5.1. Analysis of MS Results
First of all, the performance of the
REDD and
RE-LRW indices are evaluated by using the
MS metric. In
Figure 1, we investigate the impact of different dimensions
d on the
MS values of the
REDD index and
RE-LRW index. From the results, one can observe that the
MS curves of
RE-LRW index have a large variation range in 10 out of 12 graph data. The
MS curves of
RE-LRW index are relatively flat in MEET and USAI, while its
MS curves are considerably low. Moreover, it can be seen that the
MS value of the
RE-LRW index is almost close to 0 in MEET. This may be because there are some non-connected subgraphs in MEET, which will interrupt the random walk between nodes, resulting in the poor performance of the
RE-LRW index. In contrast, the
REDD index is not affected by the disconnection between nodes and can achieve good results in MEET. It can also be found that the
REDD index can maintain high
MS curves in most graph data, while keeping the variation of
MS curves small. In particular, the
MS curves of the
REDD index are clearly higher than that of the
RE-LRW index in FOBA, MEET, and USAI. From the above discussion, the
REDD index owns a greater performance in the most similar node mining.
To compare the effectiveness between the
REDD index and the seven benchmark indices,
Table 4 lists the
MS results of all indices in 12 weighted graph data. Note that the best
MS value of each row is highlighted by using boldface. Furthermore, the
RE- and
are used to represent the
RE-LRW and
REDD indices with the optimal
MS value in different dimensions
d, respectively. From the results, it can be found that the
MS values of the relative entropy-based indices are higher than those of the local structure-based indices and the random walk-based index. This indicates that it is indeed effective for the similarity measures based on relative entropy in the similarity calculation.
In these local structure-based indices, the MS values of CN and WCN indices have a greater difference in FOBA, MEET, FWFW, REHA, CELE, and USAI. For instance, the MS values of CN index are lower than those of the WCN index in FOBA and REHA, while the MS values of the CN index are higher than those of the WCN index in MEET, WFW, CELE, and USAI. This may be caused by the strong and weak ties in the weighted graph data. This phenomenon is also true for AA and WAA indices. Thus, it is necessary to comprehensively consider the degree and strength of nodes to avoid the influence of strong and weak ties.
Compared with the RE-LRW indices, the LRW index has lower MS values in most graph data, except MEET. Thus, there are many general similar nodes when the LRW index is used for the similarity calculation. At the same time, It reflects that the similarity measure based on the relative entropy can reduce the dependence on the large-degree nodes, and so the similarity of nodes can be better characterized.
For LRE and REDD indices, they can maintain higher MS values in 12 graph data. From the results, one can find that there are no general similar nodes when LRE and REDD indices are applied to calculate the similarity of nodes. Despite there being some non-connected subgraphs in MEET, the LRE and REDD indices still perform well. The MS values of the LRE and REDD indices are more than 0.4000, but the MS values of the REDD index can reach 0.7048 in USAI and 0.8000 in FOBA. Taken together, the REDD index has better performance during the most similar node mining.
5.2. Analysis of Scatter Diagram
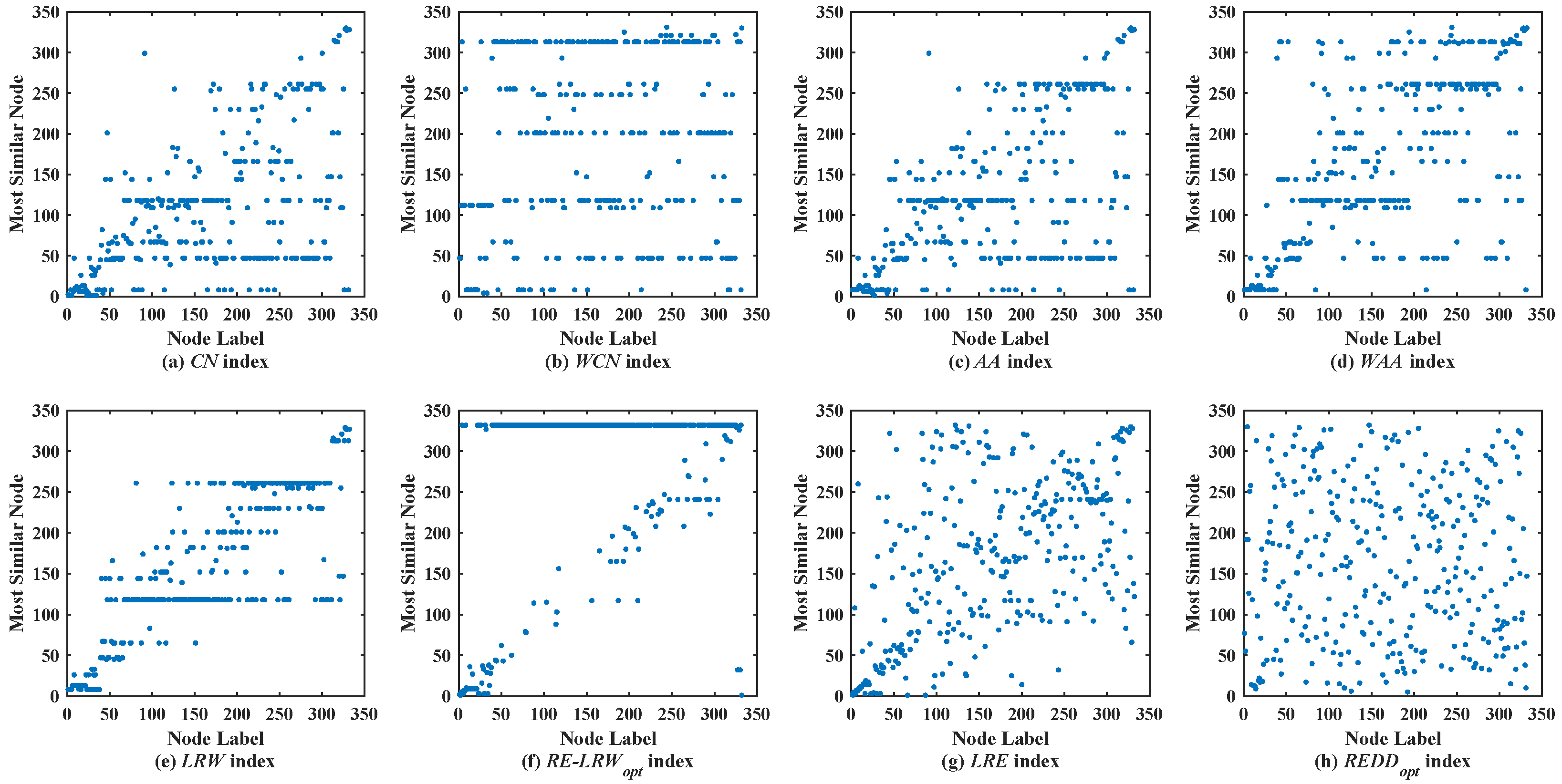
In the most similar nodes mining, the are also used to validate the performance of the similarity measure. To make the experimental results distinguishable, the scatter diagrams of all indices are merely given in the graph data with more than 100 nodes. In scatter diagrams, the horizontal ordinate represents the label of nodes, and the vertical coordinates denote the label of the most similar nodes for the node in the horizontal ordinate. Therefore, the nodes should be scattered on the two-dimensional plane as much as possible in the scatter diagram. If the nodes are concentrated near the diagonal line or present a straight line (i.e., a large number of nodes are most similar to the same node), then the performance of this similarity measure is poor.
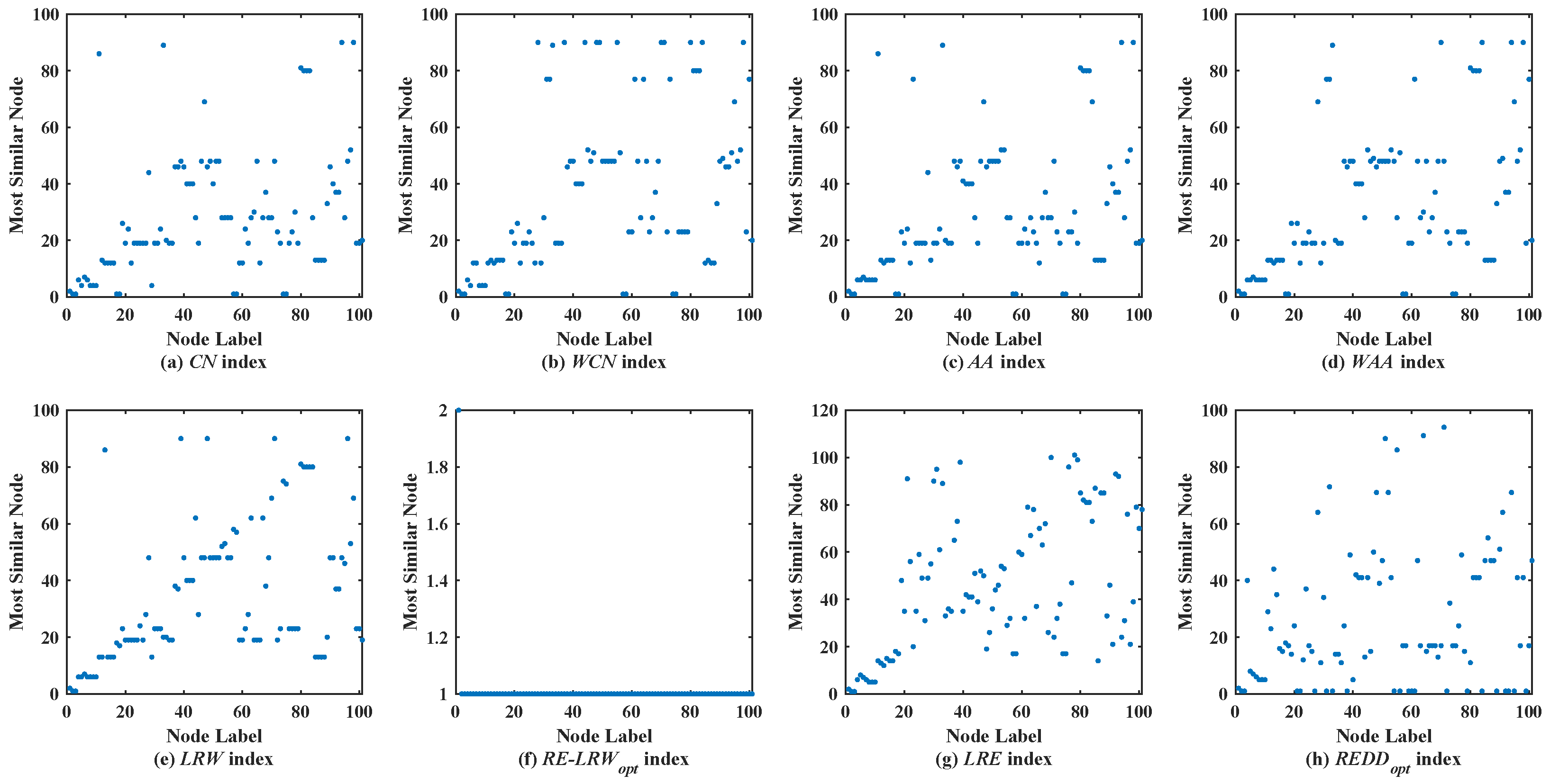
Figure 2 shows the scatter diagrams of eight indices in MMET, where the degree of node
is the largest, and the degree of node
is the second largest. From the scatter diagrams of the
CN,
WCN,
AA,
WAA, and
LRW indices, one can see that many nodes are similar to the nodes
and
. It indicates that there are generally similar nodes in the measurement results of these indices. Additionally, there are no generally similar nodes in the measurement results of the
RE-LRW index, but its scatter diagram has poor symmetry. From the scatter diagrams of the
LRE and
REDD indices, there are neither many nodes similar to the nodes of a large degree nor many nodes clustered on a straight line. Thus, the performance of
LRE and
REDD indices is outstanding in MEET.
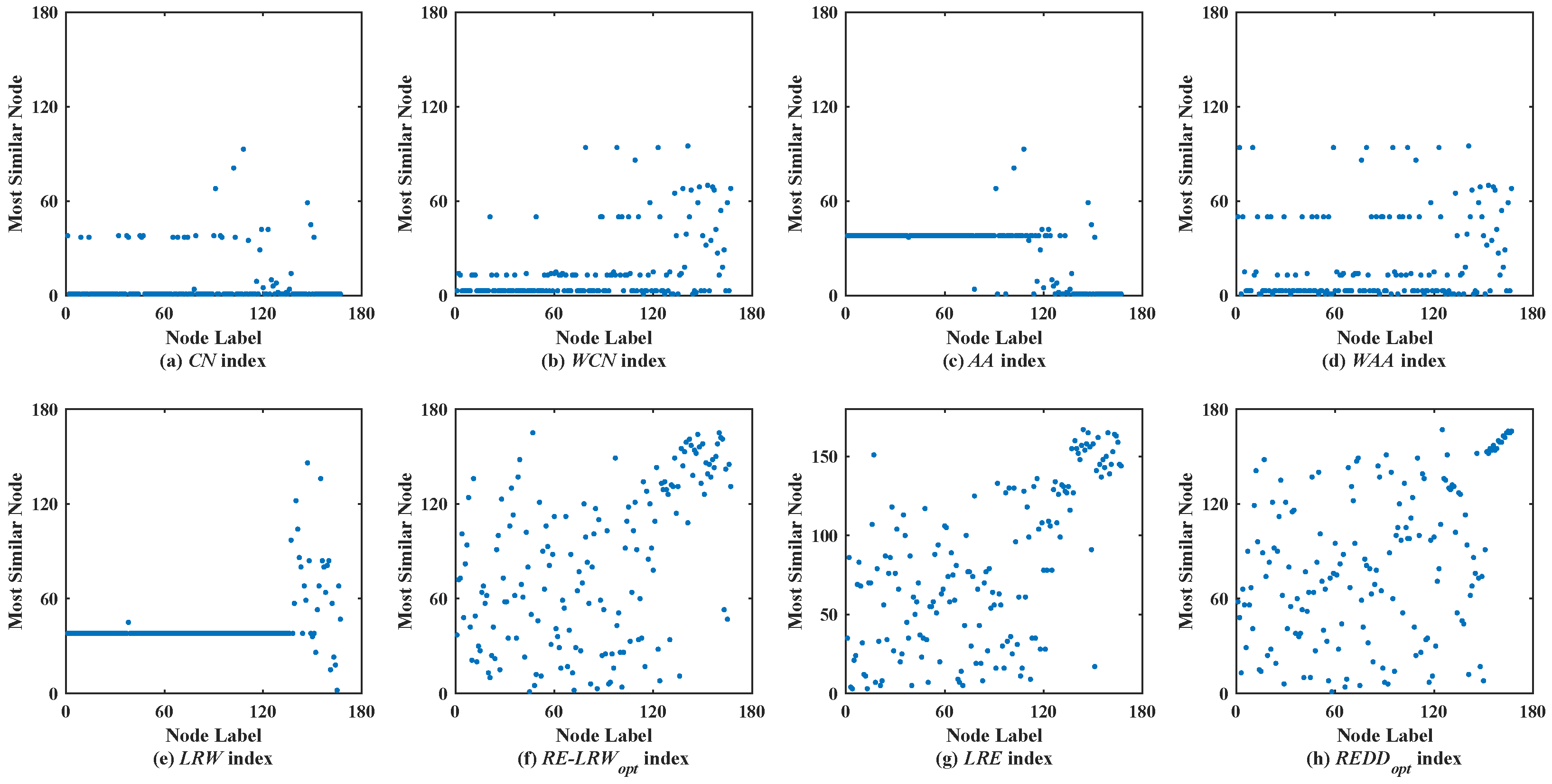
Figure 3 shows the scatter diagrams of eight indices in FWFW, where the degree of node
is the largest, the degree of node
is the second-largest. From the scatter diagrams of the
CN,
WCN,
AA,
WAA, and
LRW indices, it can be seen that there are generally similar nodes
and
. This leads to the scatter diagrams of these indices having poor symmetry. In contrast, the scatter diagrams of the
RE-LRW,
LRE, and
REDD indices show good symmetry, while in the scatter diagram of the
LRE index, one can also see that many nodes are concentrated near the diagonal line. It indicates that the performance of the
LRE index is not as good as that of the
RE-LRW and
REDD indices. Although the
MS value of the
RE-LRW and
REDD indices is equal, the scatter diagram of the former is not as well dispersed as the latter. On balance, the performance of the
REDD index is more reasonable.
Figure 4 shows the scatter diagrams of eight indices in EMAI, where the degree of node
is the largest, the degree of node
is the second largest, and the degree of node
is the third largest. Clearly, there are still generally similar nodes in the scatter diagrams of the
CN,
WCN,
AA,
WAA, and
LRW indices. From the scatter diagrams of the
RE-LRW,
LRE, and
REDD indices, one can find that these indices that use relative entropy can effectively distinguish the generally similar nodes. From the scatter diagrams of the three indices, it can be also seen that the
REDD index performed better than the
RE-LRW and
LRE indices.
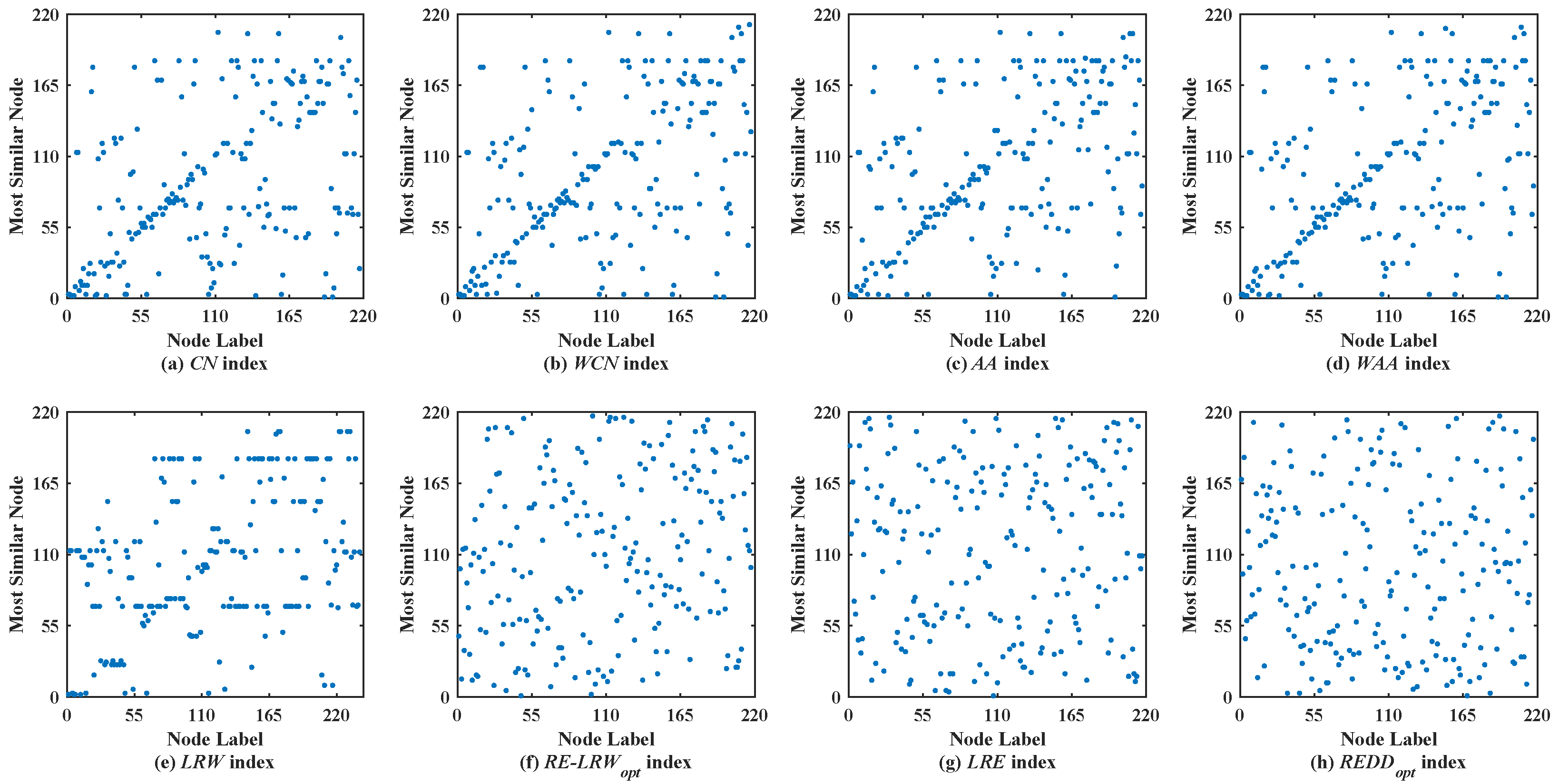
Figure 5 shows the scatter diagrams of eight indices in REHA, where the degree of node
is the largest, and the degree of node
is the second largest. From the results, one can see that there are generally similar nodes
and
in the scatter diagrams of the
CN,
WCN,
AA,
WAA, and
LRW indices. Moreover, many nodes are concentrated near the diagonal line in the scatter diagrams of the
CN,
WCN,
AA, and
WAA indices. Nevertheless, the scatter diagrams of the
RE-LRW,
LRE, and
REDD indices still maintain a better symmetry. Despite the
MS value of the
RE-LRW index being higher than that of the
LRE and
REDD indices in REHA, the symmetry is better in the scatter diagrams of the
LRE and
REDD indices.
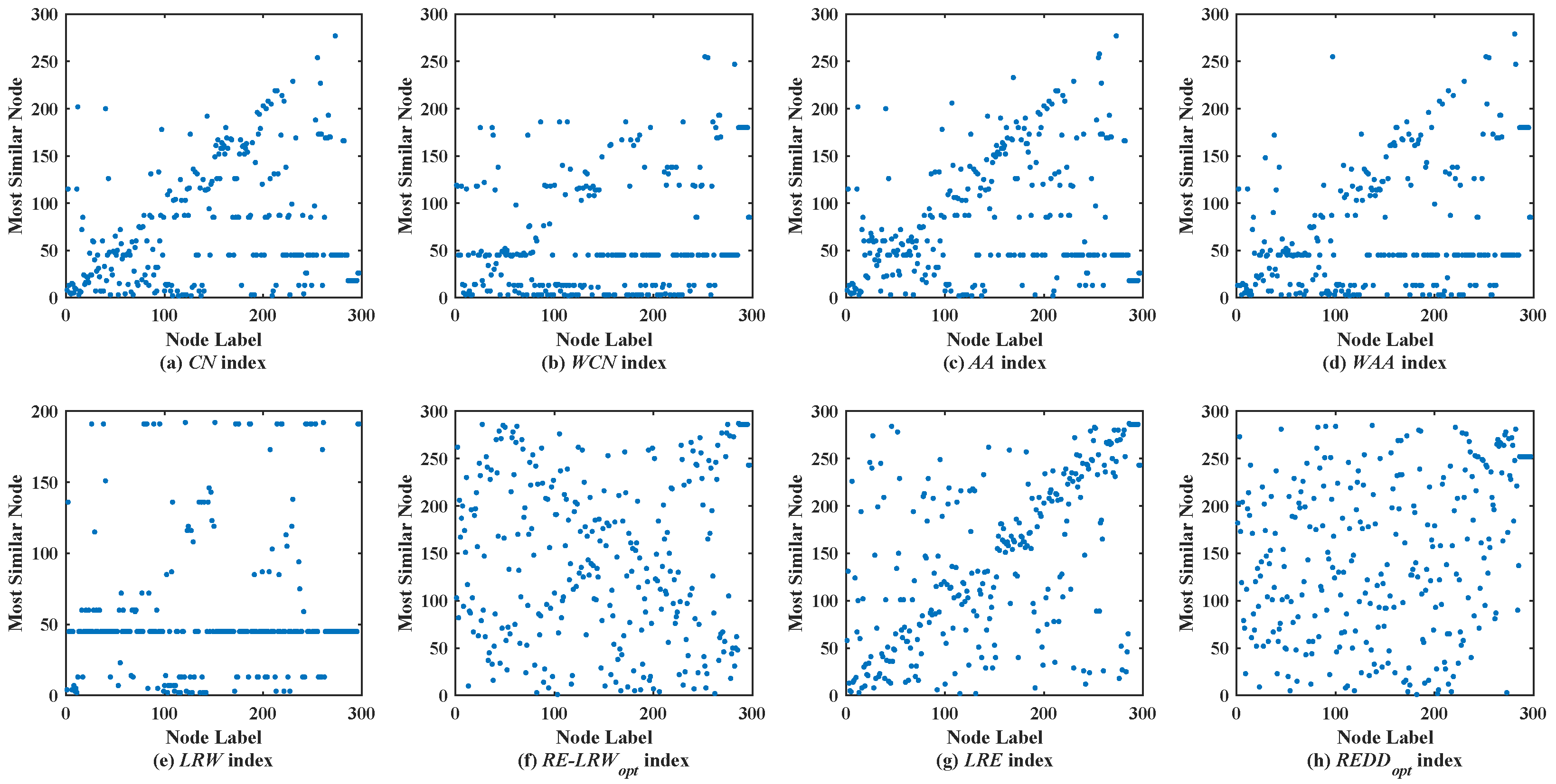
Figure 6 shows the scatter diagrams of eight indices in CELE, where the degree of node
is the largest, and the degree of node
is the second largest. From the scatter diagrams of
CN,
WCN,
AA,
WAA, and
LRW indices, it is not difficult to find that
and
become general similar nodes. Although there are not generally similar nodes in the scatter diagram of
LRE index, many nodes are concentrated near the diagonal line. Thus, the performance of
LRE index is superior to that of the
CN,
WCN,
AA,
WAA, and
LRW indices, while the symmetry of the scatter diagram of
LRE index is not as good as that of the scatter diagrams of
RE-LRW and
REDD indices. Furthermore, one can also find that the symmetry of the scatter diagram of
REDD index is better than that of the scatter diagram of the
RE-LRW index. On the whole, the
REDD index does perform better in CELE.
Figure 7 shows the scatter diagrams of eight indices in USAI, where the degree of node
is the largest, and the degree of node
is the second largest. From the results in
Figure 7, it can be observed that nodes
and
become generally similar nodes in the scatter diagrams of the
CN,
WCN,
AA,
WAA, and
LRW indices.
From
Figure 7, the
RE-LRW index can really avoid the situation that the nodes of a large degree become generally similar nodes. Unfortunately, the most similar nodes of many nodes are clustered in a straight line in the scatter diagram of the
RE-LRW index. It indicates that the symmetry of
RE-LRW index still has great room for enhancement in USAI. In the scatter diagram of the
LRE index, many nodes are rarely distributed near the diagonal line, so the
LRE index has a relatively better symmetry. As for the
REDD index, most of the nodes are not distributed near the diagonal line in its scatter diagram, and many large nodes do not become general similar nodes. Overall, the
REDD index performs better in the USAI, compared to the other benchmark indices.
In this subsection, the performance of the REDD index and seven benchmark indices is analyzed. From the results of the CN, WCN, AA, and WAA indices, one can see that these only used the degree or strength of nodes are greatly affected by the strong and weak ties. Owing to the nodes of a large degree being more likely to be visited during the random walk, there are generally similar nodes in the measurement results of the LRW index. From the results of the RE-LRW, LRE, and REDD indices, it can be found that these indices use relative entropy and their own superior performance in the most similar nodes mining. Despite the results that the RE-LRW index is performed in MMET and USAI, it still has room for improvement. The REDD index comprehensively considers the degree and strength of nodes, while the LRW index only uses the degree of nodes. Hence, one can observe that the performance of the former is better than that of the latter from their results.
5.3. Analysis of Auc Results
To test the effectiveness of similarity measure in link prediction, the
AUC metric is further used to evaluate the performance of
REDD index and seven benchmark indices.
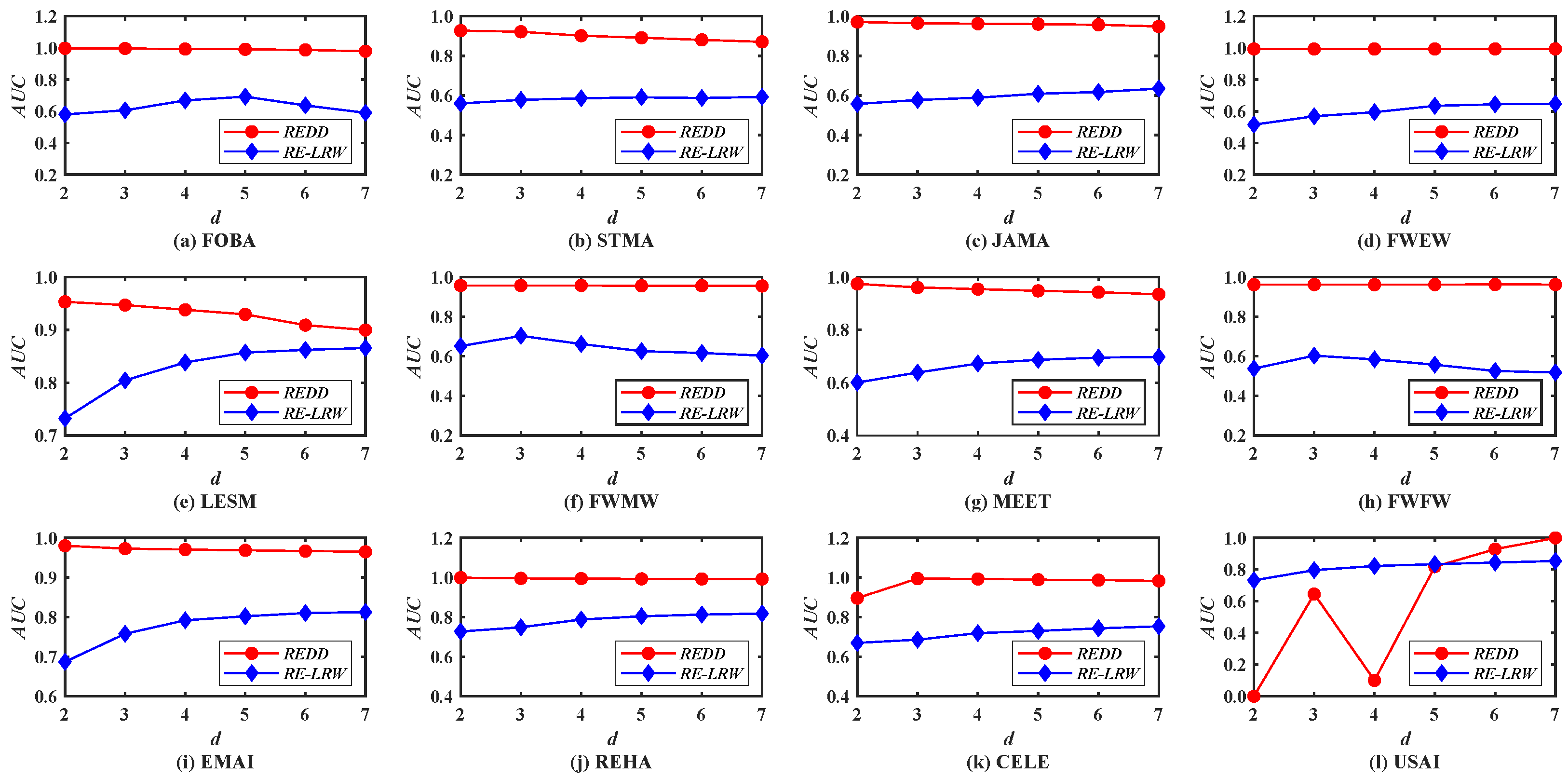
Figure 8 demonstrates the
AUC curves of the
REDD and
RE-LRW indices when the dimension
d changes from 2 to 7. From the results, one can observe that the variation amplitude of the
AUC curves of the
RE-LRW index are almost the same as that of the
REDD index in the other graph data, except for USAI, while the accuracies of the
RE-LRW index are far less than that of the
REDD index under any dimension
d. Despite the
AUC values of
RE-LRW index being higher than that of the
REDD index when
d is equal to 2, 3, 4, and 5 in USAI, the
AUC values of the
REDD index are clearly higher than that of the
RE-LRW index when
d is greater than 6. It indicates that the
REDD index owns a greater potential during the link prediction. On the whole, compared with the
RE-LRW index, the
REDD index is more suitable for link prediction.
In the following, we analyze the effectiveness of the
REDD index and seven benchmark indices during the link prediction.
Table 5 lists the
AUC results of all indices in 12 weighted graph data. From the results, one can observe that the
AUC values of the
REDD index are highest in 11 out of 12 graph data. Despite the
AUC value of the
REDD index being not as good as that of the four local indices in LESM, its
AUC value is superior to that of the
LRW,
RE-LRW, and
LRE indices.
From the results of the CN, WCN, AA, and WAA indices, one can also find that there is a great influence of the strong and weak ties on the similarity measure during the link prediction. For instance, the AUC values of the WCN and WAA indices are significantly greater than that of the CN and AA indices in FOBA, STAM, JAMA, FWEW, LESM, MEET, FWFW, EMAI, REHA, and CELE. This indicates that these similarity measures using the strength of nodes are easier to promote the formation of edges in these graph data, while the AUC values of the WCN and WAA indices are lower than that of the CN and AA indices in FWMW and USAI. It indicates that the fact of weak ties needs to be emphasized in the two graph data. Therefore, it may be more effective to construct the similarity measure by combining the degree and strength of nodes, such as the REDD index we designed.
From the results of the LRW and RE-LRW indices, although the RE-LRW index can enhance the performance of the LRW index in the most similar node mining, the effects of the RE-LRW index are inferior to those of the LRW index in link prediction. In other words, the RE-LRW index has a good performance in the most similar node mining, but its AUC results are quite poor. Therefore, the similarity measure considering only the degree of nodes might perform well only unilaterally in the most similar node mining or link prediction.
In a nutshell, the REDD index not only achieved good results in the most similar node mining, but also acquired a good application in link prediction. It further indicates that it is effective for comprehensively considering the role of the degree and strength of nodes to construct the similarity measure.
Generally, the low complexity is a vital factor in the design of an algorithm. In view of the complexity of local indices being relatively lower, we merely compare the running time of the
REDD,
RE-LRW, and
REDD indices in 12 weighted graph data. Next, the time consumption of the
LRE,
RE-LRW, and
REDD indices are compared by using the metric:
normalized time consumption [
33]. The
normalized time consumption can be understood as the running time of index
a in graph data
b being normalized in the interval [0,1]. The corresponding calculation formula is
, where
and
are the running time and the normalized time consumption of index
a in graph data
b, respectively.
Figure 9 shows the normalized time consumption of the
LRE,
RE-LRW, and
REDD indices in 12 weighted graph data. From the result, the following three phenomena can be found. The
LRE index runs the slowest in 11 out of 12 graph data. The time consumption of
RE-LRW index increases with the increase in the number of nodes. The time consumption of the
REDD index is at a medium level in LESM, MEET, and USAI. It is worth mentioning that the normalized time consumption of the
REDD index is not the highest in all graph data. Hence, it is also feasible to apply the
REDD index in large-scale weighted graph data if there is a better experimental environment. Above all, the
REDD index owns a satisfactory time complexity in the process of link prediction.
5.4. Application to Simulated Networks
As described in the process of link prediction, many real-world graph data may be incomplete. Hence, it is difficult to design a similarity measure applicable to all real-world graphic data. To further verify the effectiveness of the
REDD index, the
NW small-world model is used to construct nine simulated graph data. Therefore, these simulated graph data are similar to real-world graph data. The
NW model can establish the graph data with the different topological characteristics by adjusting the parameters
M and
P. For example, parameter
M can be applied to adjust the average degree of the network, and parameter
P can be utilized to regulate the average clustering coefficient of the network. The topological statistical characteristics of nine simulated networks are listed in
Table 6. From
Table 6, it can be observed that the node number of nine simulated graph data is specified as 100, and the topological statistical characteristics of these graph data are changed as the variation of parameters
M and
P.
From the results of
Figure 10,
Figure 11,
Figure 12 and
Figure 13, it is not difficult find that the performance of the
REDD index is hardly affected by the topological characteristics of graph data. Thus, in both the real-world graph data or in the simulated graph data, the
REDD index has better performance in the most similar node mining and link prediction.
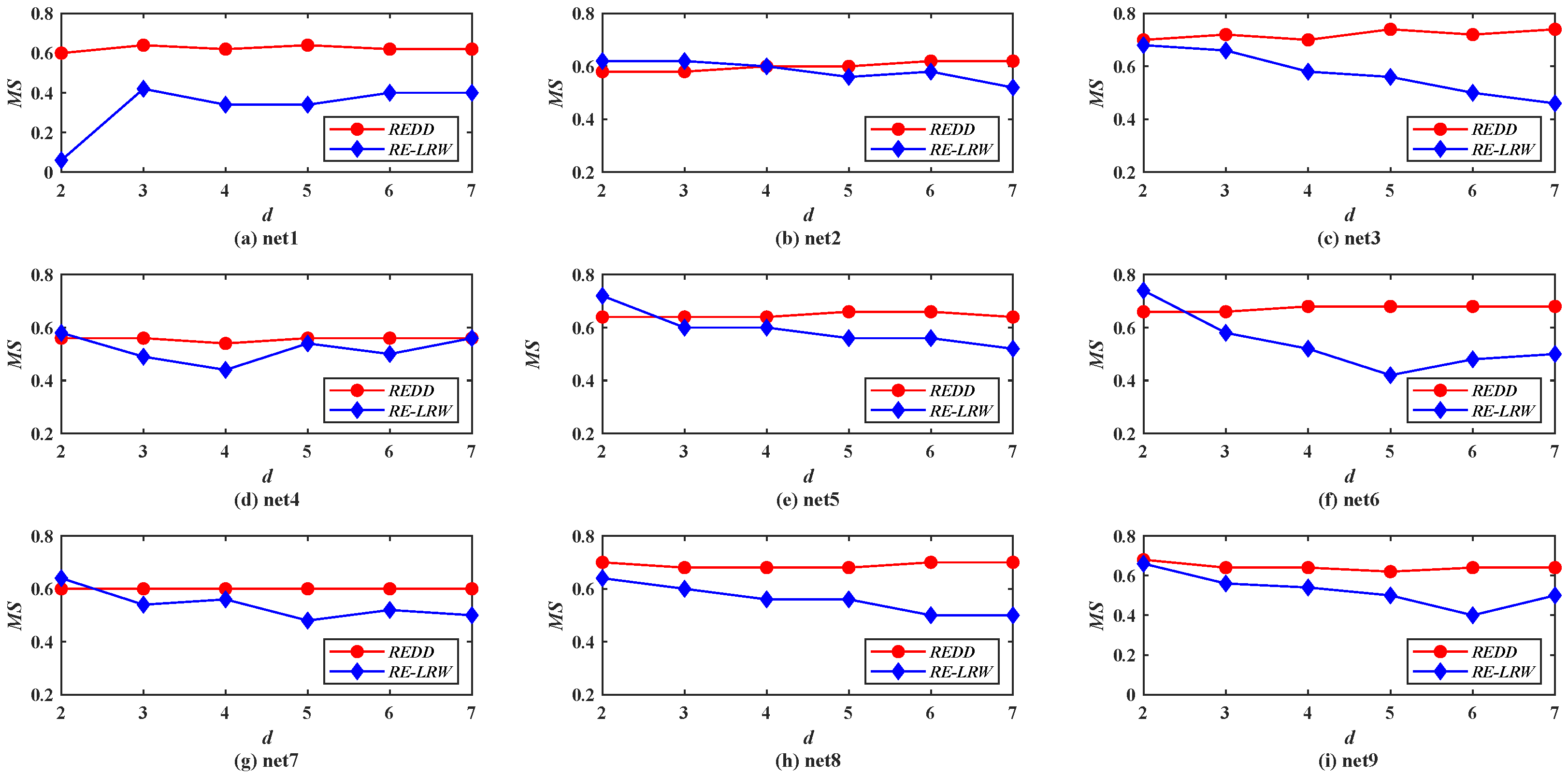
Figure 10 demonstrates the
MS curves of the
REDD and
RE-LRW indices in nine simulated networks when the dimension
d is changed from 2 to 7.
Compared with the
MS performance of the
REDD and
RE-LRW indices in real-world graph data, their
MS performance shows higher accuracy in the simulated graph data. This indicates that the performance of the corresponding algorithm will be improved to some extent if the graph data can be collected more accurately. From
Figure 10, one can observe that the
MS curves of the
RE-LRW index presents a large fluctuation range in different graph data. Thus, the robustness of the
RE-LRW index still needs to be improved in simulated graph data.
Figure 11 shows the
AUC curves of the
REDD and
RE-LRW indices in nine simulated networks when the dimension
d is changed from 2 to 7. From the results in
Figure 11, it can be observed that the
REDD index can be better than the
RE-LRW index in accuracy and robustness.
Therefore, if the similarity measure based on relative entropy is proposed by only considering the degree of nodes, its performance may have no advantage in link prediction.
Above all, these results in simulated graph data reflect that it is feasible to comprehensively take into account the degree and strength of nodes for enhancing the performance of the similarity measure based on the relative entropy once again.
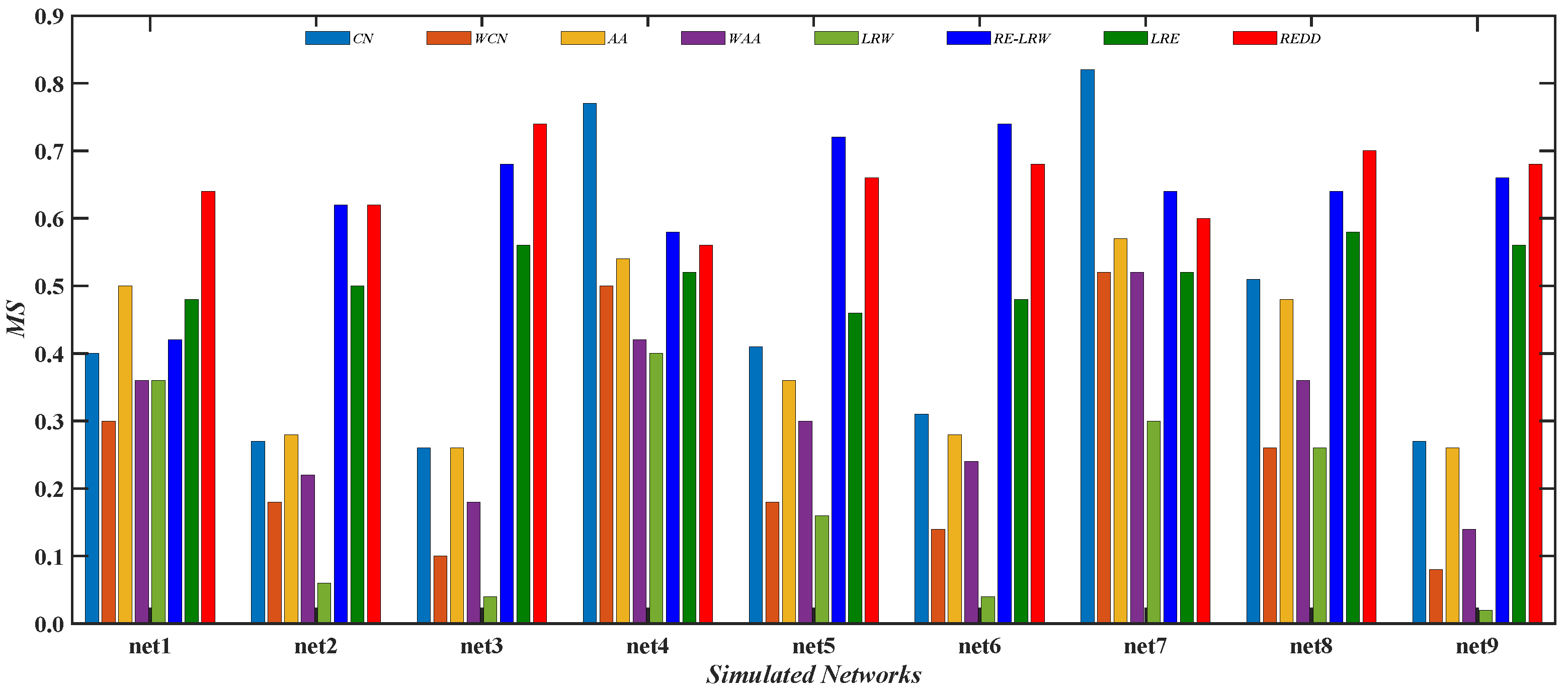
Figure 12 gives the comparison of the
MS values between the
REDD index and seven benchmark indices in nine simulated networks. From the results, it can be seen that the performance of the
CN and
AA indices is better than that of their weighted version. It indicates that the
CN and
AA indices are more suitable for performing the most similar node mining in simulated networks. Moreover, it can be also found that the
MS values of
CN index is the highest in net4 and net7. This indicates that the
CN index performs well in the graph data with a larger average clustering coefficient. Compared with the
RE-LRW index, the performance of
LRW index seems to be less than ideal in both most similar node mining cases.
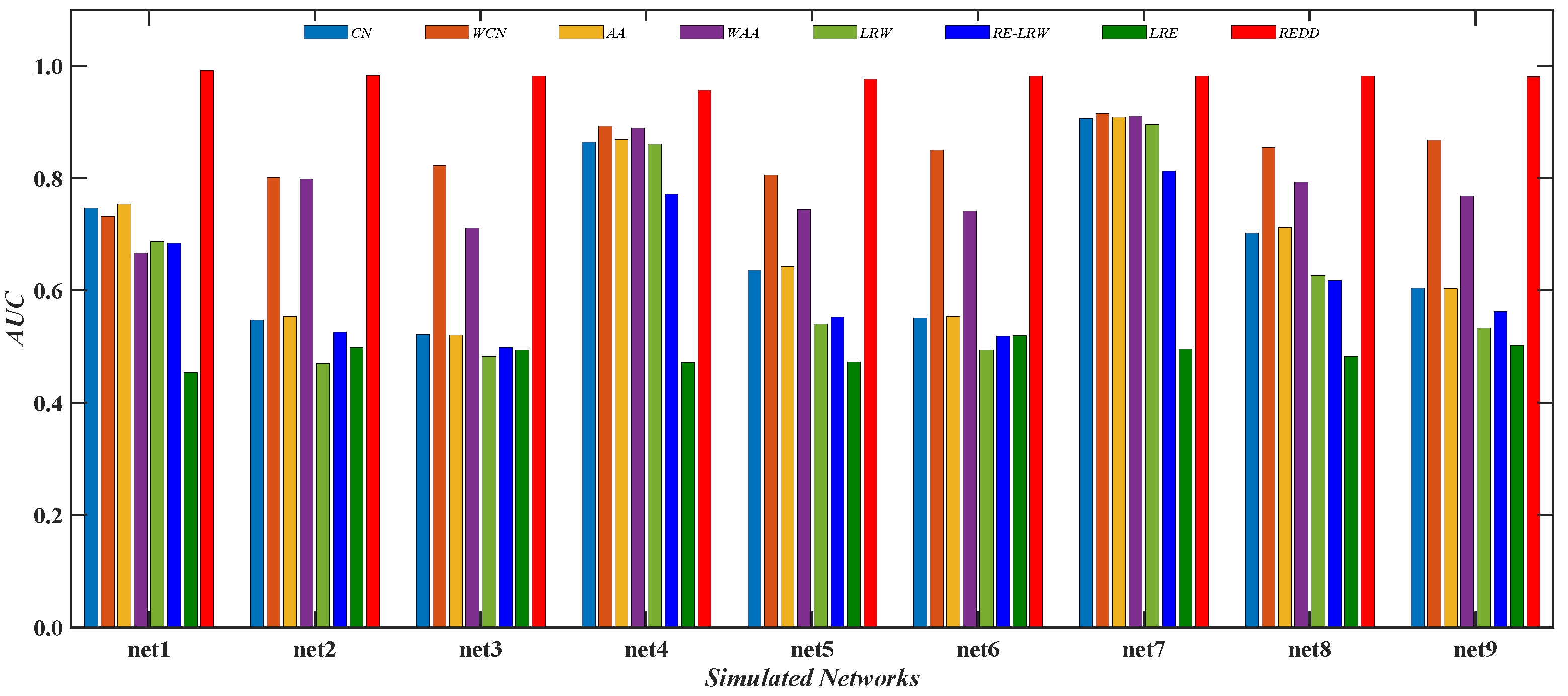
Figure 13 reports the comparison of the
AUC values between the
REDD index and seven benchmark indices in nine simulated networks. From the results, it can be seen that the performance of the
CN and
AA indices may be inferior to that of their weighted version. It indicates that these indices that only consider the degree or strength of nodes are also influenced by strong and weak ties in the weighted simulated graph data. From the results, the performance of the
LRW index may have a greater advantage than that of the
RE-LRW index in link prediction.
Thus, the performance of the
RE-LRW and
LRW indices need to be further improved in the most similar node mining and link prediction.
In addition, it can be found that despite the
MS performance of the
RE-LRW index being almost the same as that of the
REDD index, the
AUC performance of the former is far less than that of the latter. This may be because the
REDD index comprehensively utilizes the degree and strength of nodes, which results in the performance of algorithm being enhanced. From the above analysis and discussion, the
REDD index can also achieve good results in simulated networks after considering the degree and strength of nodes.