An Entropy-Based Measure of Complexity: An Application in Lung-Damage
Abstract
:1. Introduction
2. Related Works
2.1. Entropy and Complex Approaches
2.2. Artificial Intelligence
3. Preliminaries
3.1. Fractal and Information Dimensions
3.2. D-Summable Information Dimension
3.3. Entropy-Based Measure of Complexity
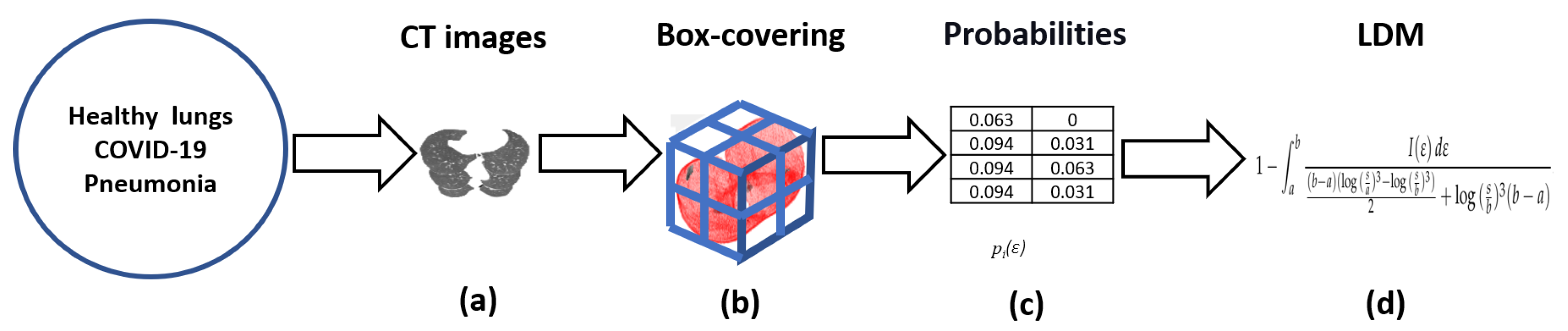
4. Method
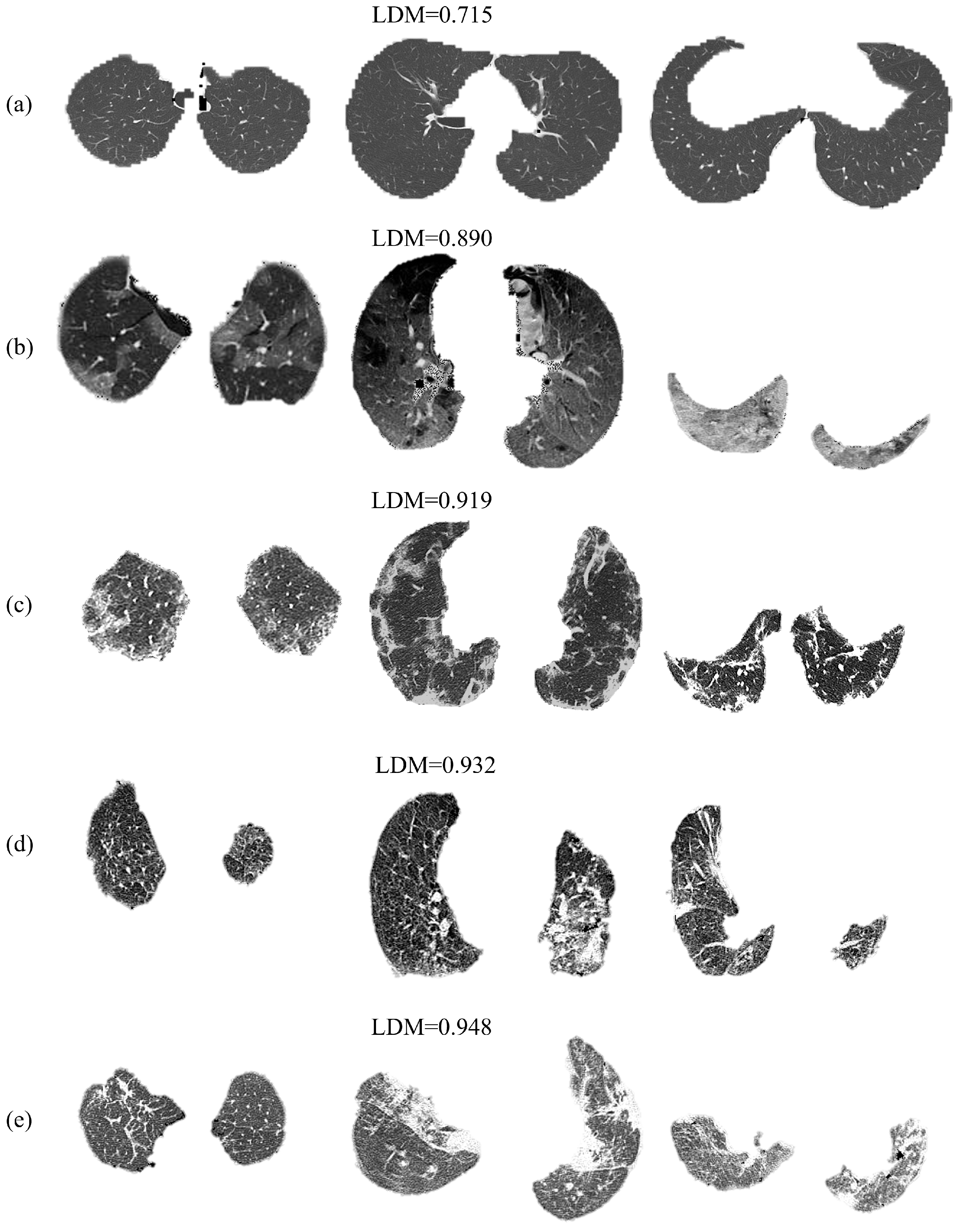
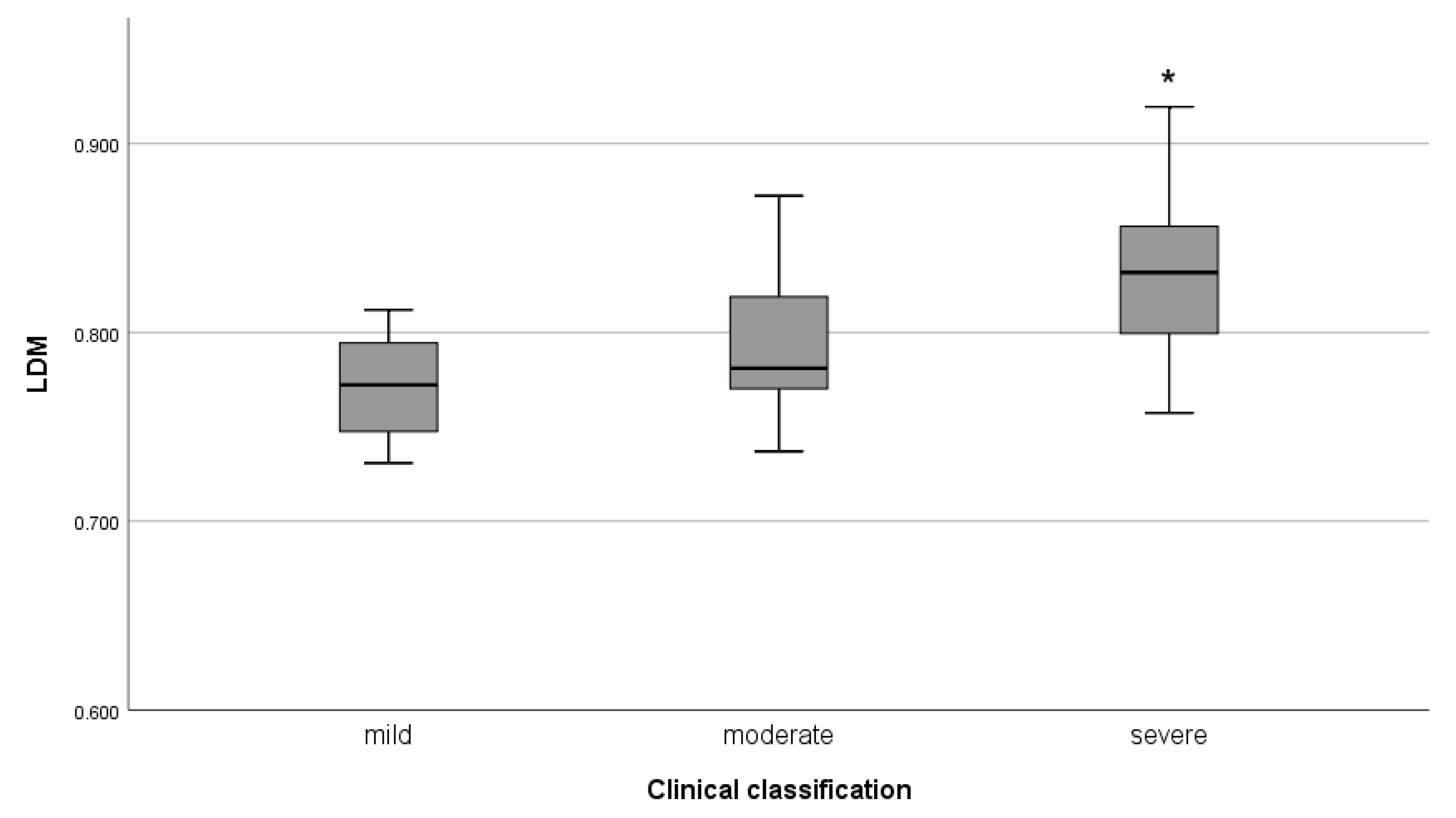
5. Applications
6. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIC | Akaike Information Criterion |
| ANOVA | Analysis of Variance |
| CAD | Computer-aided Diagnostic |
| CO-RADS | Classification of the NCCH, the COVID-19 Reporting and Data System |
| COVID-19 | Coronavirus Disease 2019 |
| CT | Computed Tomography |
| EMC | Entropy-based Measure of Complexity |
| LDM | Lung Damage Measure |
| PCR | Polymerase Chain Reaction |
| ROI | Region Of Interest |
References
- Pu, J.; Leader, J.K.; Bandos, A.; Ke, S.; Wang, J.; Shi, J.; Du, P.; Guo, Y.; Wenzel, S.E.; Fuhrman, C.R.; et al. Automated quantification of COVID-19 severity and progression using chest CT images. Eur. Radiol. 2021, 31, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Xu, X.; Zhou, L.Y.; Chen, G.; Li, Y.; Yin, H.; Sun, Z. Clinical characteristics and chest CT imaging features of critically ill COVID-19 patients. Eur. Radiol. 2020, 30, 6151–6160. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhong, Z.; Xie, X.; Yu, Q.; Liu, J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: A multicenter study. AJR Am. J. Roentgenol. 2020, 214, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.C.; Xu, P.; Lv, W.F.; Qiu, X.H.; Yao, J.L.; Gu, J.F.; Wei, W. CT manifestations of coronavirus disease-2019: A retrospective analysis of 73 cases by disease severity. Eur. J. Radiol. 2020, 126, 108941. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.; Haven, K.; Reed, P.; Bissielo, A.; Harvey, D.; McArthur, C.; Bringans, C.; Freundlich, S.; Ingram, R.J.H.; Perry, D.; et al. A chest radiograph scoring system in patients with severe acute respiratory infection: A validation study. BMC Med. Imaging 2015, 15, 61. [Google Scholar] [CrossRef]
- Salahshour, F.; Mehrabinejad, M.M.; Nassiri Toosi, M.; Gity, M.; Ghanaati, H.; Shakiba, M.; Nosrat Sheybani, S.; Komaki, H.; Kolahi, S. Clinical and chest CT features as a predictive tool for COVID-19 clinical progress: Introducing a novel semi-quantitative scoring system. Eur. Radiol. 2021, 31, 5178–5188. [Google Scholar] [CrossRef]
- De Smet, K.; De Smet, D.; Ryckaert, T.; Laridon, E.; Heremans, B.; Vandenbulcke, R.; Demedts, I.; Bouckaert, B.; Gryspeerdt, S.; Martens, G.A. Diagnostic performance of chest CT for SARS-CoV-2 infection in individuals with or without COVID-19 symptoms. Radiology 2021, 298, E30–E37. [Google Scholar] [CrossRef]
- Rossi, G.; Bauckneht, M.; Genova, C.; Rijavec, E.; Biello, F.; Mennella, S.; Dal Bello, M.G.; Cittadini, G.; Bruzzi, P.; Piva, R.; et al. Comparison between 18F-FDG PET–based and CT-based criteria in non–small cell lung cancer Patients treated with nivolumab. J. Nucl. Med. 2020, 61, 990–998. [Google Scholar] [CrossRef]
- Doubková, M.; Švancara, J.; Svoboda, M.; Šterclová, M.; Bartoš, V.; Plačková, M.; Lacina, L.; Žurková, M.; Binková, I.; Bittenglová, R.; et al. EMPIRE registry, Czech part: Impact of demographics, pulmonary function and HRCT on survival and clinical course in idiopathic pulmonary fibrosis. Clin. Respir. J. 2018, 12, 1526–1535. [Google Scholar] [CrossRef]
- Flechsig, P.; Rastgoo, R.; Kratochwil, C.; Martin, O.; Holland-Letz, T.; Harms, A.; Kauczor, H.U.; Haberkorn, U.; Giesel, F.L. Impact of computer-aided CT and PET analysis on non-invasive T staging in patients with lung cancer and atelectasis. Mol. Imaging Biol. 2018, 20, 1044–1052. [Google Scholar] [CrossRef]
- Hooda, R.; Mittal, A.; Sofat, S. Tuberculosis detection from chest radiographs: A comprehensive survey on computer-aided diagnosis techniques. Curr. Med. Imaging 2018, 14, 506–520. [Google Scholar] [CrossRef]
- Mahomed, N.; van Ginneken, B.; Philipsen, R.H.; Melendez, J.; Moore, D.P.; Moodley, H.; Sewchuran, T.; Mathew, D.; Madhi, S.A. Computer-aided diagnosis for World Health Organization-defined chest radiograph primary-endpoint pneumonia in children. Pediatr. Radiol. 2020, 50, 482–491. [Google Scholar] [CrossRef]
- Carvalho, A.R.S.; Guimarães, A.; Werberich, G.M.; de Castro, S.N.; Pinto, J.S.F.; Schmitt, W.R.; França, M.; Bozza, F.A.; Guimarães, B.L.d.S.; Zin, W.A.; et al. COVID-19 Chest Computed Tomography to Stratify Severity and Disease Extension by Artificial Neural Network Computer-Aided Diagnosis. Front. Med. 2020, 7, 577609. [Google Scholar] [CrossRef]
- Khan, M.A.; Majid, A.; Akram, T.; Hussain, N.; Nam, Y.; Kadry, S.; Wang, S.H.; Alhaisoni, M. Classification of COVID-19 CT scans via extreme learning machine. Comput. Mater. Contin. 2021, 68, 1003–1019. [Google Scholar]
- Al-Antari, M.A.; Hua, C.H.; Bang, J.; Lee, S. Fast deep learning computer-aided diagnosis of COVID-19 based on digital chest X-ray images. Appl. Intell. 2021, 51, 2890–2907. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, C.; Hu, Y.; Li, C.; Ren, Q.; Zhang, X.; Shi, H.; Zhou, M. Temporal Changes of CT Findings in 90 Patients with COVID-19 Pneumonia: A Longitudinal Study. Radiology 2020, 296, E55–E64. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wu, X.; Zeng, W.; Guo, D.; Fang, Z.; Chen, L.; Huang, H.; Li, C. Chest CT findings in patients with coronavirus disease 2019 and its relationship with clinical features. Investig. Radiol. 2020, 55, 257. [Google Scholar] [CrossRef] [PubMed]
- Abin, D.; Thepade, S.D.; Mankar, H.; Raut, S.; Yadav, A. Blending of Contrast Enhancement Techniques for Chest X-ray Pneumonia Images. In Proceedings of the 2022 International Conference on Electronics and Renewable Systems (ICEARS), Tuticorin, India, 16–18 March 2022; pp. 981–985. [Google Scholar]
- Wang, S.H.; Zhou, J.; Zhang, Y.D. Community-acquired pneumonia recognition by wavelet entropy and cat swarm optimization. Mob. Netw. Appl. 2022, 1, 1–18. [Google Scholar] [CrossRef]
- Santos, A.M.; de Carvalho Filho, A.O.; Silva, A.C.; de Paiva, A.C.; Nunes, R.A.; Gattass, M. Automatic detection of small lung nodules in 3D CT data using Gaussian mixture models, Tsallis entropy and SVM. Eng. Appl. Artif. Intell. 2014, 36, 27–39. [Google Scholar] [CrossRef]
- Brochet, T.; Lapuyade-Lahorgue, J.; Huat, A.; Thureau, S.; Pasquier, D.; Gardin, I.; Modzelewski, R.; Gibon, D.; Thariat, J.; Grégoire, V.; et al. A Quantitative Comparison between Shannon and Tsallis–Havrda–Charvat Entropies Applied to Cancer Outcome Prediction. Entropy 2022, 24, 436. [Google Scholar] [CrossRef]
- Postolache, P.; Borsos, Z.; Paun, V.A.; Paun, V.P. New way in fractal analysis of pulmonary medical images. Univ. Politeh. Buchar. Sci. Bull.-Ser. A-Appl. Math. Phys. 2018, 80, 313–322. [Google Scholar]
- Bodduluri, S.; Puliyakote, A.S.K.; Gerard, S.E.; Reinhardt, J.M.; Hoffman, E.A.; Newell, J.D.; Nath, H.P.; Han, M.K.; Washko, G.R.; Estépar, R.S.J.; et al. Airway fractal dimension predicts respiratory morbidity and mortality in COPD. J. Clin. Investig. 2018, 128, 5374–5382. [Google Scholar] [CrossRef] [PubMed]
- Van de Moortele, T.; Goerke, U.; Wendt, C.H.; Coletti, F. Airway morphology and inspiratory flow features in the early stages of Chronic Obstructive Pulmonary Disease. Clin. Biomech. 2019, 66, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Arellano, A.; Ortiz-Vilchis, P.; Bory-Reyes, J. The role of D-summable information dimension in differentiating COVID-19 disease. Fractals 2021, 29, 2150255. [Google Scholar] [CrossRef]
- Namazi, H.; Kulish, V.V. Complexity-based classification of the coronavirus disease (COVID-19). Fractals 2020, 28, 2050114. [Google Scholar] [CrossRef]
- Al-Kadi, O.S. Prediction of FDG-PET stage and uptake for non-small cell lung cancer on non-contrast enhanced CT scans via fractal analysis. Clin. Imaging 2020, 65, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Astinchap, B.; Ghanbaripour, H.; Amuzgar, R. Multifractal analysis of chest CT images of patients with the 2019 novel coronavirus disease (COVID-19). Chaos Solitons Fractals 2022, 156, 111820. [Google Scholar] [CrossRef]
- Subramanian, N.; Elharrouss, O.; Al-Maadeed, S.; Chowdhury, M. A review of deep learning-based detection methods for COVID-19. Comput. Biol. Med. 2022, 143, 105233. [Google Scholar] [CrossRef] [PubMed]
- Alafif, T.; Tehame, A.M.; Bajaba, S.; Barnawi, A.; Zia, S. Machine and Deep Learning towards COVID-19 Diagnosis and Treatment: Survey, Challenges, and Future Directions. Int. J. Environ. Res. Public Health 2021, 18, 1117. [Google Scholar] [CrossRef] [PubMed]
- Alyasseri, Z.A.A.; Al-Betar, M.A.; Doush, I.A.; Awadallah, M.A.; Abasi, A.K.; Makhadmeh, S.N.; Alomari, O.A.; Abdulkareem, K.H.; Adam, A.; Damasevicius, R.; et al. Review on COVID-19 diagnosis models based on machine learning and deep learning approaches. Expert Syst. 2022, 39, e12759. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Siegel, E.; Shen, D. Deep Learning and Medical Image Analysis for COVID-19 Diagnosis and Prediction. Annu. Rev. Biomed. Eng. 2022, 24, 179–201. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Karray, F.; Alhajj, R.; Zeng, J. A Review on Deep Learning Techniques for the Diagnosis of Novel Coronavirus (COVID-19). IEEE Access 2021, 9, 30551–30572. [Google Scholar] [CrossRef] [PubMed]
- Skandha, S.S.; Gupta, S.K.; Saba, L.; Koppula, V.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; et al. 3-D optimized classification and characterization artificial intelligence paradigm for cardiovascular/stroke risk stratification using carotid ultrasound-based delineated plaque: Atheromatic™ 2.0. Comput. Biol. Med. 2020, 125, 103958. [Google Scholar] [CrossRef] [PubMed]
- Malkawi, A.; Almarzooq, Z.; Al-Mallah, M.H.; Al’Aref, S.J. Artificial Intelligence-Based Cardiovascular Risk Stratification. In Artificial Intelligence in Cardiothoracic Imaging; Springer: Cham, Switzerland, 2022; pp. 403–419. [Google Scholar]
- Suri, J.S.; Paul, S.; Maindarkar, M.A.; Puvvula, A.; Saxena, S.; Saba, L.; Turk, M.; Laird, J.R.; Khanna, N.N.; Viskovic, K.; et al. Cardiovascular/Stroke Risk Stratification in Parkinson’s Disease Patients Using Atherosclerosis Pathway and Artificial Intelligence Paradigm: A Systematic Review. Metabolites 2022, 12, 312. [Google Scholar] [CrossRef] [PubMed]
- Willan, J.; Katz, H.; Keeling, D. The use of artificial neural network analysis can improve the risk-stratification of patients presenting with suspected deep vein thrombosis. Br. J. Haematol. 2019, 185, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.S.; Ge, P.; Jiang, C.; Zhang, Y.; Li, X.; Zhao, Z.; Zhang, L.; Duong, T.Q. Deep-learning artificial intelligence analysis of clinical variables predicts mortality in COVID-19 patients. J. Am. Coll. Emerg. Physicians Open 2020, 1, 1364–1373. [Google Scholar] [CrossRef]
- Thomas, J.; Haertling, T. AIBx, artificial intelligence model to risk stratify thyroid nodules. Thyroid 2020, 30, 878–884. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, P.; Wang, Q.; Li, P.; Zeng, J.; Qin, P.; Meng, Y. Separability of acute cerebral infarction lesions in CT based radiomics: Toward artificial intelligence-assisted diagnosis. BioMed Res. Int. 2020, 2020, 8864756. [Google Scholar] [CrossRef]
- Handa, T.; Tanizawa, K.; Oguma, T.; Uozumi, R.; Watanabe, K.; Tanabe, N.; Niwamoto, T.; Shima, H.; Mori, R.; Nobashi, T.W.; et al. Novel artificial intelligence-based technology for chest computed tomography analysis of idiopathic pulmonary fibrosis. Ann. Am. Thorac. Soc. 2022, 19, 399–406. [Google Scholar] [CrossRef]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M.; Gertych, A.; San Tan, R. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Arsalan, M.; Owais, M.; Mahmood, T.; Choi, J.; Park, K.R. Artificial intelligence-based diagnosis of cardiac and related diseases. J. Clin. Med. 2020, 9, 871. [Google Scholar] [CrossRef] [PubMed]
- Dunn, A.J.; ElRefai, M.H.; Roberts, P.R.; Coniglio, S.; Wiles, B.M.; Zemkoho, A.B. Deep learning methods for screening patients’ S-ICD implantation eligibility. Artif. Intell. Med. 2021, 119, 102139. [Google Scholar] [CrossRef] [PubMed]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Kumar, V.; Kaur, M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1379–1389. [Google Scholar] [CrossRef]
- Wang, S.H.; Govindaraj, V.V.; Górriz, J.M.; Zhang, X.; Zhang, Y.D. COVID-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inf. Fusion 2021, 67, 208–229. [Google Scholar] [CrossRef]
- Yan, T.; Wong, P.K.; Ren, H.; Wang, H.; Wang, J.; Li, Y. Automatic distinction between COVID-19 and common pneumonia using multi-scale convolutional neural network on chest CT scans. Chaos Solitons Fractals 2020, 140, 110153. [Google Scholar] [CrossRef]
- Sharifrazi, D.; Alizadehsani, R.; Roshanzamir, M.; Joloudari, J.H.; Shoeibi, A.; Jafari, M.; Hussain, S.; Sani, Z.A.; Hasanzadeh, F.; Khozeimeh, F.; et al. Fusion of convolution neural network, support vector machine and Sobel filter for accurate detection of COVID-19 patients using X-ray images. Biomed. Signal Process. Control 2021, 68, 102622. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.D. A comprehensive study on classification of COVID-19 on computed tomography with pretrained convolutional neural networks. Sci. Rep. 2020, 10, 16942. [Google Scholar] [CrossRef] [PubMed]
- Gunraj, H.; Wang, L.; Wong, A. Covidnet-ct: A tailored deep convolutional neural network design for detection of covid-19 cases from chest ct images. Front. Med. 2020, 7, 608525. [Google Scholar] [CrossRef] [PubMed]
- Mandelbrot, B.B. The Fractal Geometry of Nature; Freeman: San Francisco, CA, USA, 1982. [Google Scholar]
- Lapidus, M.L. Fractal drum, inverse spectral problems for elliptic operators and a partial resolution of the Weyl–Berry conjeture. Trans. Am. Math. Soc. 1991, 325, 465–529. [Google Scholar] [CrossRef]
- Wei, D.; Wei, B.; Hu, Y.; Zhang, H.; Deng, Y. A new information dimension of complex networks. Phys. Lett. A 2014, 378, 1091–1094. [Google Scholar] [CrossRef]
- Beck, C. Generalized information and entropy measures in physics. Contemp. Phys. 2009, 50, 495–510. [Google Scholar] [CrossRef]
- Namazi, H.; Daneshi, A.; Shirzadian, T.; Azarnoush, H.; Jafari, S. Information-Based Analysis of the Relation Between Visual Stimuli and Human Eye Movements. Fluct. Noise Lett. 2019, 18, 1950010. [Google Scholar] [CrossRef]
- Borodich, F.M.; Volovikov, A.Y. Surface integrals for domains with fractal boundaries and some applications to elasticity. Proc. R. Soc. Lond. A Math. Phys. Eng. Sci. 2000, 456, 1–24. [Google Scholar] [CrossRef]
- Ramirez-Arellano, A.; Bermúdez-Gómez, S.; Hernández-Simón, L.M.; Bory-Reyes, J. D-summable fractal dimensions of complex networks. Chaos Solitons Fractals 2019, 119, 210–214. [Google Scholar] [CrossRef]
- Rosenberg, E. Maximal entropy coverings and the information dimension of a complex network. Phys. Lett. A 2017, 381, 574–580. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, X.; Shen, J.; Li, Z.; Sang, Y.; Wu, X.; Zha, Y.; Liang, W.; Wang, C.; Wang, K.; et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell 2020, 181, 1423–1433. [Google Scholar] [CrossRef]
- Radiopaedia. 1999. Available online: https://radiopaedia.org/ (accessed on 12 May 2021).
- Wei, P.F. Diagnosis and treatment protocol for novel coronavirus pneumonia (trial version 7). Chin. Med. J. 2020, 133, 1087–1095. [Google Scholar]
- Prokop, M.; Van Everdingen, W.; van Rees Vellinga, T.; Quarles van Ufford, H.; Stöger, L.; Beenen, L.; Geurts, B.; Gietema, H.; Krdzalic, J.; Schaefer-Prokop, C.; et al. CO-RADS: A categorical CT assessment scheme for patients suspected of having COVID-19—Definition and evaluation. Radiology 2020, 296, E97–E104. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Seber, G.; Wild, C. Nonlinear Regression; Wiley Series in Probability and Statistics; Wiley: Hoboken, NJ, USA, 2003. [Google Scholar]
- Ennos, A. Statistical and Data Handling Skills in Biology; Pearson/Prentice Hall: Upper Saddle River, NJ, USA, 2007. [Google Scholar]
- Burnham, K.P.; Anderson, D.R. Multimodel inference: Understanding AIC and BIC in model selection. Sociol. Methods Res. 2004, 33, 261–304. [Google Scholar] [CrossRef]
- Burnham, K.; Selection, A.M.; Inference, M. A Practical Information-Theoretic Approach; Springer: New York, NY, USA, 2010. [Google Scholar]

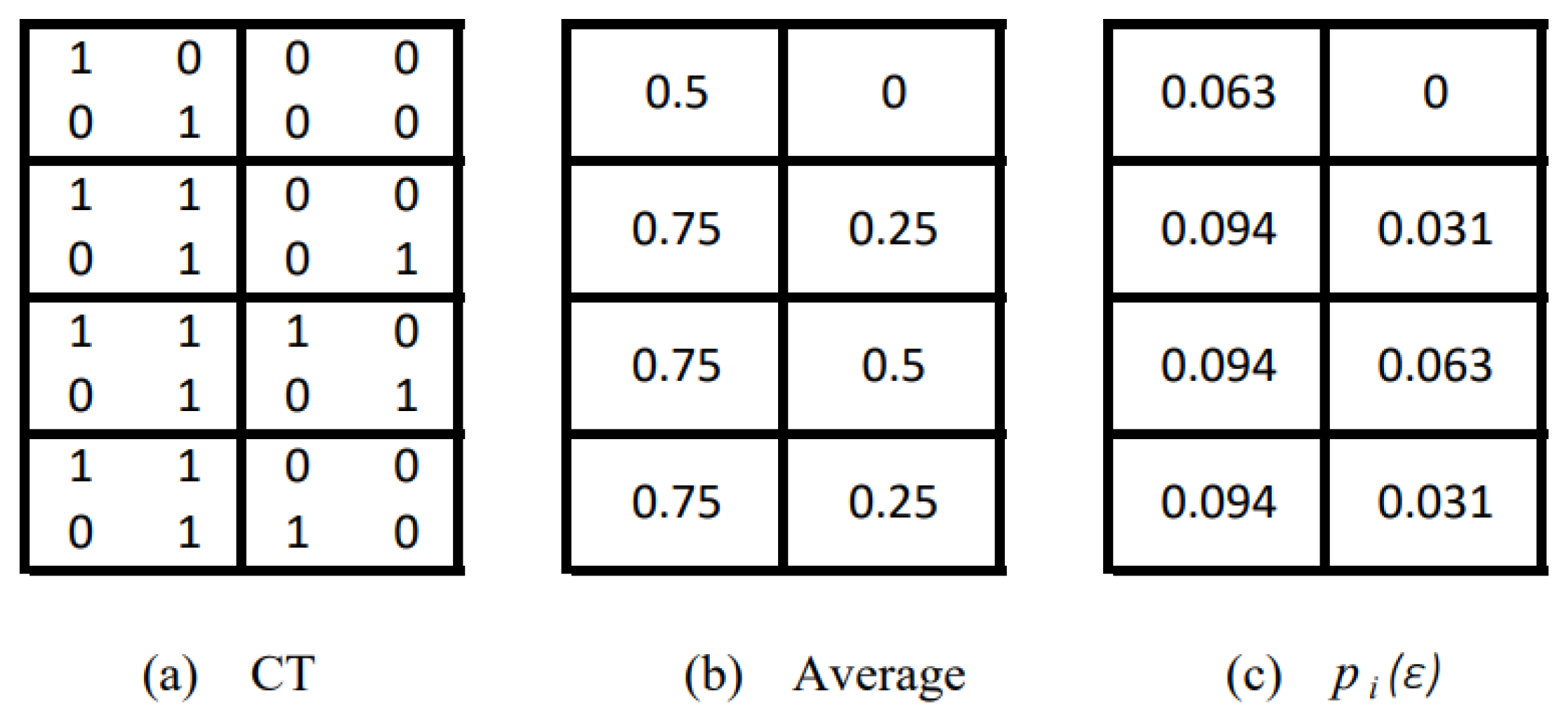
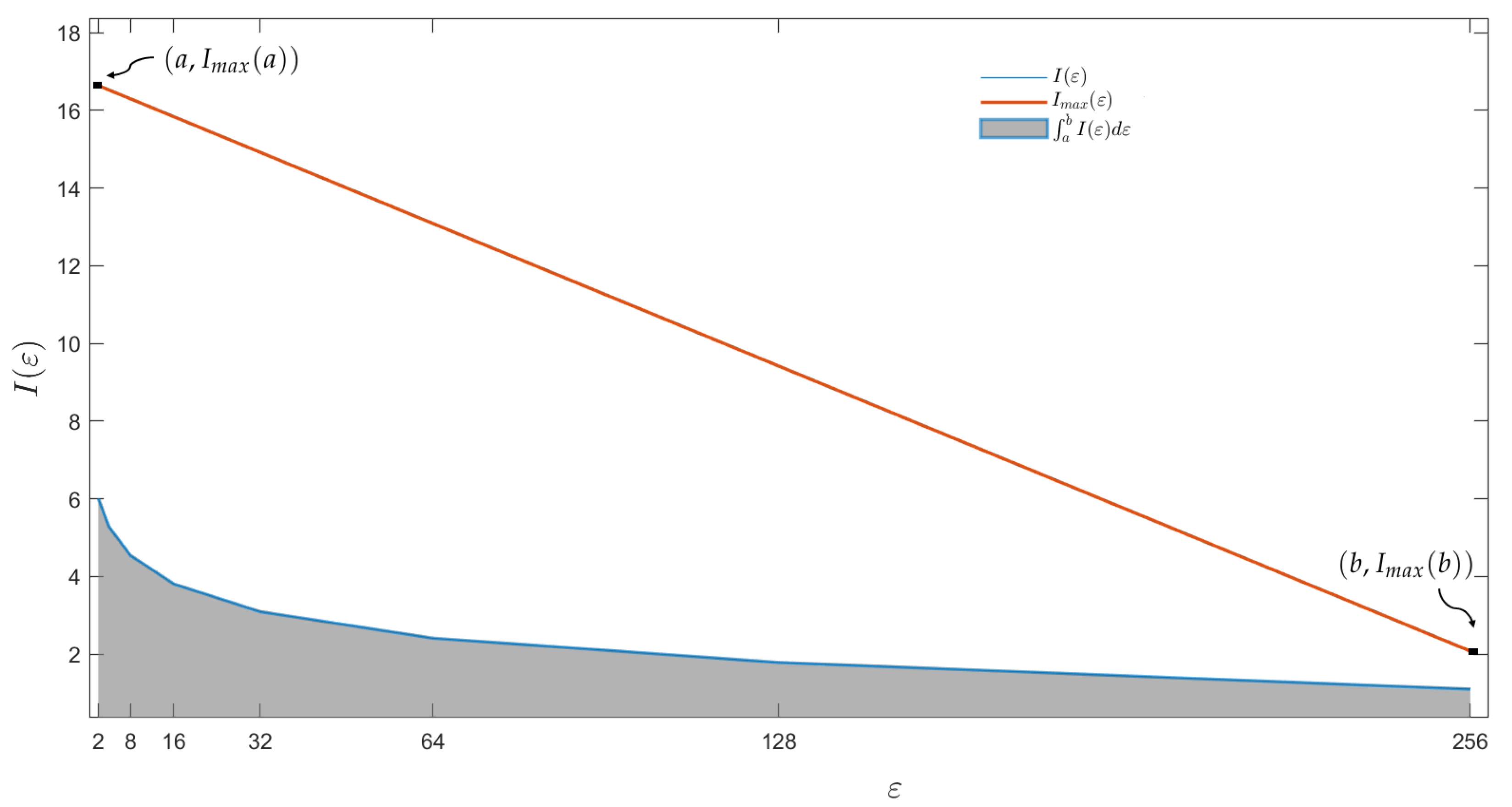

| Disease | CT Number | Slice Number | LDM |
|---|---|---|---|
| Healthy lungs | 486 | 90.300 (8.703) | 0.779 (0.031) |
| COVID-19 | 263 | 65.027 (16.084) | 0.814 (0.042) |
| Common pneumonia | 329 | 86.611 (14.262) | 0.852 (0.045) |
| Disease | |||||||
|---|---|---|---|---|---|---|---|
| Healthy lungs | −24.182 (3.255) | −42.927 (1.760) | 15.14 (2.782) | 0 (0) | 1.016 (0.164) | 1.007 (0.165) | 1.019 (0.004) |
| COVID-19 | −23.085 (2.53) | −42.273 (3.1384) | 15.58 (2.601) | 0 (0) | 0.838 (0.211) | 0.826 (0.213) | 1.027 (0.009) |
| Common pneumonia | −30.146 (9.990) | −46.358 (6.035) | 12.986 (5.601) | 0.08 (0.352) | 0.637 (0.231) | 0.628 (0.230) | 1.024 (0.001) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz-Vilchis, P.; Ramirez-Arellano, A. An Entropy-Based Measure of Complexity: An Application in Lung-Damage. Entropy 2022, 24, 1119. https://doi.org/10.3390/e24081119
Ortiz-Vilchis P, Ramirez-Arellano A. An Entropy-Based Measure of Complexity: An Application in Lung-Damage. Entropy. 2022; 24(8):1119. https://doi.org/10.3390/e24081119
Chicago/Turabian StyleOrtiz-Vilchis, Pilar, and Aldo Ramirez-Arellano. 2022. "An Entropy-Based Measure of Complexity: An Application in Lung-Damage" Entropy 24, no. 8: 1119. https://doi.org/10.3390/e24081119
APA StyleOrtiz-Vilchis, P., & Ramirez-Arellano, A. (2022). An Entropy-Based Measure of Complexity: An Application in Lung-Damage. Entropy, 24(8), 1119. https://doi.org/10.3390/e24081119







