A Family of Fitness Landscapes Modeled through Gene Regulatory Networks
Abstract
1. Introduction
2. Methods
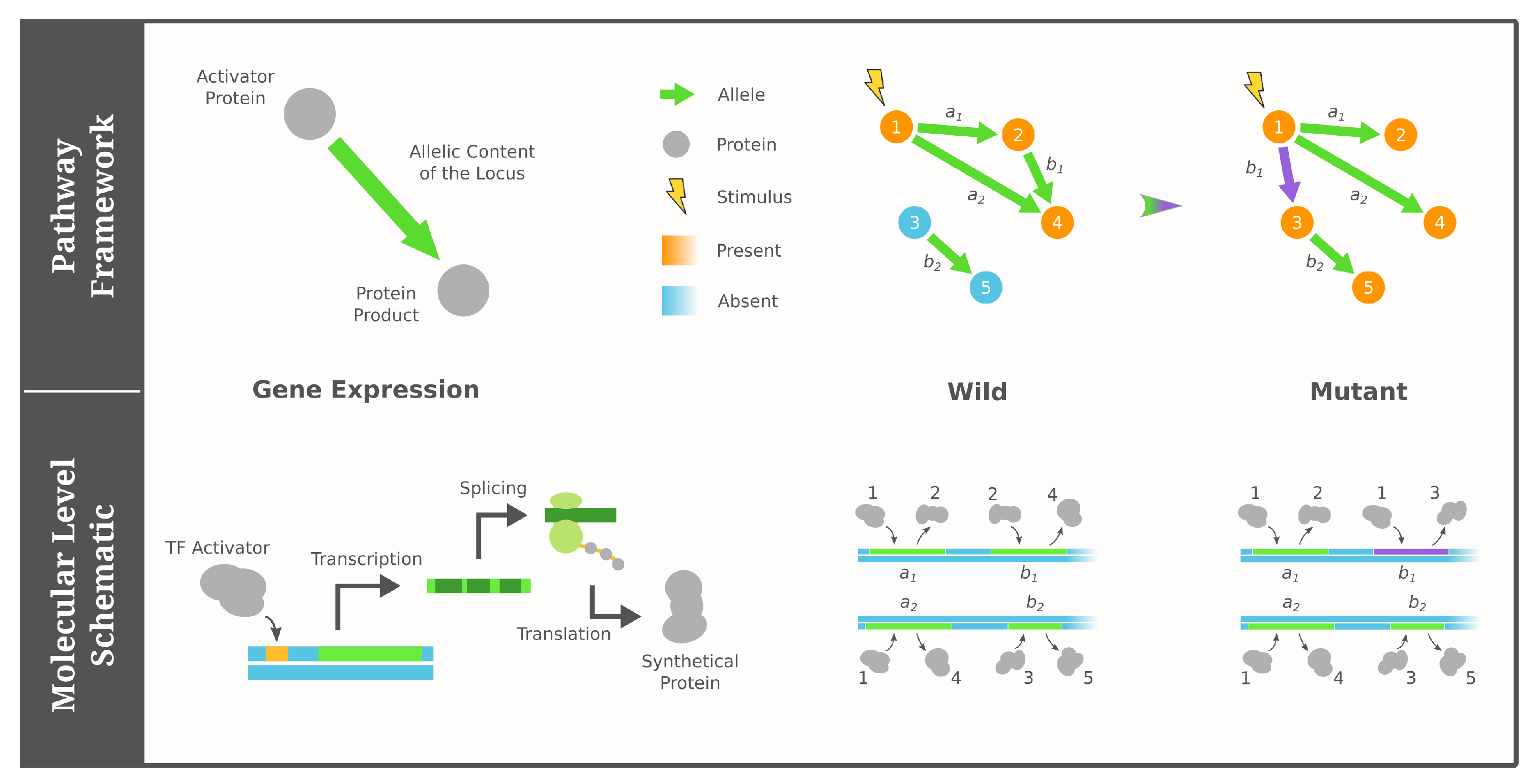
2.1. Pathway Framework of GRNs
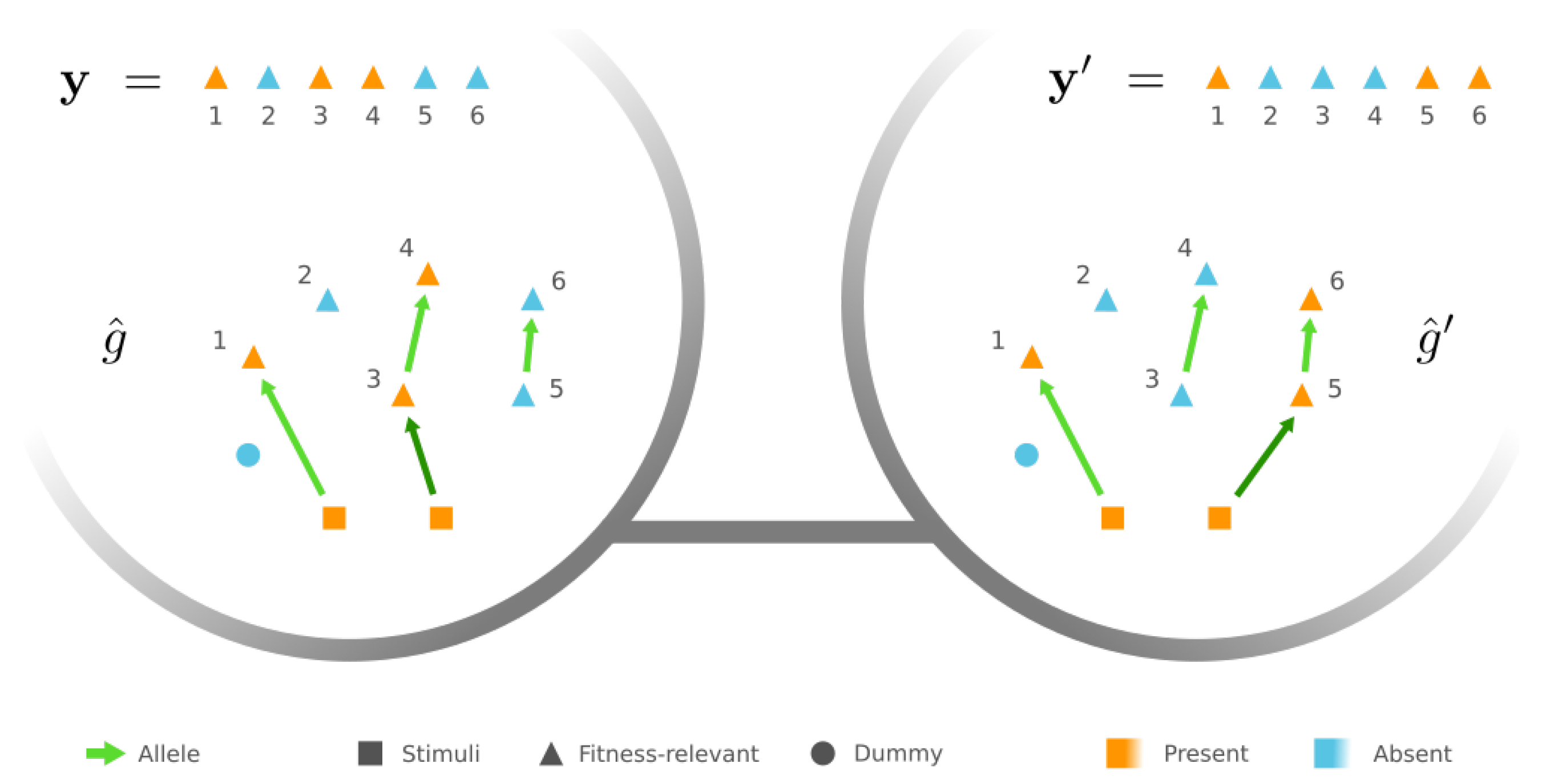
2.2. Fitness Landscape of GRNs under the Pathway Framework
3. Results
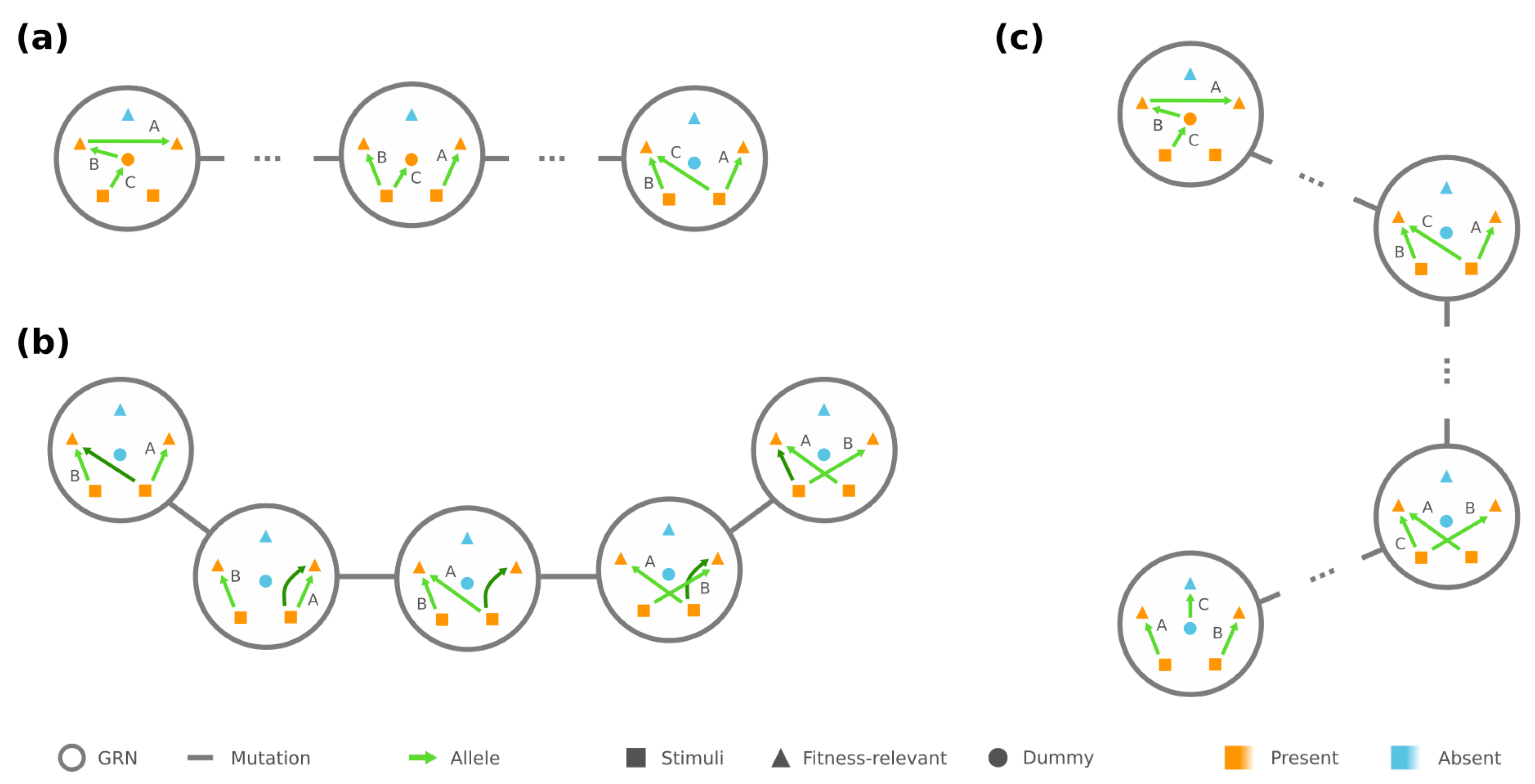
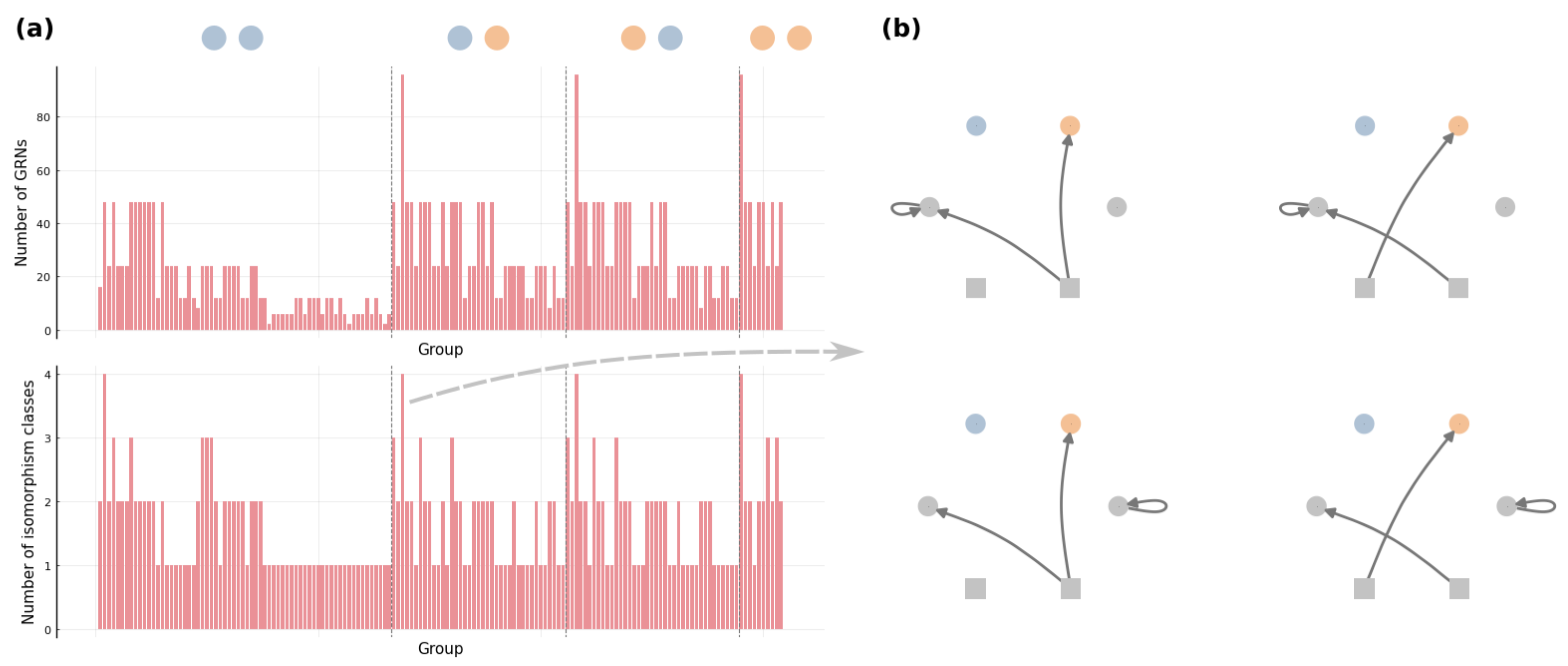
3.1. Connectivity and Accessibility in a Fitness Landscape of GRNs
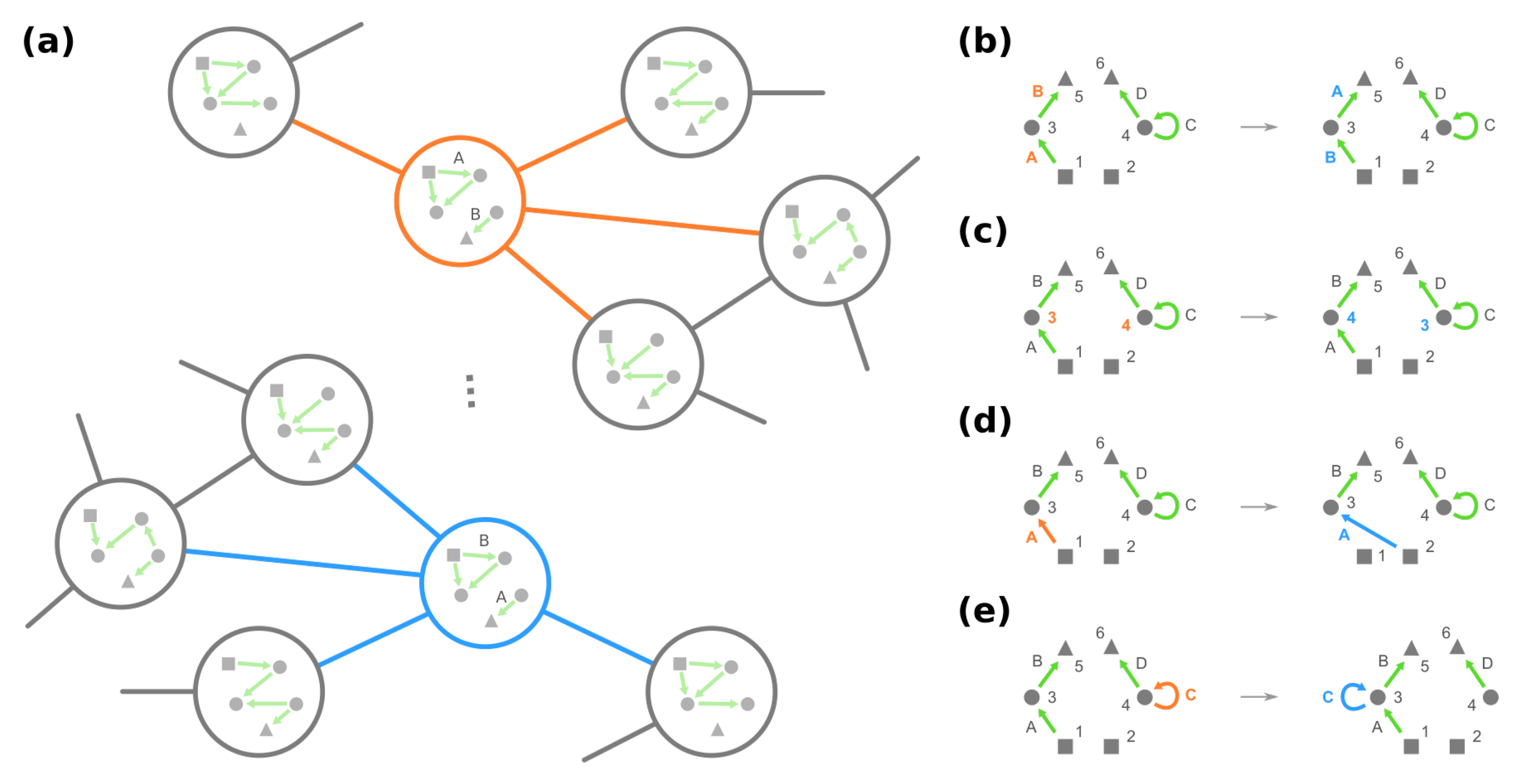
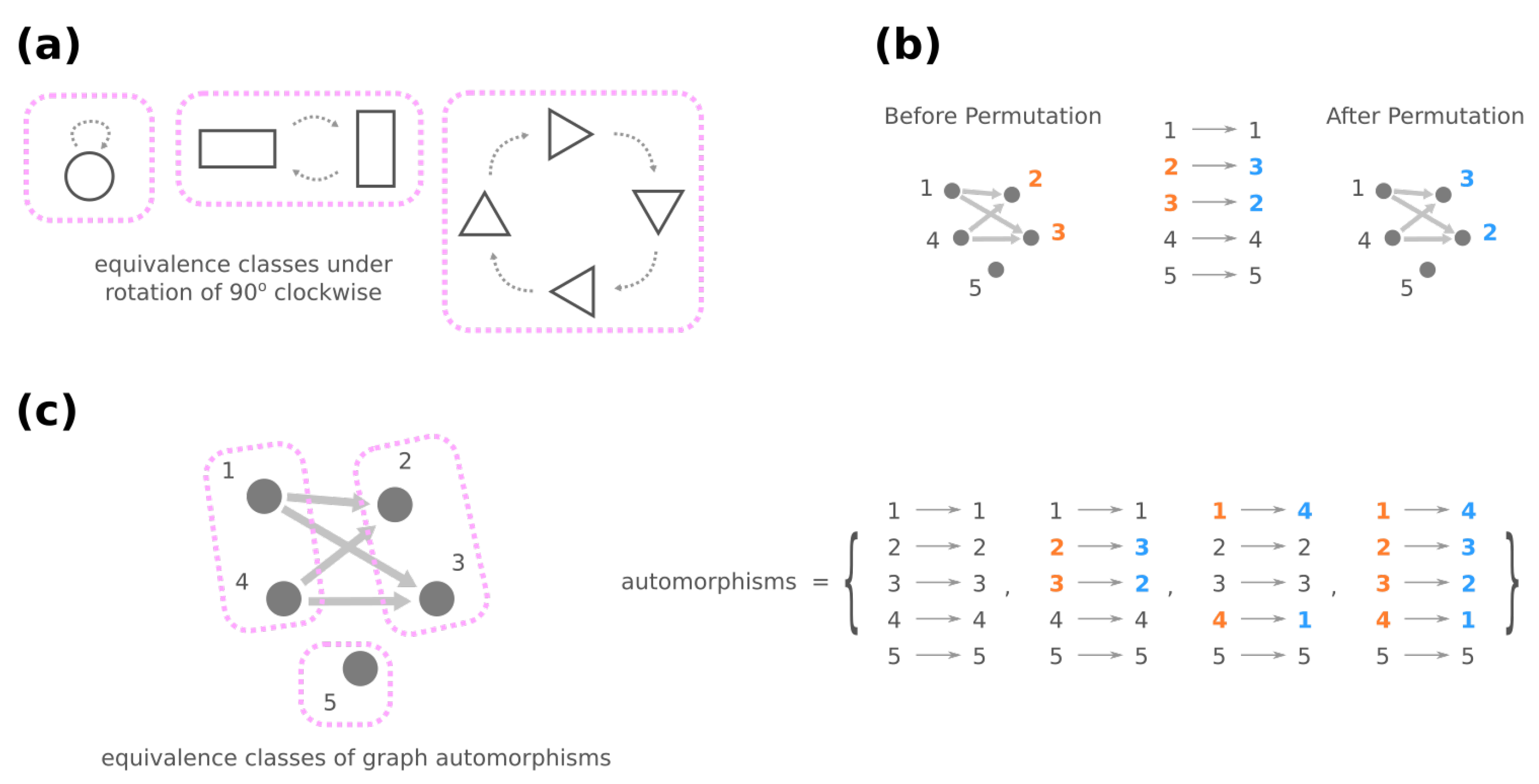
3.2. Mesoscopic Skeleton Derived from “Symmetries” in the Genotype Network of GRNs
- (iii)
- Change the source node of an edge from one stimulus to another stimulus and vice versa, e.g., in Figure 4d, moving an edge pointing from node 1 to node 3 to pointing from node 2. (Note that this operation is not necessarily equivalent to permuting the identities of stimuli since at most only the single focal edge will be affected.)
- (iv)
- Move a self-loop at one node to another node and vice versa, for example, re-allocating a self-loop at node 3 to node 4 in Figure 4e.
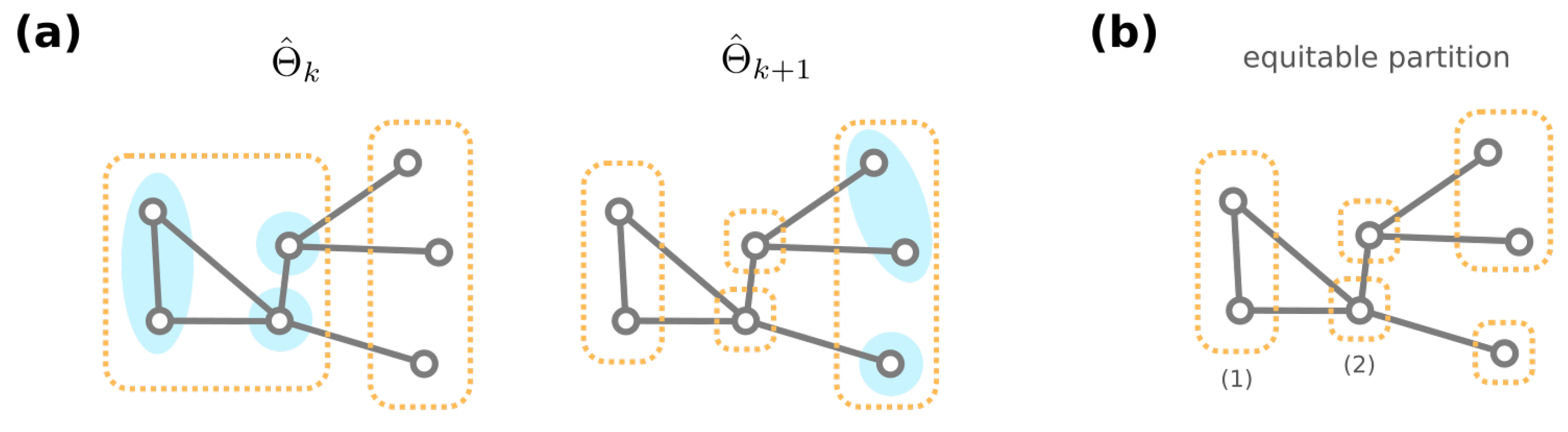
3.3. Algorithmic Construction of the Mesoscopic Backbone of GRN Fitness Landscape
- If has one more non-self-loop edge than g, then ;
- If has one less non-self-loop edge than g, then we have ;
- If has the same number of non-self-loop edges as g, and then they share a common mutational neighbor , where the only different edge between g and is rewired to a self-loop and thus .
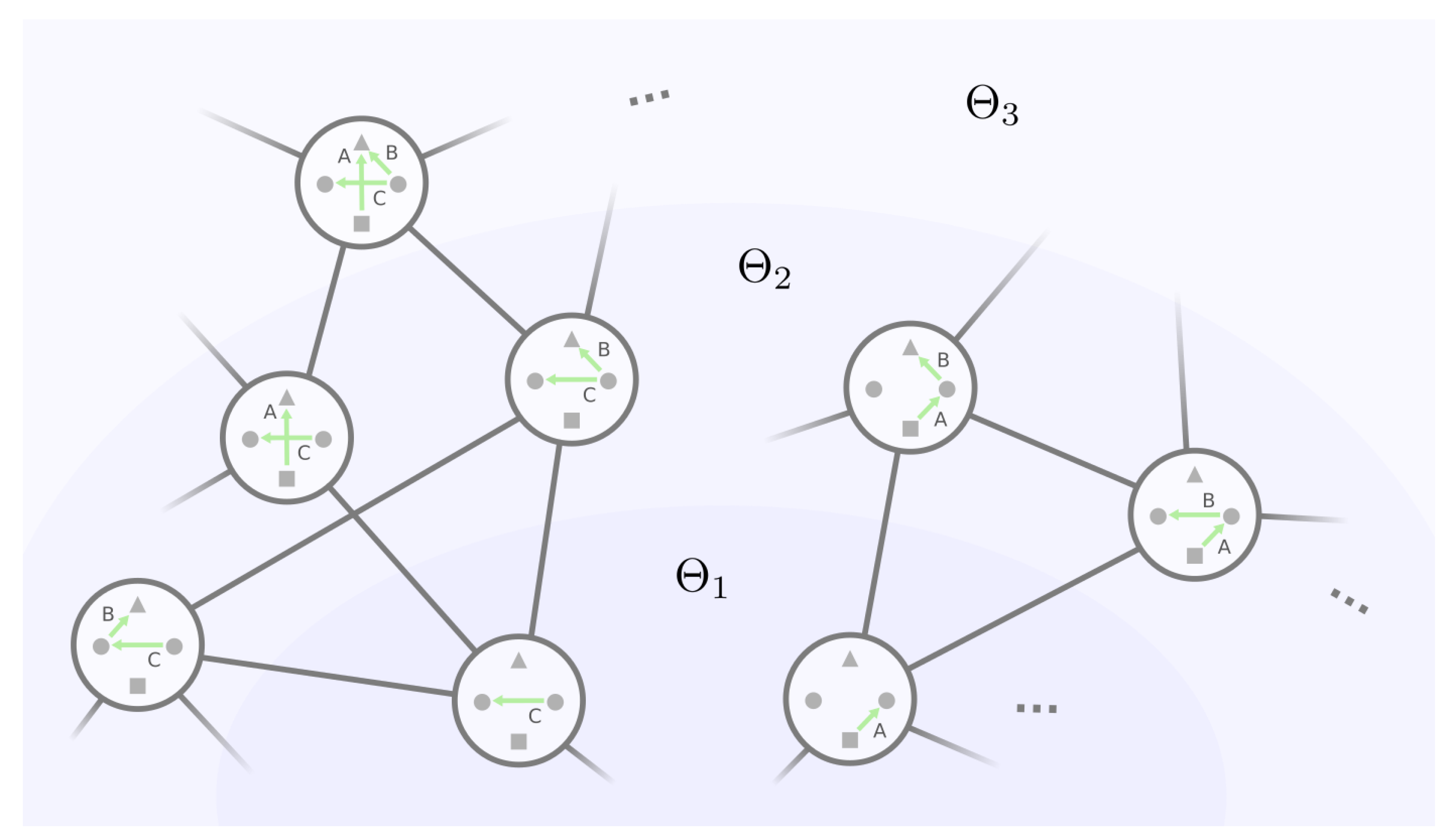
- (A)
- For an equivalence class and its representative GRN , under what condition will belong to the same equivalence class in layer ?
- (B)
- For two distinct equivalence classes and their representative GRNs and , under what condition will and belong to the same equivalence class in layer ?
- There is an integer p such that and ;
- There is another integer such that and ;
- for ;
- for ;
- For any locus and non-self-loop source–target pair such that for , we have if and only if .
- (I)
- For every representative GRN g in and every phenotype-preserving automorphism of g, there is an operation that joins together the groups of and , where and ;
- (II)
- For every representative GRN g in and every phenotype-preserving automorphism of each subgraph of g such that the edge differences are sequentially connected via , there is an operation that joins together the groups of and , where automorphism consecutively transforms edge into through ;
- (III)
- For every representative GRN in and each and in two different equivalence classes and , such that we have phenotype-preserving isomorphisms / from / to the representative GRN / after self-loop removal, there is an operation that joins together the groups of , and .
| Algorithm 1 Constructing the underlying space of a fitness landscape of GRNs |
| Require: The fixed underlying collections of loci and proteins of GRNs Ensure: The representative GRN of each equivalence class , and its number of mutational neighbors in any equivalence class
|
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Central and Peripheral GRNs Where No Regulation Presents
Appendix B. Phenotype-Preserving Automorphisms of the Genotype Network of GRNs
Appendix C. Combining Mutational Neighbors into Equivalence Classes
- (A)
- For an equivalence class and its representative GRN , under what condition will belong to the same equivalence class in layer ?
- (B)
- For two distinct equivalence classes and their representative GRNs and , under what condition will and belong to the same equivalence class in layer ?
- i.
- ;
- ii.
- ;
- iii.
- and for ;
- iv.
- for .
- i.
- and belong to the same equivalence class;
- ii.
- ;
- iii.
- .
Appendix D. Size of an Equivalence Class of GRNs
References
- Wright, S. The roles of mutation, inbreeding, crossbreeding, and selection in evolution. In Proceedings of the Sixth International Congress on Genetics, Ithaca, NY, USA, 24–31 August 1932; pp. 356–366. [Google Scholar]
- De Visser, J.A.G.M.; Krug, J. Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 2014, 15, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Fragata, I.; Blanckaert, A.; Louro, M.A.D.; Liberles, D.A.; Bank, C. Evolution in the light of fitness landscape theory. Trends Ecol. Evol. 2019, 34, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Szendro, I.G.; Schenk, M.F.; Franke, J.; Krug, J.; De Visser, J.A.G. Quantitative analyses of empirical fitness landscapes. J. Stat. Mech. Theory Exp. 2013, 2013, P01005. [Google Scholar] [CrossRef]
- Wagner, A. The Origins of Evolutionary Innovations: A Theory of Transformative Change in Living Systems; Oxford University Press: Oxford, UK, 2011. [Google Scholar]
- Jain, K.; Krug, J. Deterministic and stochastic regimes of asexual evolution on rugged fitness landscapes. Genetics 2007, 175, 1275–1288. [Google Scholar] [CrossRef] [PubMed]
- Kryazhimskiy, S.; Tkačik, G.; Plotkin, J.B. The dynamics of adaptation on correlated fitness landscapes. Proc. Natl. Acad. Sci. USA 2009, 106, 18638–18643. [Google Scholar] [CrossRef]
- Draghi, J.A.; Parsons, T.L.; Wagner, G.P.; Plotkin, J.B. Mutational robustness can facilitate adaptation. Nature 2010, 463, 353–355. [Google Scholar] [CrossRef]
- Wu, N.C.; Dai, L.; Olson, C.A.; Lloyd-Smith, J.O.; Sun, R. Adaptation in protein fitness landscapes is facilitated by indirect paths. eLife 2016, 5, e16965. [Google Scholar] [CrossRef]
- Gavrilets, S. Fitness Landscapes and the Origin of Species; Princeton University Press: Princeton, NY, USA, 2004. [Google Scholar]
- Fraïsse, C.; Gunnarsson, P.A.; Roze, D.; Bierne, N.; Welch, J.J. The genetics of speciation: Insights from fisher’s geometric model. Evolution 2016, 70, 1450–1464. [Google Scholar] [CrossRef]
- de Visser, J.A.G.; Park, S.C.; Krug, J. Exploring the effect of sex on empirical fitness landscapes. Am. Nat. 2009, 174, S15–S30. [Google Scholar] [CrossRef]
- Otto, S.P. The evolutionary enigma of sex. Am. Nat. 2009, 174, S1–S14. [Google Scholar] [CrossRef]
- Watson, R.A.; Weinreich, D.M.; Wakeley, J. Genome structure and the benefit of sex. Evol. Int. J. Org. Evol. 2011, 65, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Orr, H.A. The genetic theory of adaptation: A brief history. Nat. Rev. Genet. 2005, 6, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Lobkovsky, A.E.; Wolf, Y.I.; Koonin, E.V. Predictability of evolutionary trajectories in fitness landscapes. PLoS Comput. Biol. 2011, 7, e1002302. [Google Scholar] [CrossRef] [PubMed]
- Salverda, M.L.; Dellus, E.; Gorter, F.A.; Debets, A.J.; Van Der Oost, J.; Hoekstra, R.F.; Taw, D.S.; de Visser, J.A.G. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 2011, 7, e1001321. [Google Scholar] [CrossRef] [PubMed]
- Bank, C.; Matuszewski, S.; Hietpas, R.T.; Jensen, J.D. On the (un) predictability of a large intragenic fitness landscape. Proc. Natl. Acad. Sci. USA 2016, 113, 14085–14090. [Google Scholar] [CrossRef]
- Van Nimwegen, E.; Crutchfield, J.P.; Huynen, M. Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. USA 1999, 96, 9716–9720. [Google Scholar] [CrossRef]
- Van Nimwegen, E.; Crutchfield, J.P. Metastable evolutionary dynamics: Crossing fitness barriers or escaping via neutral paths? Bull. Math. Biol. 2000, 62, 799–848. [Google Scholar] [CrossRef][Green Version]
- Wilke, C.O. Adaptive evolution on neutral networks. Bull. Math. Biol. 2001, 63, 715–730. [Google Scholar] [CrossRef]
- Smith, T.; Husbands, P.; O’Shea, M. Neutral networks and evolvability with complex genotype-phenotype mapping. In European Conference on Artificial Life; Springer: Berlin/Heidelberg, Germany, 2001; pp. 272–281. [Google Scholar]
- Aguirre, J.; Catalán, P.; Cuesta, J.A.; Manrubia, S. On the networked architecture of genotype spaces and its critical effects on molecular evolution. Open Biol. 2018, 8, 180069. [Google Scholar] [CrossRef]
- Manrubia, S.; Cuesta, J.A.; Aguirre, J.; Ahnert, S.E.; Altenberg, L.; Cano, A.V.; Catalán, P.; Diaz-Uriarte, R.; Elena, S.F.; García-Martín, J.A.; et al. From genotypes to organisms: State-of-the-art and perspectives of a cornerstone in evolutionary dynamics. Phys. Life Rev. 2021, 38, 55–106. [Google Scholar] [CrossRef]
- Tenaillon, O. The utility of fisher’s geometric model in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 179–201. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The Genetical Theory of Natural Selection; Oxford University Press: Oxford, UK, 1930. [Google Scholar]
- Kingman, J.F.C. A simple model for the balance between selection and mutation. J. Appl. Probab. 1978, 15, 1–12. [Google Scholar] [CrossRef]
- Kauffman, S.; Levin, S. Towards a general theory of adaptive walks on rugged landscapes. J. Theor. Biol. 1987, 128, 11–45. [Google Scholar] [CrossRef]
- Kauffman, S.A.; Weinberger, E.D. The nk model of rugged fitness landscapes and its application to maturation of the immune response. J. Theor. Biol. 1989, 141, 211–245. [Google Scholar] [CrossRef]
- Aita, T.; Uchiyama, H.; Inaoka, T.; Nakajima, M.; Kokubo, T.; Husimi, Y. Analysis of a local fitness landscape with a model of the rough mt. fuji-type landscape: Application to prolyl endopeptidase and thermolysin. Biopolym. Orig. Res. Biomol. 2000, 54, 64–79. [Google Scholar] [CrossRef]
- Neidhart, J.; Szendro, I.G.; Krug, J. Adaptation in tunably rugged fitness landscapes: The rough mount fuji model. Genetics 2014, 198, 699–721. [Google Scholar] [CrossRef]
- Weinreich, D.M.; Delaney, N.F.; DePristo, M.A.; Hartl, D.L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 2006, 312, 111–114. [Google Scholar] [CrossRef]
- Chou, H.-H.; Chiu, H.-C.; NDelaney, N.F.; Segrè, D.; Marx, C.J. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 2011, 332, 1190–1192. [Google Scholar] [CrossRef]
- Khan, A.I.; Dinh, D.M.; Schneider, D.; Lenski, R.E.; Cooper, T.F. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 2011, 332, 1193–1196. [Google Scholar] [CrossRef]
- De Visser, J.A.G.M.; Hoekstra, R.F.; van den Ende, H. Test of interaction between genetic markers that affect fitness in aspergillus niger. Evolution 1997, 51, 1499–1505. [Google Scholar] [CrossRef]
- Hall, D.W.; Agan, M.; Pope, S.C. Fitness epistasis among 6 biosynthetic loci in the budding yeast saccharomyces cerevisiae. J. Hered. 2010, 101 (Suppl. S1), S75–S84. [Google Scholar] [CrossRef] [PubMed]
- Whitlock, M.C.; Bourguet, D. Factors affecting the genetic load in drosophila: Synergistic epistasis and correlations among fitness components. Evolution 2000, 54, 1654–1660. [Google Scholar] [CrossRef] [PubMed]
- Hinkley, T.; Martins, J.; Chappey, C.; Haddad, M.; Stawiski, E.; Whitcomb, J.M.; Petropoulos, C.J.; Bonhoeffer, S. A systems analysis of mutational effects in hiv-1 protease and reverse transcriptase. Nat. Genet. 2011, 43, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Kouyos, R.D.; Leventhal, G.E.; Hinkley, T.; Haddad, M.; Whitcomb, J.M.; Petropoulos, C.J.; Bonhoeffer, S. Exploring the complexity of the hiv-1 fitness landscape. PLoS Genet. 2012, 8, e1002551. [Google Scholar] [CrossRef] [PubMed]
- Hietpas, R.T.; Jensen, J.D.; Bolon, D.N.A. Experimental illumination of a fitness landscape. Proc. Natl. Acad. Sci. USA 2011, 108, 7896–7901. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, J.; Nemenman, I. Genotype to phenotype mapping and the fitness landscape of the E. coli lac promoter. PLoS ONE 2013, 8, e61570. [Google Scholar] [CrossRef]
- Sarkisyan, K.S.; Bolotin, D.A.; Meer, M.V.; Usmanova, D.R.; Mishin, A.S.; Sharonov, G.V.; Ivankov, D.N.; Bozhanova, N.G.; Baranov, M.S.; Soylemez, O.; et al. Local fitness landscape of the green fluorescent protein. Nature 2016, 533, 397–401. [Google Scholar] [CrossRef]
- Rogers, Z.N.; McFarland, C.D.; Winters, I.P.; Seoane, J.A.; Brady, J.J.; Yoon, S.; Curtis, C.; Petrov, D.A.; Winslow, M.M. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat. Genet. 2018, 50, 483–486. [Google Scholar] [CrossRef]
- Watson, C.J.; Papula, A.L.; Poon, G.Y.P.; Wong, W.H.; Young, A.L.; Druley, T.E.; Fisher, D.S.; Blundell, J.R. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 2020, 367, 1449–1454. [Google Scholar] [CrossRef]
- Rowe, W.; Platt, M.; Wedge, D.C.; Day, P.J.; Kell, D.B.; Knowles, J. Analysis of a complete DNA–protein affinity landscape. J. R. Soc. Interface 2010, 7, 397–408. [Google Scholar] [CrossRef]
- Pitt, J.N.; Ferré-D’Amaré, A.R. Rapid construction of empirical rna fitness landscapes. Science 2010, 330, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, J.I.; Xulvi-Brunet, R.; Campbell, G.W.; Turk-MacLeod, R.; Chen, I.A. Comprehensive experimental fitness landscape and evolutionary network for small rna. Proc. Natl. Acad. Sci. USA 2013, 110, 14984–14989. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qian, W.; Maclean, C.J.; Zhang, J. The fitness landscape of a trna gene. Science 2016, 352, 837–840. [Google Scholar] [CrossRef] [PubMed]
- Bendixsen, D.P.; Østman, B.; Hayden, E.J. Negative epistasis in experimental rna fitness landscapes. J. Mol. Evol. 2017, 85, 159–168. [Google Scholar] [CrossRef]
- Aguilar-Rodríguez, J.; Payne, J.L.; Wagner, A. A thousand empirical adaptive landscapes and their navigability. Nat. Ecol. Evol. 2017, 1, 0045. [Google Scholar] [CrossRef]
- Martin, C.H. Context dependence in complex adaptive landscapes: Frequency and trait-dependent selection surfaces within an adaptive radiation of caribbean pupfishes. Evolution 2016, 70, 1265–1282. [Google Scholar] [CrossRef]
- Boucher, J.I.; Cote, P.; Flynn, J.; Jiang, L.; Laban, A.; Mishra, P.; Roscoe, B.P.; Bolon, D.N.A. Viewing protein fitness landscapes through a next-gen lens. Genetics 2014, 198, 461–471. [Google Scholar] [CrossRef]
- Aita, T.; Iwakura, M.; Husimi, Y. A cross-section of the fitness landscape of dihydrofolate reductase. Protein Eng. 2001, 14, 633–638. [Google Scholar] [CrossRef]
- Carneiro, M.; Hartl, D.L. Adaptive landscapes and protein evolution. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. S1), 1747–1751. [Google Scholar] [CrossRef]
- Poelwijk, F.J.; Tănase-Nicola, S.; Kiviet, D.J.; Tans, S.J. Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. J. Theor. Biol. 2011, 272, 141–144. [Google Scholar] [CrossRef]
- Lozovsky, E.R.; Chookajorn, T.; Brown, K.M.; Imwong, M.; Shaw, P.J.; Kamchonwongpaisan, S.; Neafsey, D.E.; Weinreich, D.M.; Hartl, D.L. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc. Natl. Acad. Sci. USA 2009, 106, 12025–12030. [Google Scholar] [CrossRef] [PubMed]
- Lunzer, M.; Miller, S.P.; Felsheim, R.; Dean, A.M. The biochemical architecture of an ancient adaptive landscape. Science 2005, 310, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Bridgham, J.T.; Carroll, S.M.; Thornton, J.W. Evolution of hormone-receptor complexity by molecular exploitation. Science 2006, 312, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Poelwijk, F.J.; Kiviet, D.J.; Tans, S.J. Evolutionary potential of a duplicated repressor-operator pair: Simulating pathways using mutation data. PLoS Comput. Biol. 2006, 2, e58. [Google Scholar] [CrossRef] [PubMed]
- Poelwijk, F.J.; Kiviet, D.J.; Weinreich, D.M.; Tans, S.J. Empirical fitness landscapes reveal accessible evolutionary paths. Nature 2007, 445, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Franke, J.; Klözer, A.; de Visser, J.A.G.; Krug, J. Evolutionary accessibility of mutational pathways. PLoS Comput. Biol. 2011, 7, e1002134. [Google Scholar] [CrossRef]
- Hegarty, P.; Martinsson, A. On the existence of accessible paths in various models of fitness landscapes. Ann. Appl. Probab. 2014, 24, 1375–1395. [Google Scholar] [CrossRef]
- Zagorski, M.; Burda, Z.; Waclaw, B. Beyond the hypercube: Evolutionary accessibility of fitness landscapes with realistic mutational networks. PLoS Comput. Biol. 2016, 12, e1005218. [Google Scholar] [CrossRef]
- Kell, D.B. Genotype–phenotype mapping: Genes as computer programs. Trends Genet. 2002, 18, 555–559. [Google Scholar] [CrossRef]
- Kondrashov, D.A.; Kondrashov, F.A. Topological features of rugged fitness landscapes in sequence space. Trends Genet. 2015, 31, 24–33. [Google Scholar] [CrossRef]
- Chan, H.S.; Bornberg-Bauer, E. Perspectives on protein evolution from simple exact models. Appl. Bioinform. 2002, 50, 121–144. [Google Scholar]
- Cowperthwaite, M.C.; Economo, E.P.; Harcombe, W.R.; Miller, E.L.; Meyers, L.A. The ascent of the abundant: How mutational networks constrain evolution. PLoS Comput. Biol. 2008, 4, e1000110. [Google Scholar] [CrossRef] [PubMed]
- Stich, M.; Lázaro, E.; Manrubia, S.C. Phenotypic effect of mutations in evolving populations of rna molecules. BMC Evol. Biol. 2010, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.E.; Moudgil, A.; Feldman, M.W. Long-term evolution is surprisingly predictable in lattice proteins. J. R. Soc. Interface 2013, 10, 20130026. [Google Scholar] [CrossRef]
- Bershtein, S.; Serohijos, A.W.R.; Shakhnovich, E.I. Bridging the physical scales in evolutionary biology: From protein sequence space to fitness of organisms and populations. Curr. Opin. Struct. Biol. 2017, 42, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Perfeito, L.; Ghozzi, S.; Berg, J.; Schnetz, K.; Lässig, M. Nonlinear fitness landscape of a molecular pathway. PLoS Genet. 2011, 7, e1002160. [Google Scholar] [CrossRef]
- Chou, H.-H.; Delaney, N.F.; Draghi, J.A.; Marx, C.J. Mapping the fitness landscape of gene expression uncovers the cause of antagonism and sign epistasis between adaptive mutations. PLoS Genet. 2014, 10, e1004149. [Google Scholar] [CrossRef]
- Friedlander, T.; Prizak, R.; Barton, N.H.; Tkačik, G. Evolution of new regulatory functions on biophysically realistic fitness landscapes. Nat. Commun. 2017, 8, 216. [Google Scholar] [CrossRef]
- Cuypers, T.D.; Rutten, J.P.; Hogeweg, P. Evolution of evolvability and phenotypic plasticity in virtual cells. BMC Evol. Biol. 2017, 17, 60. [Google Scholar] [CrossRef]
- Yubero, P.; Manrubia, S.; Aguirre, J. The space of genotypes is a network of networks: Implications for evolutionary and extinction dynamics. Sci. Rep. 2017, 7, 13813. [Google Scholar] [CrossRef]
- Harmand, N.; Gallet, R.; Jabbour-Zahab, R.; Martin, G.; Lenormand, T. Fisher’s geometrical model and the mutational patterns of antibiotic resistance across dose gradients. Evolution 2017, 71, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-H.; Scarpino, S.V. Reproductive barriers as a byproduct of gene network evolution. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yang, C.-H.; Scarpino, S.V. The ensemble of gene regulatory networks at mutation-selection balance. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ciliberti, S.; Martin, O.C.; Wagner, A. Innovation and robustness in complex regulatory gene networks. Proc. Natl. Acad. Sci. USA 2007, 104, 13591–13596. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.L.; Wagner, A. Latent phenotypes pervade gene regulatory circuits. BMC Syst. Biol. 2014, 8, 64. [Google Scholar] [CrossRef]
- Godsil, C.D. Compact graphs and equitable partitions. Linear Algebra Its Appl. 1997, 255, 259–266. [Google Scholar] [CrossRef]
- Das, S.G.; Direito, S.O.L.; Waclaw, B.; Allen, R.J.; Krug, J. Predictable properties of fitness landscapes induced by adaptational tradeoffs. eLife 2020, 9, e55155. [Google Scholar] [CrossRef]
- Wagner, A. Neutralism and selectionism: A network-based reconciliation. Nat. Rev. Genet. 2008, 9, 965–974. [Google Scholar] [CrossRef]
- Bendixsen, D.P.; Collet, J.; Østman, B.; Hayden, E.J. Genotype network intersections promote evolutionary innovation. PLoS Biol. 2019, 17, e3000300. [Google Scholar] [CrossRef]
- Hunt, G.; Hopkins, M.J.; Lidgard, S. Simple versus complex models of trait evolution and stasis as a response to environmental change. Proc. Natl. Acad. Sci. USA 2015, 112, 4885–4890. [Google Scholar] [CrossRef]
- Heasley, L.R.; Sampaio, N.M.V.; Argueso, J.L. Systemic and rapid restructuring of the genome: A new perspective on punctuated equilibrium. Curr. Genet. 2020, 67, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Rodríguez, J.; Peel, L.; Stella, M.; Wagner, A.; Payne, J.L. The architecture of an empirical genotype-phenotype map. Evolution 2018, 72, 1242–1260. [Google Scholar] [CrossRef] [PubMed]
- López-Presa, J.L.; Chiroque, L.F.; Anta, A.F. Novel techniques to speed up the computation of the automorphism group of a graph. J. Appl. Math. 2014, 2014, 934637. [Google Scholar] [CrossRef]
- Stoichev, S.D. New exact and heuristic algorithms for graph automorphism group and graph isomorphism. J. Exp. Algorithmics (JEA) 2019, 24, 1–27. [Google Scholar] [CrossRef]
- Battiston, F.; Cencetti, G.; Iacopini, I.; Latora, V.; Lucas, M.; Patania, A.; Young, J.-G.; Petri, G. Networks beyond pairwise interactions: Structure and dynamics. Phys. Rep. 2020, 874, 1–92. [Google Scholar] [CrossRef]
- Maheshwari, P.; Albert, R. A framework to find the logic backbone of a biological network. BMC Syst. Biol. 2017, 11, 122. [Google Scholar] [CrossRef]
- Yang, G.; Gómez Tejeda Zañudo, J.; Albert, R. Target control in logical models using the domain of influence of nodes. Front. Physiol. 2018, 9, 454. [Google Scholar] [CrossRef]
- Hu, T.; Tomassini, M.; Banzhaf, W. A network perspective on genotype–phenotype mapping in genetic programming. Genet. Program. Evolvable Mach. 2020, 21, 375–397. [Google Scholar] [CrossRef]
- Greenbury, S.F.; Louis, A.A.; Ahnert, S.E. The structure of genotype-phenotype maps makes fitness landscapes navigable. bioRxiv 2021. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-H.; Scarpino, S.V. A Family of Fitness Landscapes Modeled through Gene Regulatory Networks. Entropy 2022, 24, 622. https://doi.org/10.3390/e24050622
Yang C-H, Scarpino SV. A Family of Fitness Landscapes Modeled through Gene Regulatory Networks. Entropy. 2022; 24(5):622. https://doi.org/10.3390/e24050622
Chicago/Turabian StyleYang, Chia-Hung, and Samuel V. Scarpino. 2022. "A Family of Fitness Landscapes Modeled through Gene Regulatory Networks" Entropy 24, no. 5: 622. https://doi.org/10.3390/e24050622
APA StyleYang, C.-H., & Scarpino, S. V. (2022). A Family of Fitness Landscapes Modeled through Gene Regulatory Networks. Entropy, 24(5), 622. https://doi.org/10.3390/e24050622





