T-cell Receptor Is a Threshold Detector: Sub- and Supra-Threshold Stochastic Resonance in TCR-MHC Clusters on the Cell Surface
Abstract
1. Introduction
2. Properties of the TCR-MHC System Analyzed from the Viewpoint of SR
2.1. TCR Behaves as a Threshold Detector
2.1.1. Structure and Signaling of TCR
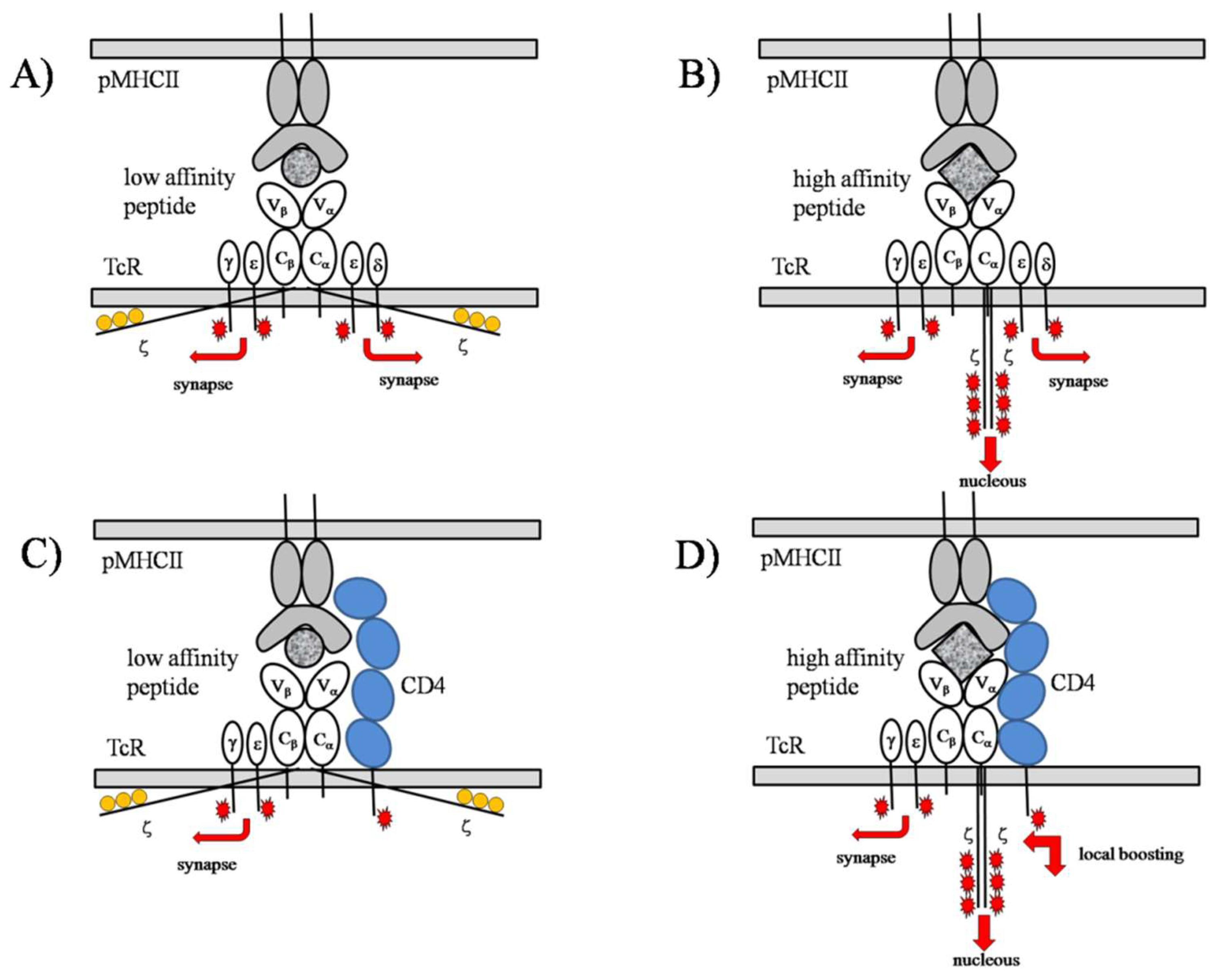
2.1.2. The Noncognate–Cognate Dichotomy Is Mirrored by the TCR Structure
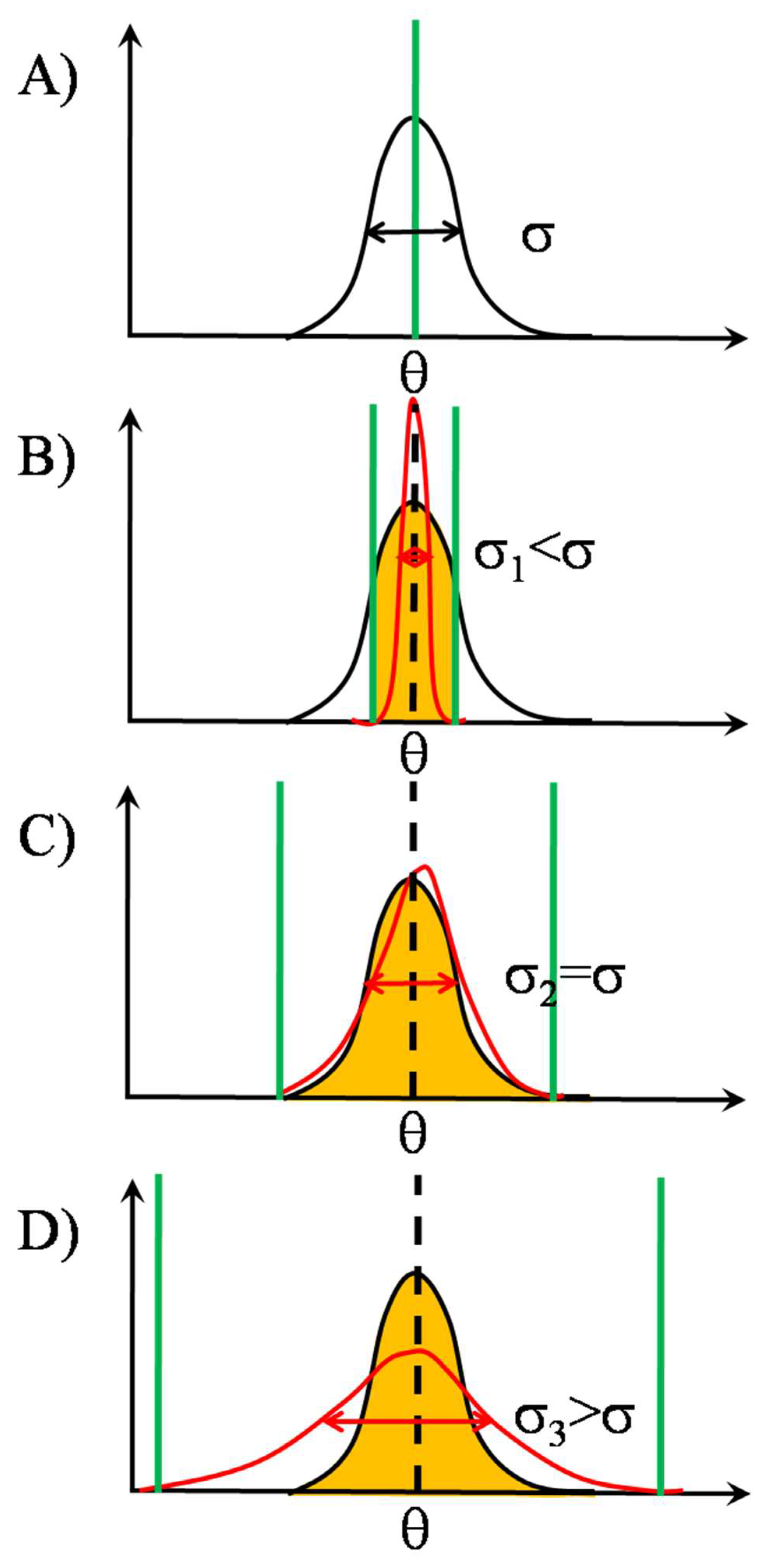
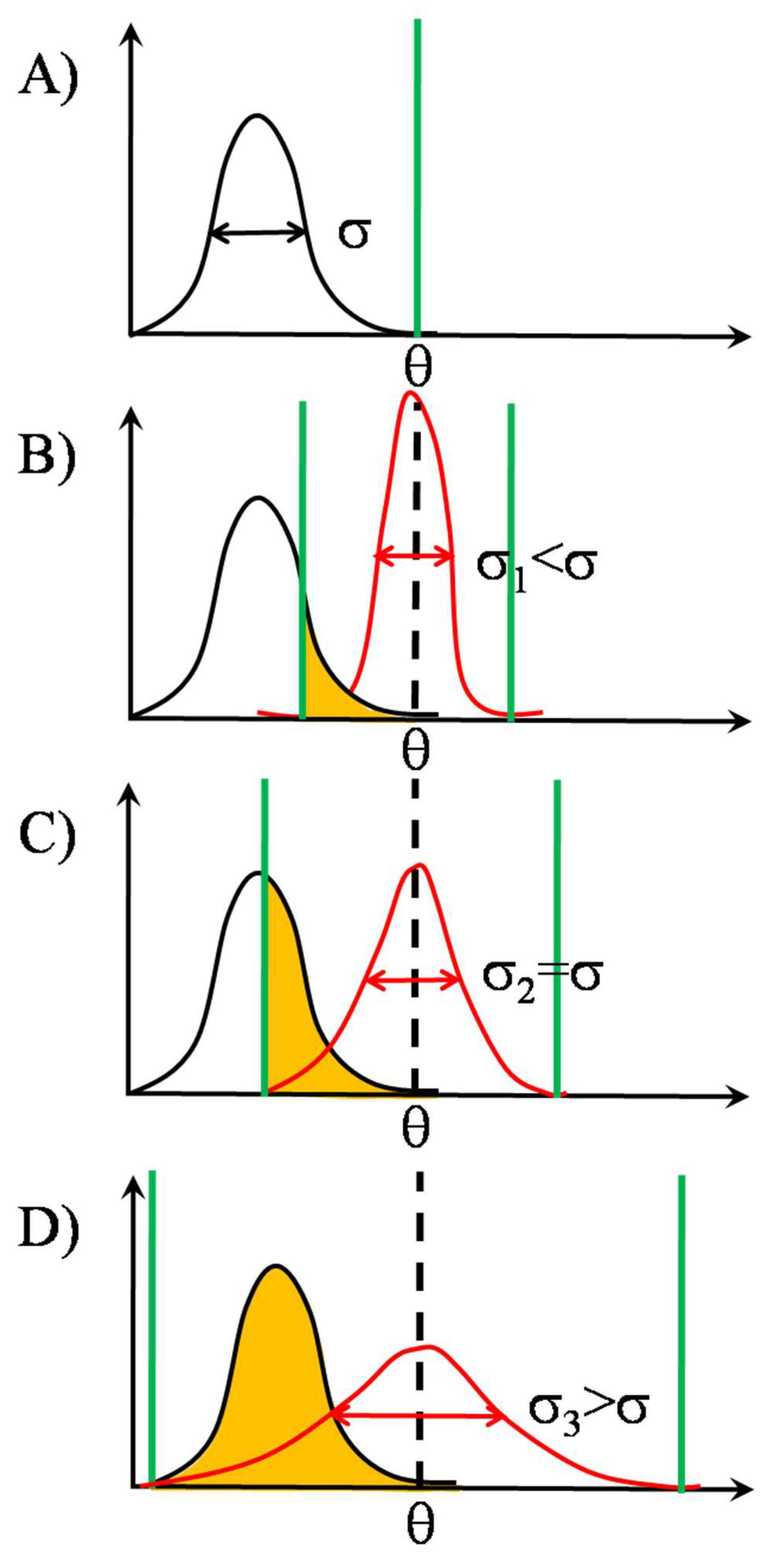
2.1.3. TCR Triggering as a Biphasic Process: Adaptive Thresholding
2.2. SSR in TCR Clusters
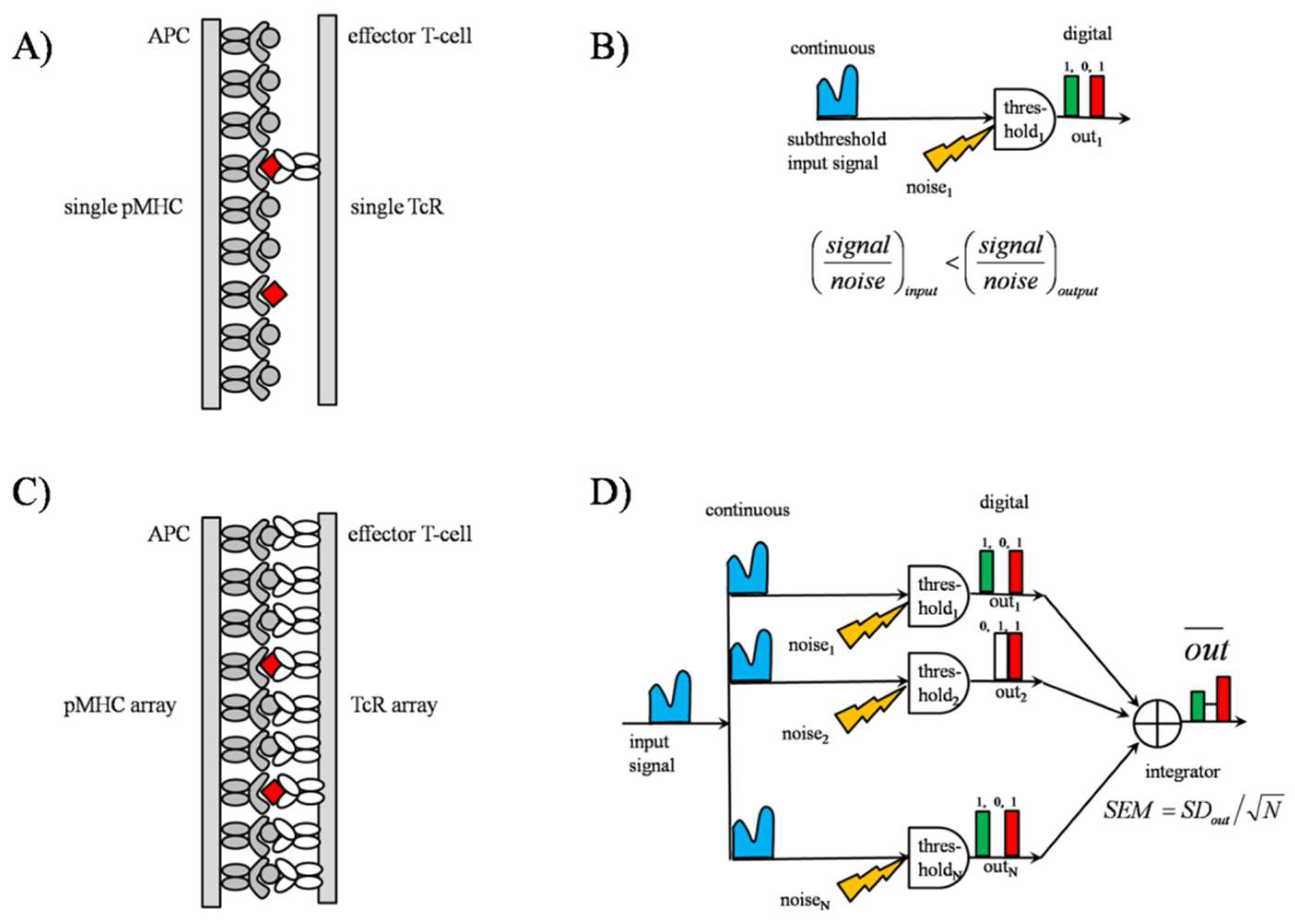
2.2.1. TCR Array as an “Equivalent Pooling Network”
2.2.2. Signal Transmission by SSR
2.2.3. Noise’s Nature
2.3. Qualifying CARs with SR
3. Discussion
3.1. Two Levels of SR in Immune Recognition
3.2. SR in TCR and MHC Arrays: SSR
3.3. Exploiting SSR to Optimize Molecular Prostheses
3.4. Förster (Fluorescence) Resonance Energy Transfer (FRET) as an Information Channel
3.5. SSR as an Analogue-to-Digital Signal Converter (ADC)
3.6. Future Directions and Limitations of the SR Model
3.7. Metastases Finding as a Problem of Weak “Fault Signal” Detection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AFM | atomic force microscopy, |
| APC | antigen presenting cell, e.g., dendritic cell, |
| CAR | genetically engineered chimeric antigen receptor, composed of the variable and constant regions of the antigen recognising Fab portion of antibody connected by a linker region, as well as a spacer and transmembrane region, and an intracytoplasmic domain containing 3–5 (signaling) sites, which can be phosphorylated, depending on the generation of CAR (1th–4th), |
| CD28 | coreceptor (signal amplifier) on the side of TcR on a T cell in apposition to an APC, its counter receptor is CD80, |
| CD45/CD148/CD137 | phophatases, dephosphorilate sites, which can be phophorylated, act against kinases, |
| CD8/CD4 | coreceptors (signal amplifiers) CD8 and CD4 on T cells—killer and helper, respectively–, binding to MHCI and MHCII, respectively on an APC in apposition to a T cell, |
| cSMAC/pSMAC/dSMAC | central, peripheral, and distal supramolecular activation clusters, concentric zones—centripetally from inside to outside—constituting the immune synapse (IS) on T cells, rich in non-signaling TcRs |
| (cSMAC) | microclusters of signaling TcR and LFA-1 (pSMAC), and rich in CD45 (dSMAC), |
| FRET | Förster (or fluorescence) resonance energy transfer, occuring between an energy donor and an energy acceptor when their distance is between 1 nm and 10 nm, |
| ICAM-1/ICAM-2 | intracellular adhesion molecules type 1 and type 2, on the surface of an APC, binding to LFA-1/LFA-3 on the recognising T cell, |
| IS | immune synapse, consisting of concentric zones called cSMAC, pSMAC, and dSMAC, |
| ITAM | tyrosine based activation motif, which can be phosphorylated, or docking site for tyrosine kinases Lck, Src, |
| Lck | Src tyrosine kinase p65lck, |
| LFA-1/LFA-3 | leukocyte function associated antigen type 1 and type 3 on the side of TCR on the recognising T cell, its counter receptor on the APC is ICAM-1/ICAM-2, capable also for signaling, |
| MHCI/MHCII | Major Hystocompatibility Gene Complex products I and II, presenting antigen of the surface of an APC towards effector cells, killer and helper T cells, respectively, |
| pMHC | MHC molecule presenting short—8–9 amino acids long—peptide fragment of antigen on the surface of APC towards the recognising receptor TCR on the surface of an effector (killer, helper, memory) T-cell, |
| SR | stochastic resonance, |
| SSR | supra-threshold stochastic resonance, |
| TcR | T-cell receptor—composed of a heterodimer of TCRα and TCRβ (briefly αβ) recognising the antigen, a long signaling domain TCRζζ (briefly ζζ) containing 6 phosphorilatable sites (ITAMs) and CD3γε (briefly γε), CD3δε (briefly δε) signaling domains each containing 2 sites, which can be phosphorilated—binding to peptide-loaded MHCs on the surface of an APC, |
| TEM | transmission electron microscopy, |
| TM4SF | transmembrane-4 family protein, accessorical, adaptor protein spanning the membrane four times. |
References
- Bacsó, Z.; Bene, L.; Damjanovich, L.; Damjanovich, S. IFN-γ rearranges membrane topography of MHC-I and ICAM-1 in colon carcinoma cells. Biochem. Biophys. Res. Commun. 2002, 290, 635–640. [Google Scholar] [CrossRef]
- Bene, L.; Bodnár, A.; Damjanovich, S.; Vámosi, G.; Bacsó, Z.; Aradi, J.; Berta, A.; Damjanovich, J. Membrane topography of HLA I, HLA II, and ICAM-1 is affected by IFN-γin lipid rafts of uveal melanomas. Biochem. Biophys. Res. Commun. 2004, 322, 678–683. [Google Scholar] [CrossRef]
- Bene, L.; Kanyári, Z.; Bodnár, A.; Kappelmayer, J.; Waldmann, T.A.; Vámosi, G.; Damjanovich, L. Colorectal carcinoma rearranges cell surface protein topology and density in CD4+ T cells. Biochem. Biophys. Res. Commun. 2007, 361, 202–207. [Google Scholar] [CrossRef]
- Damjanovich, L.; Volkó, J.; Forgács, A.; Hohenberger, W.; Bene, L. Crohn’s disease alters MHC-rafts in CD4+ T-cells. Cytom. Part A 2012, 81, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Damjanovich, S.; Bene, L.; Matkó, J.; Mátyus, L.; Krasznai, Z.; Szabó, G., Jr.; Pieri, C.; Gáspár, R., Jr.; Szöllősi, J. Two-dimensional receptor patterns in the plasma membrane of cells. A critical evaluation of their identification, origin and information content. Biophys. Chem. 1999, 82, 99–108. [Google Scholar] [CrossRef]
- Ojcius, D.M.; Delarbre, C.; Kourilsky, P.; Gachelin, G. MHC and MHC-related proteins as pleiotropic signal molecules. FASEB J. 2001, 16, 202–206. [Google Scholar] [CrossRef][Green Version]
- Fishman, D.; Elhyany, S.; Segal, S. Non-immune functions of MHC class I glycoproteins in normal and malignant cells. Folia Biol. 2004, 50, 35–42. [Google Scholar]
- Skov, S.; Nielsen, M.; Bregenholt, S.; Ødum, N.; Claesson, M.H. Activation of Stat-3 is involved in the induction of apoptosis after ligation of Major Histocompatibility Complex class I molecules on human Jurkat T cells. Blood 1998, 91, 3566–3573. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, A.E.; Skov, S.; Bregenholt, S.; Ruhwald, M.; Claesson, M.H. Signal transduction by the major histocompatibility complex class I molecule. APMIS 1999, 107, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Gáspár, R., Jr.; Bagossi, P.; Bene, L.; Matkó, J.; Szöllősi, J.; Tőzsér, J.; Fésüs, L.; Waldmann, T.A.; Damjanovich, S. Clustering of class I HLA oligomers with CD8 and TCR: Three-dimensional models based on fluorescence resonance energy transfer and crystallographic data. J. Immunol. 2001, 166, 5078–5086. [Google Scholar] [CrossRef]
- Garcia, E.; Ismail, S. Spatiotemporal regulation of signaling: Focus on T cell activation and the immunological synapse. Int. J. Mol. Sci. 2020, 21, 3283. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, H.; Ladell, K.; Miners, K.; Laugel, B.; Llewellyn-Lacey, S.; Clement, M.; Cole, D.K.; Gostick, E.; Wooldridge, L.; Sewell, A.K.; et al. Cellular-level versus receptor-level response threshold hierarchies in T-cell activation. Front. Immunol. 2013, 4, 250. [Google Scholar] [CrossRef] [PubMed]
- Benzi, R.; Sutera, A.; Vulpiani, A. The mechanism of stochastic resonance. J. Phys. A Math. Gen. 1981, 14, L453–L457. [Google Scholar] [CrossRef]
- Nicolis, C. Stochastic aspects of climatic transitions—Response to a periodic forcing. Tellus 1982, 34, 1–9. [Google Scholar] [CrossRef]
- Gammaitoni, L.; Hänggi, P.; Jung, P.; Marchesoni, F. Stochastic resonance. Rev. Mod. Phys. 1998, 70, 223–287. [Google Scholar] [CrossRef]
- Hänggi, P. Stochastic resonance in biology. ChemPhysChem 2002, 3, 285–290. [Google Scholar] [CrossRef]
- Gingl, Z.; Kiss, L.B.; Moss, F. Non-dynamical stochastic resonance: Theory and experiments with white and arbitrarily coloured noise. Europhys. Lett. 1995, 29, 191–196. [Google Scholar] [CrossRef]
- Moss, F. Stochastic resonance: Looking forward. Ch. 10. In Self-Organized Biological Dynamics and Nonlinear Control, 1st ed.; Wallaczek, J., Ed.; Cambridge University Press: New York, NY, USA, 2000; pp. 236–256. [Google Scholar]
- McDonnell, M.D.; Abbott, D. What is stochastic resonance? Definitions, misconceptions, debates, and its relevance to biology. PLoS Comp. Biol. 2009, 5, e1000348. [Google Scholar] [CrossRef]
- Bezrukov, S.M.; Vodyanoy, I. Stochastic resonance and small-amplitude signal transduction in voltage-gated ion channels. Ch. 11. In Self-Organized Biological Dynamics and Nonlinear Control, 1st ed.; Wallaczek, J., Ed.; Cambridge University Press: New York, NY, USA, 2000; pp. 257–280. [Google Scholar]
- Moss, F.; Pei, X. Neurons in parallel. Nature 1995, 376, 211–212. [Google Scholar] [CrossRef]
- Stacey, W.C.; Durand, D.M. Stochastic resonance improves signal detection in hippocampal CA1 neurons. J. Neurophysiol. 2000, 83, 1394–1402. [Google Scholar] [CrossRef]
- Lindner, J.F.; Meadows, B.K.; Ditto, W.L.; Inchiosa, M.E.; Bulsara, A.R. Array enhanced stochastic resonance and spatiotemporal synchronization. Phys. Rev. Lett. 1995, 75, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Simonotto, E.; Riani, M.; Seife, C.; Roberts, M.; Twitty, J.; Moss, F. Visual perception of stochastic resonance. Phys. Rev. Lett. 1997, 78, 1186–1189. [Google Scholar] [CrossRef]
- Ryu, C.; Kong, S.G.; Kim, H. Enhancement of feature extraction for low-quality fingerprint images using stochastic resonance. Pattern Recog. Lett. 2011, 32, 107–113. [Google Scholar] [CrossRef]
- Babinec, P. Stochastic resonance in the Weidlich model of public opinion formation. Phys. Lett. A 1997, 225, 179–181. [Google Scholar] [CrossRef]
- Citovsky, G.; Focardi, S. A novel view of suprathreshold stochastic resonance and its applications to financial markets. Front. Appl. Math. Stat. 2015, 1, 10. [Google Scholar] [CrossRef]
- Stocks, N.G. Suprathreshold stochastic resonance in multilevel threshold systems. Phys. Rev. Lett. 2000, 84, 2310–2313. [Google Scholar] [CrossRef]
- Stocks, N.G. Suprathreshold stochastic resonance: An exact result for uniformly distributed signal and noise. Phys. Lett. A 2001, 279, 308–312. [Google Scholar] [CrossRef]
- Stocks, N.G. Information transmission in parallel threshold arrays. Phys. Rev. E 2001, 63, 041114. [Google Scholar] [CrossRef]
- McDonnell, M.; Stocks, N.G.; Pearce, C.E.M.; Abbott, D. From Suprathreshold Stochastic Resonance to Stochastic Signal Quantization. In Stochastic Resonance; Cambridge University Press: New York, NY, USA, 2008; pp. 1–420. [Google Scholar]
- Guedan, S.; Calderon, H.; Posey, A.D., Jr.; Maus, M.V. Engineering and design of chimeric antigen receptors. Methods Clin. Dev. 2019, 12, 145–156. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Zhong, J.F.; Zhang, X. Engineering CAR-T cells. Biomark. Res. 2017, 5, 22. [Google Scholar] [CrossRef]
- Sebestyén, Z.; Schooten, E.; Sals, T.; Zaldivar, I.; San José, E.; Alarcón, B.; Bobisse, S.; Rosato, A.; Szöllősi, J.; Gratama, J.W.; et al. Human TCR that incorporate CD3ζ induce highly preferred pairing between TCRα and β chains following gene transfer. J. Immunol. 2008, 180, 7736–7746. [Google Scholar] [CrossRef] [PubMed]
- Roszik, J.; Sebestyén, Z.; Covers, C.; Guri, Y.; Szöőr, Á.; Pályi-Krekk, Z.; Vereb, G.; Nagy, P.; Szöllősi, J.; Debets, R. T-cell synapse formation depends on antigen recognition but not CD3 interaction: Studies with TCR:ζ, a candidate transgene for TCR gene therapy. Eur. J. Immunol. 2011, 41, 1288–1297. [Google Scholar] [CrossRef] [PubMed]
- Bridgeman, J.S.; Hawkins, R.E.; Bagley, S.; Blaylock, M.; Holland, M.; Gilham, D.E. The optimal antigen response of chimeric antigen receptors harboring the CD3ζ transmembrane domain is dependent upon incorporation of the receptor into the endogenous TCR/CD3 complex. J. Immunol. 2010, 184, 6938–6949. [Google Scholar] [CrossRef] [PubMed]
- Bridgeman, J.S.; Ladell, K.; Sheard, V.E.; Miners, K.; Hawkins, R.E.; Price, D.A.; Gilham, D.E. CD3ζ-based chimeric antigen receptors mediate T cell activation via cis- and trans-signalling mechanisms: Implications for optimization of receptor structure for adoptive cell therapy. Clin. Exp. Immunol. 2013, 175, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.-G.; Fu, Q.-J.; Morse, R. Human hearing enhanced by noise. Brain Res. 2000, 869, 251–255. [Google Scholar] [CrossRef]
- Dustin, M.L. Signaling at neuro/immune synapses. J. Clin. Investig. 2012, 122, 1149–1155. [Google Scholar] [CrossRef]
- Rodrigo, G.; Stocks, N.G. Suprathreshold stochastic resonance behind cancer. Trends Biochem. Sci. 2018, 43, 483–485. [Google Scholar] [CrossRef]
- Itzcovich, E.; Massimo, R.; Sannita, W.G. Stochastic resonance improves vision in the severely impaired. Sci. Rep. 2017, 7, 12840. [Google Scholar] [CrossRef]
- Paffi, A.; Camera, F.; Apollonio, F.; d’Inzeo, G.; Liberti, M. Restoring the encoding properties of a stochastic neuron model by an exogeneous noise. Front. Comput. Neurosci. 2015, 9, 42. [Google Scholar] [CrossRef][Green Version]
- Wülfing, C.; Sumen, C.; Sjaastad, M.D.; Wu, L.C.; Dustin, M.L.; Davis, M.M. Costimulation and endogeneous MHC ligands contribute to T cell recognition. Nat. Immunol. 2002, 3, 42–47. [Google Scholar] [CrossRef]
- Bakker, T.; van der Merwe, P.A. Self-help in T cell recognition? Nat. Immunol. 2002, 3, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Štefanová, I.; Dorfman, J.R.; Germain, R.N. Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature 2002, 420, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Sankaran, S.; Yap, J.; Too, C.T.; Ho, Z.Z.; Dolton, G.; Legut, M.; Ren, E.C.; Sewell, A.K.; Bertoletti, A.; et al. Nonstimulatory peptide-MHC enhances human T-cell antigen-specific responses by amplifying proximal TCR signaling. Nat. Commun. 2018, 9, 2716. [Google Scholar] [CrossRef] [PubMed]
- Krogsgaard, M.; Li, Q.-J.; Sumen, C.; Huppa, J.B.; Huse, M.; Davis, M.M. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature 2005, 434, 238–243. [Google Scholar] [CrossRef]
- Anikeeva, N.; Gakamsky, D.; Scholler, J.; Sykulev, Y. Evidence that the density of self peptide-MHC ligands regulates T-cell receptor signaling. PLoS ONE 2012, 7, e41466. [Google Scholar] [CrossRef]
- Altan-Bonnet, G.; Germain, R.N. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005, 3, e356. [Google Scholar] [CrossRef]
- François, P.; Altan-Bonnet, G. The case for absolute ligand discrimination: Modeling information processing and decision by immune T cells. J. Stat. Phys. 2016, 162, 1130–1152. [Google Scholar] [CrossRef]
- Bene, L.; Bagdány, M.; Damjanovich, L. Adaptive threshold-stochastic resonance (AT-SR) in MHC clusters on the cell surface. Immun. Lett. 2020, 217, 65–71. [Google Scholar] [CrossRef]
- Courtney, A.H.; Lo, W.-L.; Weiss, A. TCR signaling: Mechanisms of initiation and propagation. Trends. Biochem. Sci. 2019, 43, 2. [Google Scholar] [CrossRef]
- Chang, V.T.; Fernandes, R.A.; Ganzinger, K.A.; Lee, S.F.; Siebold, C.; McColl, J.; Jöhnsson, P.; Palayret, M.; Harlos, K.; Coles, C.H.; et al. Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol. 2016, 17, 574–582. [Google Scholar] [CrossRef]
- Zhang, M.; Moran, M.; Round, J.; Low, T.A.; Patel, V.P.; Tomassian, T.; Hernandez, J.D.; Miceli, M.C. CD45 signals outside of lipid rafts to promote ERK activation, synaptic raft clustering, and IL-2 production. J. Immunol. 2005, 174, 1479–1490. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.; Nadarajah, V.; Felsovalyi, K.; Wang, W.; Jeyachandran, V.R.; Wasson, R.A.; Cardozo, T.; Bracken, C.; Krogsgaard, M. Structural model of the extracellular assambley of the TCR-CD3 complex. Cell Rep. 2016, 14, 2833–2845. [Google Scholar] [CrossRef] [PubMed]
- Pielak, R.M.; O’Donoghue, G.P.; Lin, J.J.; Alfieri, K.N.; Fay, N.C.; Low-Nam, S.T.; Groves, J.T. Early T cell receptor signals globally modulate ligand:receptor affinities during antigen discrimination. Proc. Natl. Acad. Sci. USA 2017, 114, 12190–12195. [Google Scholar] [CrossRef] [PubMed]
- Tailor, P.; Tsai, S.; Shameli, A.; Serra, P.; Wang, J.; Robbins, S.; Nagata, M.; Szymczak-Workman, A.L.; Vignali, D.A.A.; Santamaria, P. The proline-rich sequence of CD3ε as an amplifier of low-avidity TCR signaling. J. Immunol. 2008, 181, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Mingueneau, M.; Sansoni, A.; Grégoire, C.; Roncagalli, R.; Aguado, E.; Weiss, A.; Malissen, M.; Malissen, B. The proline-rich sequence of CD3ε controls T cell antigen receptor expression on and signaling potency in preselection CD4+CD8+ thymocytes. Nat. Immunol. 2008, 9, 522–532. [Google Scholar] [CrossRef]
- Xu, C.; Gagnon, E.; Call, M.E.; Schnell, J.R.; Schwieters, C.D.; Carman, C.V.; Chou, J.J.; Wucherpfenning, K.W. Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell 2008, 135, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Aivazian, D.; Stern, L.J. Phosphorylation of T cell receptor ζ is regulated by a lipid dependent folding transition. Nat. Struct. Biol. 2000, 7, 1023–1026. [Google Scholar]
- Brazin, K.N.; Mallis, R.J.; Das, D.K.; Feng, Y.; Hwang, W.; Wang, J.-H.; Wagner, G.; Lang, M.J.; Reinherz, E.L. Structural features of the αβTCR mechanotransduction apparatus that promote pMHC discrimination. Front. Immunol. 2015, 6, 441. [Google Scholar] [CrossRef]
- Lee, M.S.; Glassman, C.R.; Deshpande, N.R.; Badgandi, H.B.; Parrish, H.L.; Uttamapinant, C.; Stawski, P.S.; Ting, A.Y.; Kuhns, M.S. A mechanical switch couples T cell receptor triggering to the cytoplasmic juxtamembrane regions of CD3ζζ. Immunity 2015, 43, 227–239. [Google Scholar] [CrossRef]
- Glassman, C.R.; Parrish, H.L.; Lee, M.S.; Kuhns, M.S. Reciprocal TCR-CD3 and CD4 engagement of a nucleating pMHCII stabilizes a functional receptor macrocomplex. Cell Rep. 2018, 22, 1263–1275. [Google Scholar] [CrossRef]
- Artyomov, M.N.; Lis, M.; Devadas, S.; Davis, M.M.; Chakraborty, A.K. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc. Natl. Acad. Sci. USA 2010, 107, 16916–16921. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, W.; Evavold, B.D.; Zhu, C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 2014, 157, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Ayres, C.M.; Scott, D.R.; Corcelli, S.A.; Baker, B.M. Differential utilization of binding loop flexibility in T cell receptor ligand selection and cross-reactivity. Sci. Rep. 2016, 6, 25070. [Google Scholar] [CrossRef] [PubMed]
- Das, D.K.; Feng, Y.; Mallis, R.J.; Li, X.; Keskin, D.B.; Hussey, R.E.; Brady, S.K.; Wang, J.-H.; Wagner, G.; Reinherz, E.L.; et al. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl. Acad. Sci. USA 2015, 112, 1517–1522. [Google Scholar] [CrossRef] [PubMed]
- Perica, K.; Bieler, J.G.; Edidin, M.; Schneck, J. Modulation of MHC binding by lateral association of TCR and coreceptor. Biophys. J. 2012, 103, 1890–1898. [Google Scholar] [CrossRef] [PubMed]
- Glatzová, D.; Cebecauer, M. Dual role of CD4 in peripheral T lymphocytes. Front. Immunol. 2019, 10, 618. [Google Scholar] [CrossRef] [PubMed]
- Yachi, P.P.; Ampudia, J.; Gascoigne, N.R.J.; Zal, T. Non-stimulatory peptides contribute to antigen induced CD8-TCR interaction at the immunological synapse. Nat. Immunol. 2005, 6, 785–792. [Google Scholar] [CrossRef]
- Goyette, J.; Nieves, D.J.; Ma, Y.; Gauss, K. How does T cell receptor clustering impact on signal transduction? J. Cell Sci. 2019, 132, jcs226423. [Google Scholar] [CrossRef]
- Kumar, R.; Ferez, M.; Swamy, M.; Arechaga, I.; Rejas, M.T.; Valpuesta, J.M.; Schamel, W.W.A.; Alarcon, B.; van Santen, H.M. Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity 2011, 35, 375–387. [Google Scholar] [CrossRef]
- Pageon, S.V.; Tabarin, T.; Yamamoto, Y.; Ma, Y.; Nicovich, P.R.; Bridgeman, J.S.; Cohnen, A.; Benzing, C.; Gao, Y.; Crowther, M.D.; et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discriminaton. Proc. Natl. Acad. Sci. USA 2016, 113, E5454–E5463. [Google Scholar] [CrossRef]
- Levchenko, A.; Nemenman, I. Cellular noise and information transmission. Curr. Opin. Biotechnol. 2014, 28, 156–164. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, M.D.; Amblard, P.-O.; Stocks, N.G. Stochastic pooling networks. J. Stat. Mech. Theory Exp. 2009, 2009, P01012. [Google Scholar] [CrossRef]
- Davtyan, A.; Platkov, M.; Gruebele, M.; Papoian, G.A. Stochastic resonance in protein folding dynamics. ChemPhysChem 2016, 17, 1305–1313. [Google Scholar] [CrossRef]
- Dave, K.; Davtyan, A.; Papoian, G.A.; Gruebele, M.; Platkov, M. Environmental fluctuations and stochastic resonance in protein folding. ChemPhysChem 2016, 17, 1341–1348. [Google Scholar] [CrossRef]
- Kim, W.K.; Hyeon, C.; Sung, W. Weak temporal signals can synchronize and accelerate the transition dynamics of biopolymers under tension. Proc. Natl. Acad. Sci. USA 2012, 109, 14410–14415. [Google Scholar] [CrossRef] [PubMed]
- Utzny, C.; Faroudi, M.; Valitutti, S. Frequency encoding of T-cell receptor engagement dynamics in calcium time series. Biophys. J. 2005, 88, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.; Lei, Y.; Li, N. Applications of stochastic resonance to machinery fault detection: A review and tutorial. Mech. Syst. Sign. Proc. 2019, 122, 502–536. [Google Scholar] [CrossRef]
- Lu, S.; He, Q.; Wang, J. A review of stochastic resonance in rotating machine fault detection. Mech. Syst. Sign. Proc. 2019, 116, 230–260. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bene, L.; Bagdány, M.; Damjanovich, L. T-cell Receptor Is a Threshold Detector: Sub- and Supra-Threshold Stochastic Resonance in TCR-MHC Clusters on the Cell Surface. Entropy 2022, 24, 389. https://doi.org/10.3390/e24030389
Bene L, Bagdány M, Damjanovich L. T-cell Receptor Is a Threshold Detector: Sub- and Supra-Threshold Stochastic Resonance in TCR-MHC Clusters on the Cell Surface. Entropy. 2022; 24(3):389. https://doi.org/10.3390/e24030389
Chicago/Turabian StyleBene, László, Miklós Bagdány, and László Damjanovich. 2022. "T-cell Receptor Is a Threshold Detector: Sub- and Supra-Threshold Stochastic Resonance in TCR-MHC Clusters on the Cell Surface" Entropy 24, no. 3: 389. https://doi.org/10.3390/e24030389
APA StyleBene, L., Bagdány, M., & Damjanovich, L. (2022). T-cell Receptor Is a Threshold Detector: Sub- and Supra-Threshold Stochastic Resonance in TCR-MHC Clusters on the Cell Surface. Entropy, 24(3), 389. https://doi.org/10.3390/e24030389




