1. Introduction
As one of the most threatening diseases to human health, cancer can not only bring great pain and psychological pressure to patients but can also bring heavy economic burden to countless families and even the whole of society. Cancer is an immune disease caused by the uncontrolled growth and division of abnormal cells in the body and the spread to the whole body [
1]. Early diagnosis (prediction) of cancer can help physicians decide a treatment plan, which has important and positive significance for the adequate and effective treatment of cancer. Therefore, accurate prediction of cancer is very critical in the treatment of cancer. However, the early diagnosis of cancer is a very difficult task; once the symptoms of cancer appear, it is usually in advanced stages and is difficult to treat. At present, cancer recognition mainly depends on gene test or protein test, among which gene tests are inherited and static, which are mostly used in the detection of congenital genetic diseases, and cannot reflect the occurrence of diseases in the body in terms of autoimmunity and metabolism; moreover, genetic tests are difficult to interpret and are also expensive. Protein tests are dynamic and can directly reflect the occurrence and development of diseases. They can detect the development of a variety of diseases, including genetic diseases. Comparatively speaking, the practical space of protein test is wider.
Blood protein is relatively easy to be obtained; furthermore, studies show that blood protein test in the early stage can not only improve the prognosis, but also has the advantages of being non-invasive and conferring no pain to the patients [
2], so in recent years, cancer diagnosis based on blood protein markers has become a research hotspot [
3]. It is even expected that, as soon as the predicting technology is mature, with just 1 to 2 mL of blood, the protein-chip screen could know the risk of cancer one to three years in advance, giving people more time in the fight against cancer.
Since there are many types of protein markers in blood, their levels in the blood of patients for different cancers are different. Diagnostic results are limited if cancer is identified only by a single marker, which is usually inaccurate and not comprehensive. Multi-index comprehensive recognition based on protein markers should improve the efficiency of cancer prediction to some extent [
4]. With the increase in incidence and mortality of cancer in recent years, there is an urgent need for convenient and effective technology for cancer diagnosis and prediction.
Recently, machine learning methods have been widely designed in medical diagnosis and prediction because of their powerful learning and prediction capabilities in dealing with nonlinear problems [
5,
6]. There are many machine learning algorithms that have been developed for cancer diagnosis [
7,
8,
9,
10,
11], and a model with better predictive power can benefit cancer patients going through the toxic side effects and extra medical expenses related to unnecessary treatment. However, the predictive effect of a model largely depends on the algorithm and the features for a given data set [
12], and different results may be yielded with different algorithms and different feature extractions.
Because traditional classification algorithms are based on the assumption of equal misclassification costs, they ignore the sample particularity of the minority class, resulting in the inadequate recognition ability of the algorithm in dealing with imbalanced datasets. Cancer data are often uneven, which makes cancer prediction more difficult. Since the imbalance of data is usually accompanied by the imbalanced cost of sample misclassification. When dealing with the problem of imbalanced data, cost-sensitive learning often gives a large misclassification cost to minority classes and a small misclassification cost to the majority classes, so as to improve the attention to samples of the minority class and improve the classification accuracy.
In the past two decades, in order to deal with the classification tasks with different costs, many popular classification algorithms have been extended based on cost analysis, including misclassification costs and other cost classification techniques, and have been applied to the actual environment and obtained good classification results [
13,
14,
15,
16]. Among them, the most popular are decision trees, Bayes and support vector machines [
17,
18,
19]. As one of the top ten mining algorithms, K-Nearest Neighbors (KNN) is one of the simplest and most commonly used classification algorithms as it is easy to understand and also easy to implement with no parameter needed to estimate, so it is particularly suitable for multi-modal problems [
20,
21]. However, the studies on the extended KNN algorithm and its related applications are relatively few, considering the imbalance of our dataset, and encouraged by existing studies, a cost-sensitive learning technique of KNN that has a sensitive cost matrix was developed here for cancer prediction.
At the same time, for different cancers, patients have different levels of protein markers in their serum, and entropy, which is extended from the concept of thermodynamics, can not only describe the disorder of a system, but also describe the degree of dispersion of a certain index. In this paper, we attempted to take the patient’s serum protein composition as a subsystem and analyzed different entropy values of this system to study its screening values for cancer identification; moreover, entropies were also used as characteristics components of each patient’s feature sequence to construct a predictive model of cancer.
3. Results and Discussion
In order to excavate more useful information from blood protein index and effectively predict cancer, four kinds of entropy values of cancer protein sequence were analyzed firstly, and the improved algorithm and model were applied to predict cancer.
3.1. Entropy Analysis Based on Serum Proteins
Considering each sample as a sequence, the average entropies of each type of sample were calculated.
From
Table 2, we can see that there is almost no difference between the fuzzy entropies and information entropies of different types of sample, the difference of their fuzzy entropies is less than 0.002, and the difference of their information entropies is less than 0.003. However, for approximate entropy and sample entropy, their differences are relatively bigger, which means that these two types of entropy can help distinguish between different types of patients. Among the average entropies of three types of samples, those of a normal person are the biggest, and those of liver cancer patients are the smallest, which indicates that different cancer patients have different levels of serum protein complexity, approximate entropy and sample entropy can reflect the complexity of different serum protein sequence, and the complexity of serum protein in normal people is obviously higher than that in cancer patients, which means that cancer cells may reduce the complexity of serum proteins.
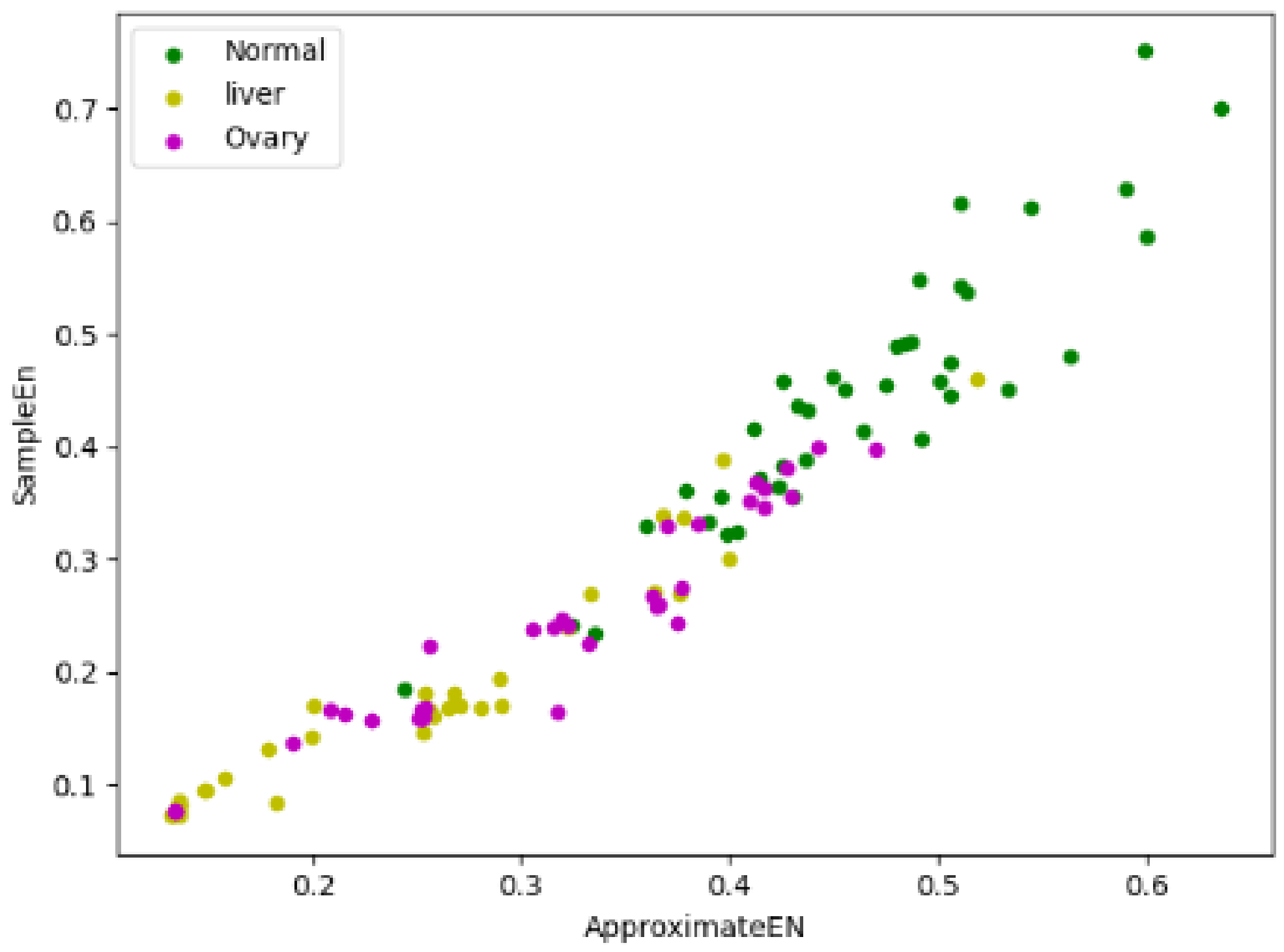
Because there is an inconspicuous difference between fuzzy entropy and information entropy in three types of samples, we only focused on their approximate entropy and sample entropy. If approximate entropy is taken as the X-axis and sample entropy is taken as the Y-axis, forty samples were randomly selected from three types of samples, respectively, and the entropy vectors of these 120 samples were presented in the form of a scatter plot, which is shown in
Figure 1:
In
Figure 1, the green dots represent the normal samples, the yellow dots represent the liver cancer samples, and the purple dots represent the ovarian cancer samples. The scatter plot shows that the entropy vectors are roughly dispersed in three different regions, although there are overlapping parts among three regions, but we can also clearly distinguish the centers of three regions are different, which provides meaningful information for our research, namely taking approximate entropy and sample entropy as classification characteristics of cancer has certain feasibility.
3.2. Results Analysis
Due to the imbalance of our dataset, the number of normal samples is much larger than that of liver cancer samples and ovarian cancer samples. When k neighbor samples are selected, the probability of the normal samples being selected is greater than those of other cancer samples, which leads to the trend of leaning toward the majority in judgement. In order to improve this situation, a cost-sensitive KNN algorithm was proposed in this paper, which improved the impact caused by the imbalance of the dataset by increasing the misclassification cost of minority classes. The main difficulty of the cost-sensitive KNN algorithm is how to determine the appropriate cost matrix. Here, we assumed that the correctly classified samples have no error cost, namely
. In this paper, 0 means normal, 1 means liver cancer, and 2 means ovarian cancer.
was determined according to the proportion of data, and the final cost matrix obtained is shown in
Table 3:
In addition, in order to reduce the errors caused by the different values of , the method of m-estimation was adopted to make the model more effective here.
Because the selection of k value in KNN is of vital importance to the classification results. It should not be too large or too small; if the k value is too small, it will cause model too complex; if the k value is too large, it will result in fuzzy classification. In the experiment, we selected parameter k value through 10-fold cross-validation. Under different k values (k = 1, 2, …, 30), we calculated the average prediction accuracy and the variance of accuracy, and finally selected the appropriate k value by compromise according to the principle of maximum accuracy and minimum variance.
Each sample has 39 serum protein marker indexes. First, we standardized these indexes and took these indexes as the eigenvectors of the sample to establish the feature space. Then, we used the jackknife test; the results of KNN algorithm and our improved algorithm (CS-KNN) are shown in
Table 4 and
Figure 2.
From the results in
Table 4, compared with KNN algorithm, the accuracy, recall and F1_score of our cost-sensitive KNN algorithm are all improved, the extent of the improvements is 0.422%, 16.64% and 6.26%, and the Auc reaches 0.9 in
Figure 2. The overall effect of the model is relatively stability and satisfactory.
In fact, in order to further verify the validity of our cost-sensitive KNN algorithm, we selected three different imbalanced datasets from the UCI database: Breast Cancer Wisconsin, Heart Disease and Speaker Accent Recognition, and the properties of these datasets are shown in
Table 5.
Because these datasets in the UCI database were only selected to further illustrate the effect and general applicability of our algorithm, here, we only used the method of 10-fold cross-validation to discuss the effect of our algorithm; the results are listed in
Table 6.
From
Table 6, for the cost-sensitive KNN algorithm, it can be seen that its average accuracy and F1 both are improved in the Breast Cancer Wisconsin dataset, and the average accuracy in Speaker Accent Recognition dataset and F1 in Heart Disease dataset are improved, respectively. No matter the average accuracy or F1, the effect of the cost-sensitive KNN algorithm on the three datasets is no less than those of KNN algorithm; moreover, their variances are lower than those of the KNN algorithm. By comprehensive analysis of evaluation indicator values of different datasets, our improved KNN algorithm is more effective than the original KNN algorithm on imbalanced datasets, and the stability of our algorithm is obviously better than that of the original KNN algorithm, which shows that the cost-sensitive matrix designed by us is reasonable, and cost-sensitive learning based on this sensitive matrix can improve the impact of imbalanced data. Our algorithm based on cost-sensitive learning is suitable for the analysis of imbalanced data.
In order to further enhance the effect of cancer prediction and make full use of the information contained by the data, in this paper we attempted to regard sample entropy and approximate entropy as characteristic attributes of samples; then, the two-dimensional dataset composed of approximate entropy and sample entropy of the samples, and the 41 dimensional dataset with approximate entropy and sample entropy added on the original 39 features were used for cancer prediction. The results are shown in
Table 7 and
Figure 3.
Table 7 and
Figure 3 shows that under the three characteristic indexes, the prediction effect of cost-sensitive KNN combining serum protein marker indexes and entropy values as a feature vector is superior to the other two characteristic indexes. Compared with the indexes only considering serum protein indexes, recall and F1 have been further improved; they are enhanced by 3.39% and 6.14%, respectively, and the AUC has reached 0.92, an increase of 2.2%. Compared with the model of KNN and original indexes, recall and F1 have been greatly improved and the rates of increase have achieved 20.55% and 6.92%, respectively. These results indicate that the prediction effects can indeed be improved by adding entropy information, and the results also indicate that the model proposed in this paper, namely the cost-sensitive KNN model based on serum protein markers and their entropy values, is suitable for cancer prediction research.
4. Conclusions
The results of this study show that there are significant differences in approximate entropy and sample entropy of serum protein indexes among normal people, patients with liver cancer and patients with ovarian cancer. The entropy values of normal people are higher than those of cancer patients, which indicates that the serum protein composition of normal people is more complex than that of cancer patients, and cancer cells are suspected to affect the dense structure of human serum protein.
Taking approximate entropy and sample entropy as the attributes of the feature vector is helpful to improve the accuracy of cancer recognition to some extent. For imbalanced datasets, cost-sensitive learning by the method of constructing a misjudgment cost matrix can improve the performance of the original KNN algorithm. The experiment results demonstrate that our improved method can improve the influence of data imbalance. Synthesizing five evaluation indexes, the model of the cost-sensitive KNN algorithm based on serum protein indexes and entropies has the best effect, which is suitable for the classification and prediction of cancer. The work of this paper can provide a research basis for the intelligence of medical treatment in the future.