Weddell Seals (Leptonychotes weddellii). Entropy 2023, 25, 403. https://doi.org/10.3390/e25030403
Powell JH, Kalinowski ST, Taper ML, Rotella JJ, Davis CS, Garrott RA. Evidence of an Absence of Inbreeding Depression in a Wild Population of Weddell Seals (Leptonychotes weddellii). Entropy. 2023; 25(3):403. https://doi.org/10.3390/e25030403
Chicago/Turabian StylePowell, John H., Steven T. Kalinowski, Mark L. Taper, Jay J. Rotella, Corey S. Davis, and Robert A. Garrott. 2023. "Evidence of an Absence of Inbreeding Depression in a Wild Population of Weddell Seals (Leptonychotes weddellii)" Entropy 25, no. 3: 403. https://doi.org/10.3390/e25030403
APA StylePowell, J. H., Kalinowski, S. T., Taper, M. L., Rotella, J. J., Davis, C. S., & Garrott, R. A. (2023). Evidence of an Absence of Inbreeding Depression in a Wild Population of Weddell Seals (Leptonychotes weddellii). Entropy, 25(3), 403. https://doi.org/10.3390/e25030403







