The Complex Structure of the Pharmacological Drug–Disease Network
Abstract
:1. Introduction
2. Data and Methods
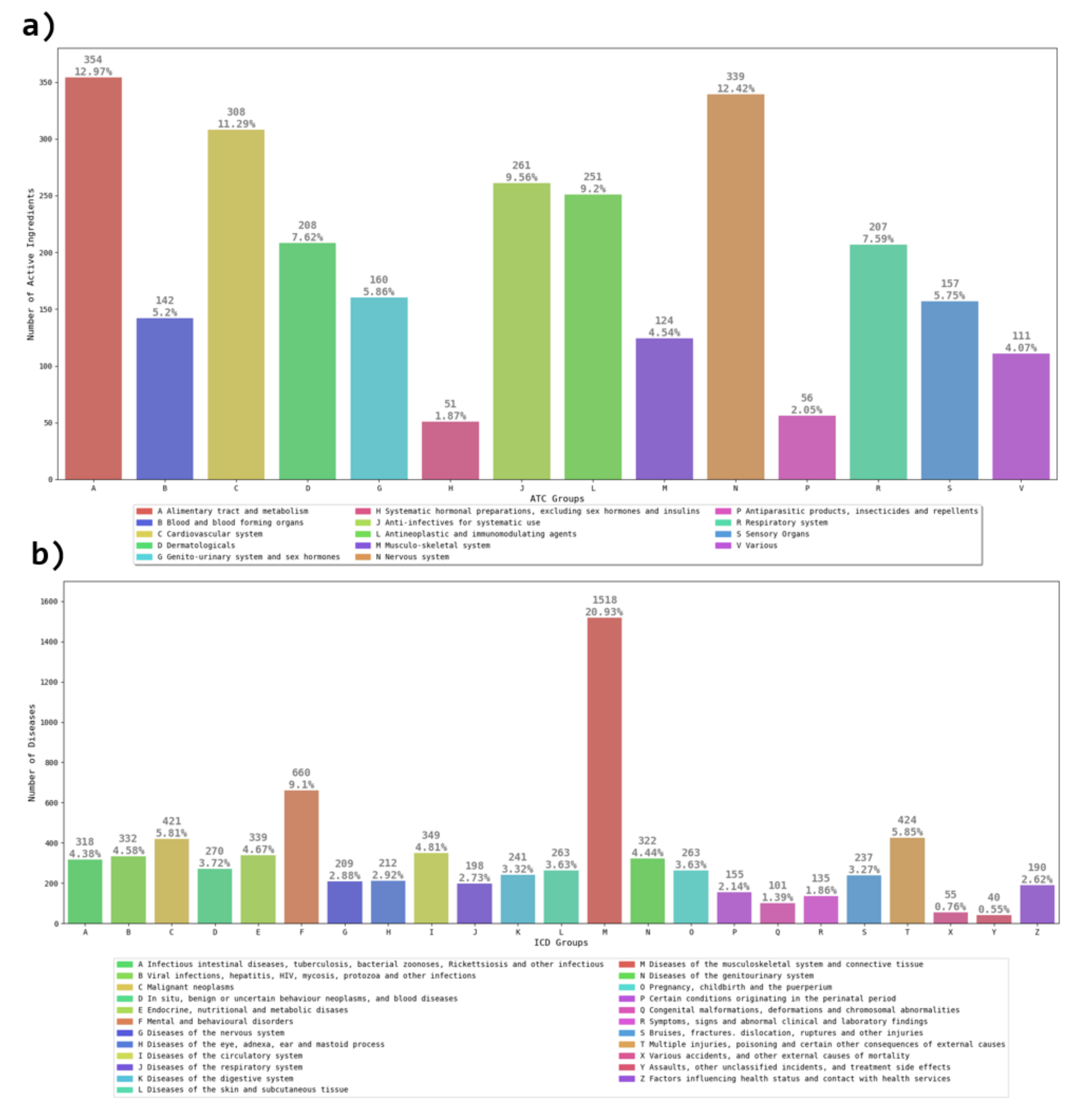
2.1. Pharmacological Dataset
2.2. Network Metrics
- Density (): the density of a network is defined as follows:where g is the number of actual connections and is the maximum number of edges with and , the number of nodes for drug and disease networks, respectively. Notice that for a bipartite network, the normalization term is given by .A value of close to 1 denotes an almost complete graph, while close to 0 indicates a poorly connected network.
- Shortest path length (ℓ): represents the shortest path between two nodes, i.e., a path with the minimum number of edges.
- Clustering coefficient (): measures the degree of transitivity in connectivity amongst the nearest neighbors of a node i [27]:where is the number of links between the neighbors of the node i. The average clustering is the mean value from all nodes.
- Assortative mixing coefficient by degree (): measure associated to the tendency frequently observed in networks, where nodes with a large number of neighbors are connected to other nodes with many (or a few) connections [27]. Formally, the coefficient is given by the following:where . For perfectly assortative networks, the coefficient reaches a maximum value of 1, whereas a minimum value of is observed for perfectly disassortative ones.
3. Results
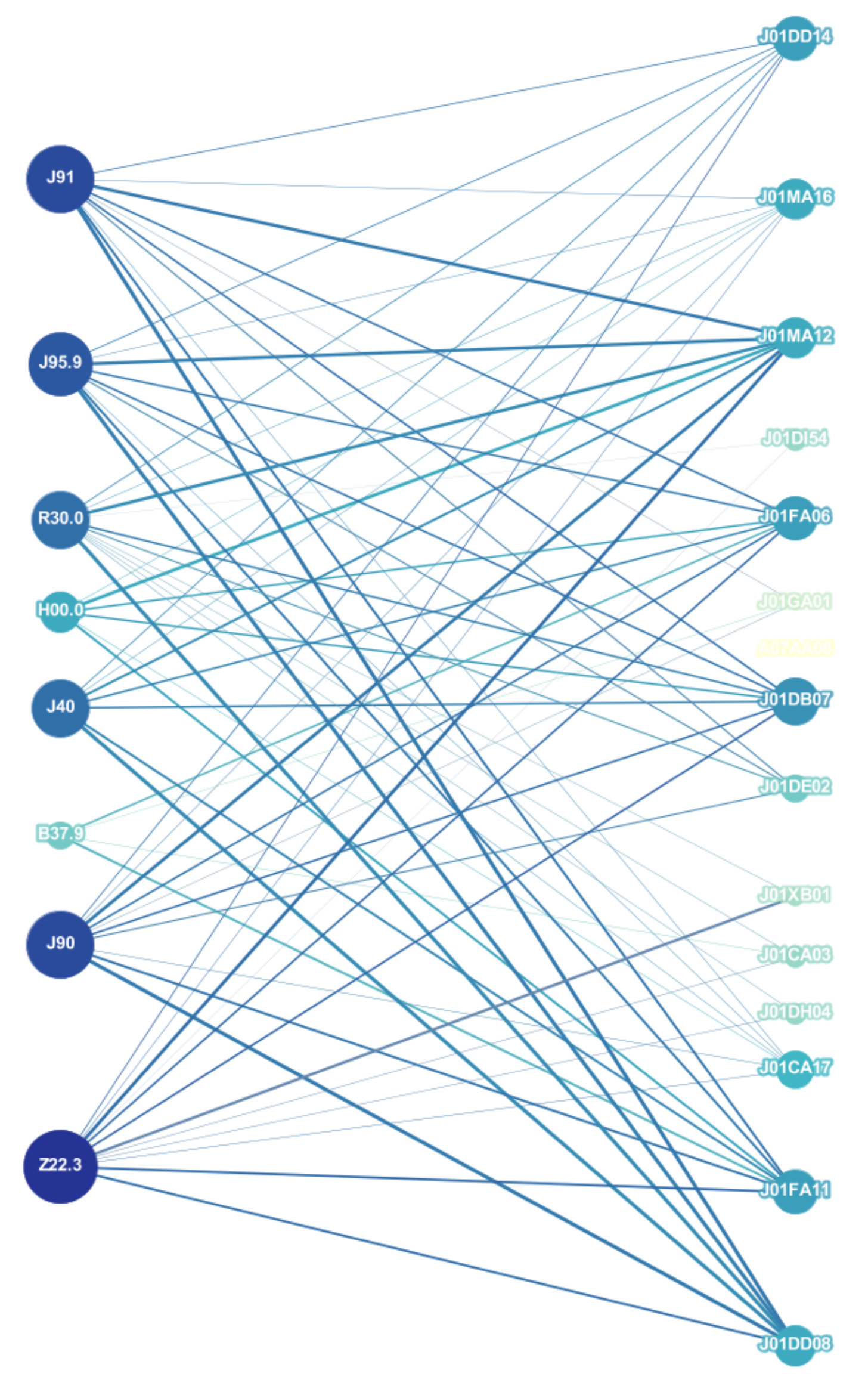
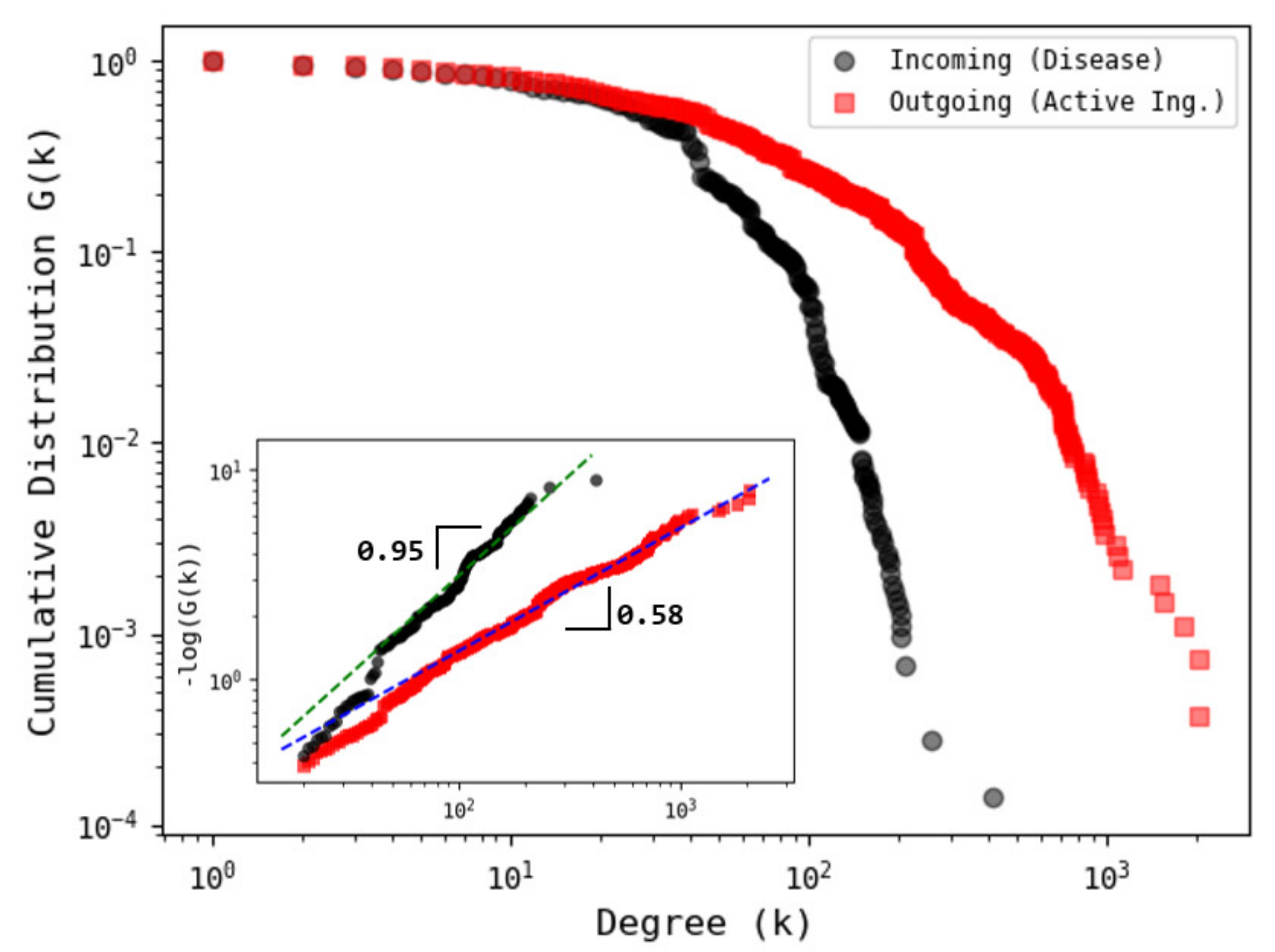
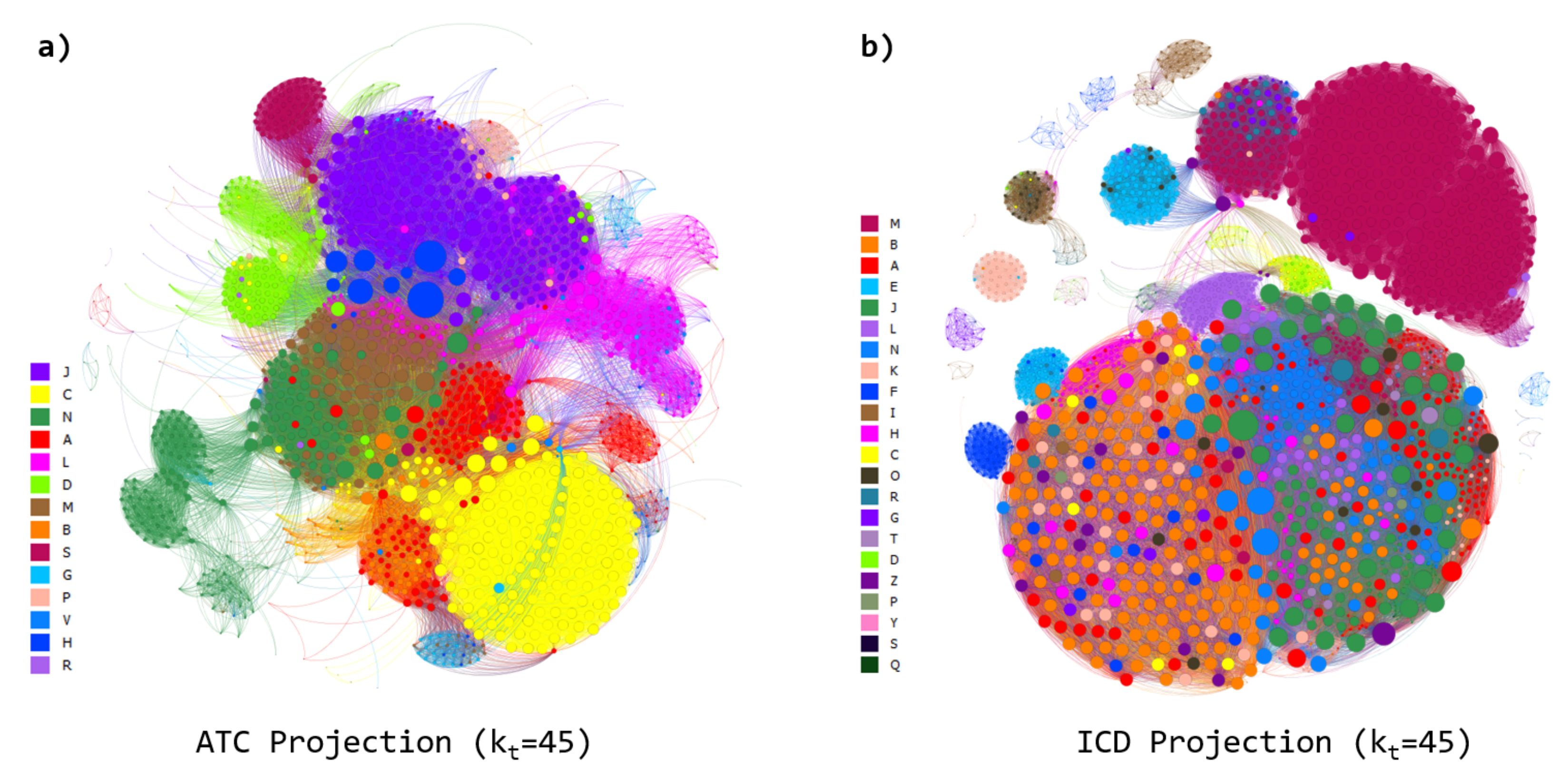
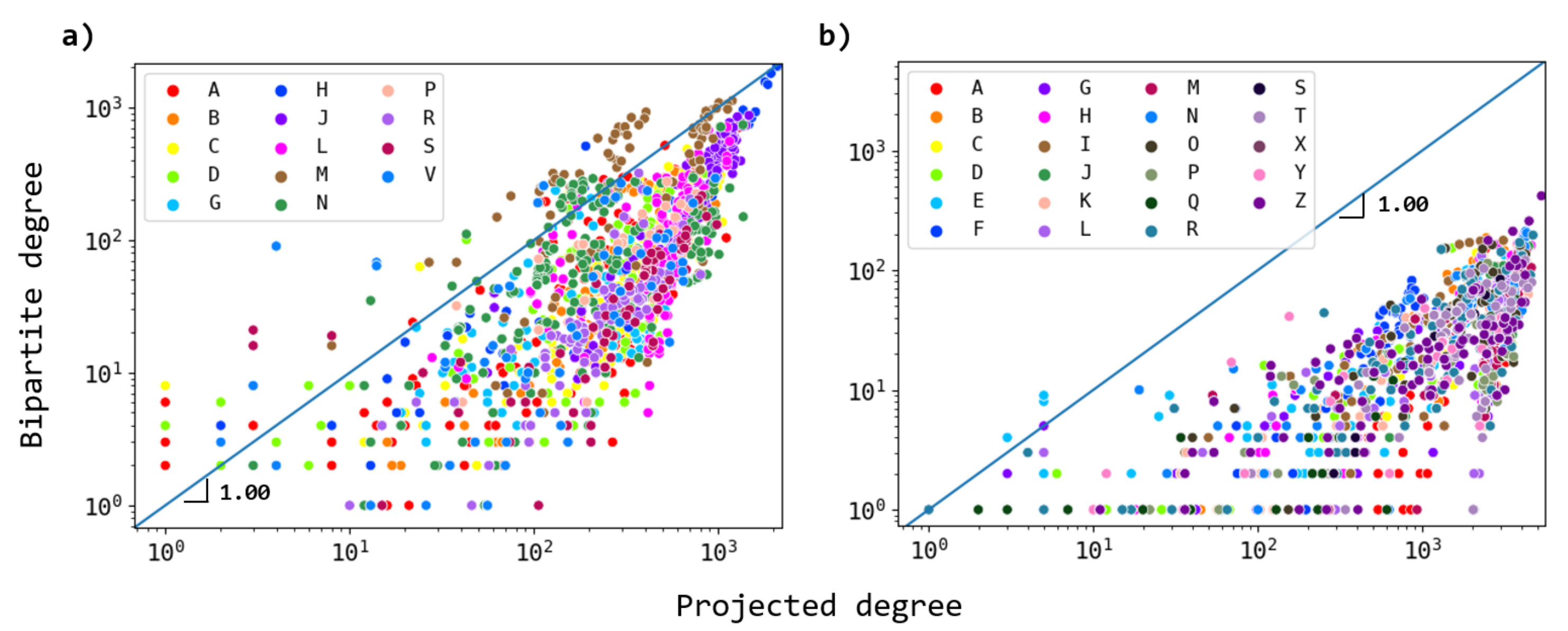
3.1. Network Analysis
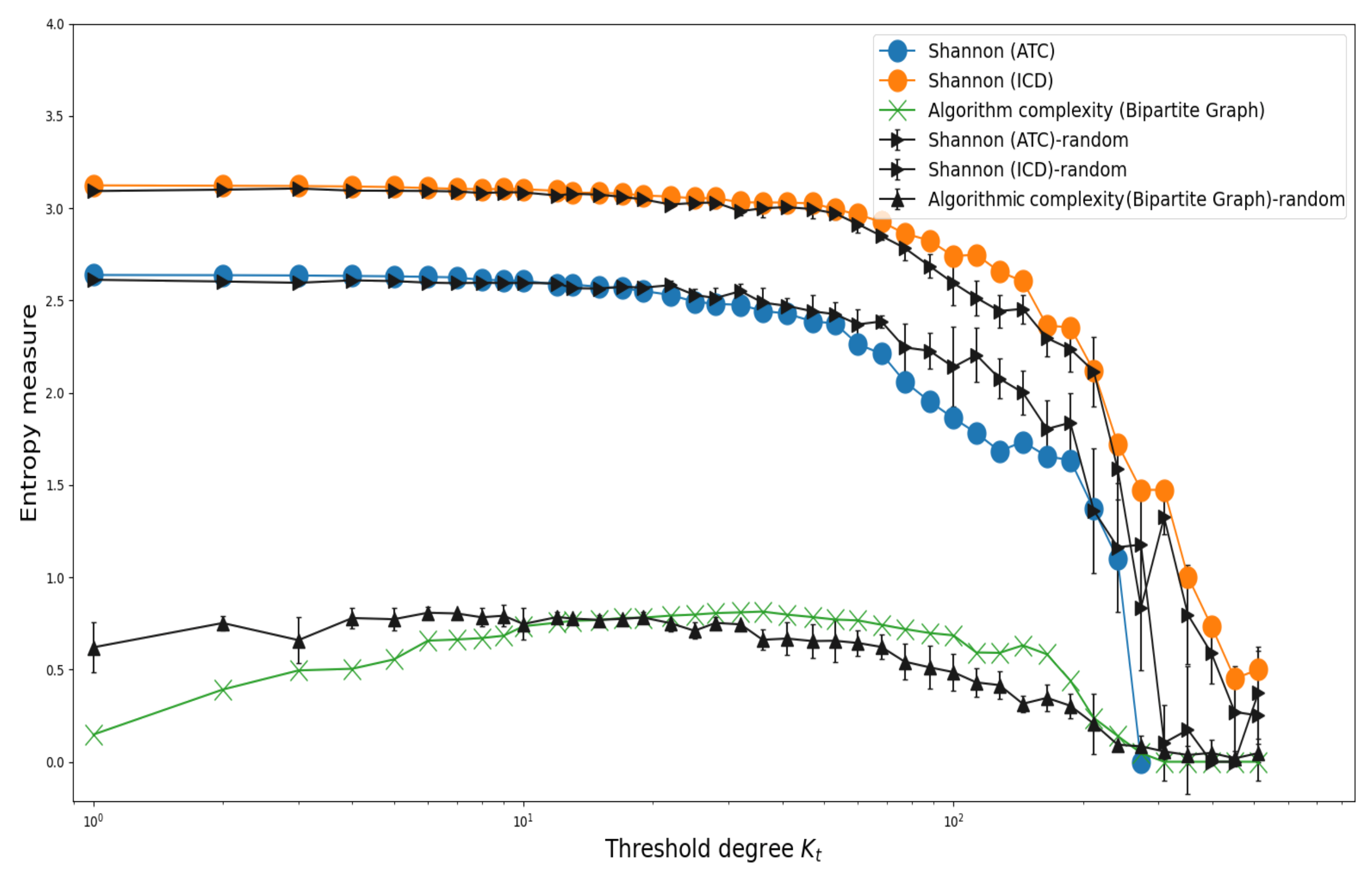
3.2. Shannon’s Entropy and Algorithmic Complexity
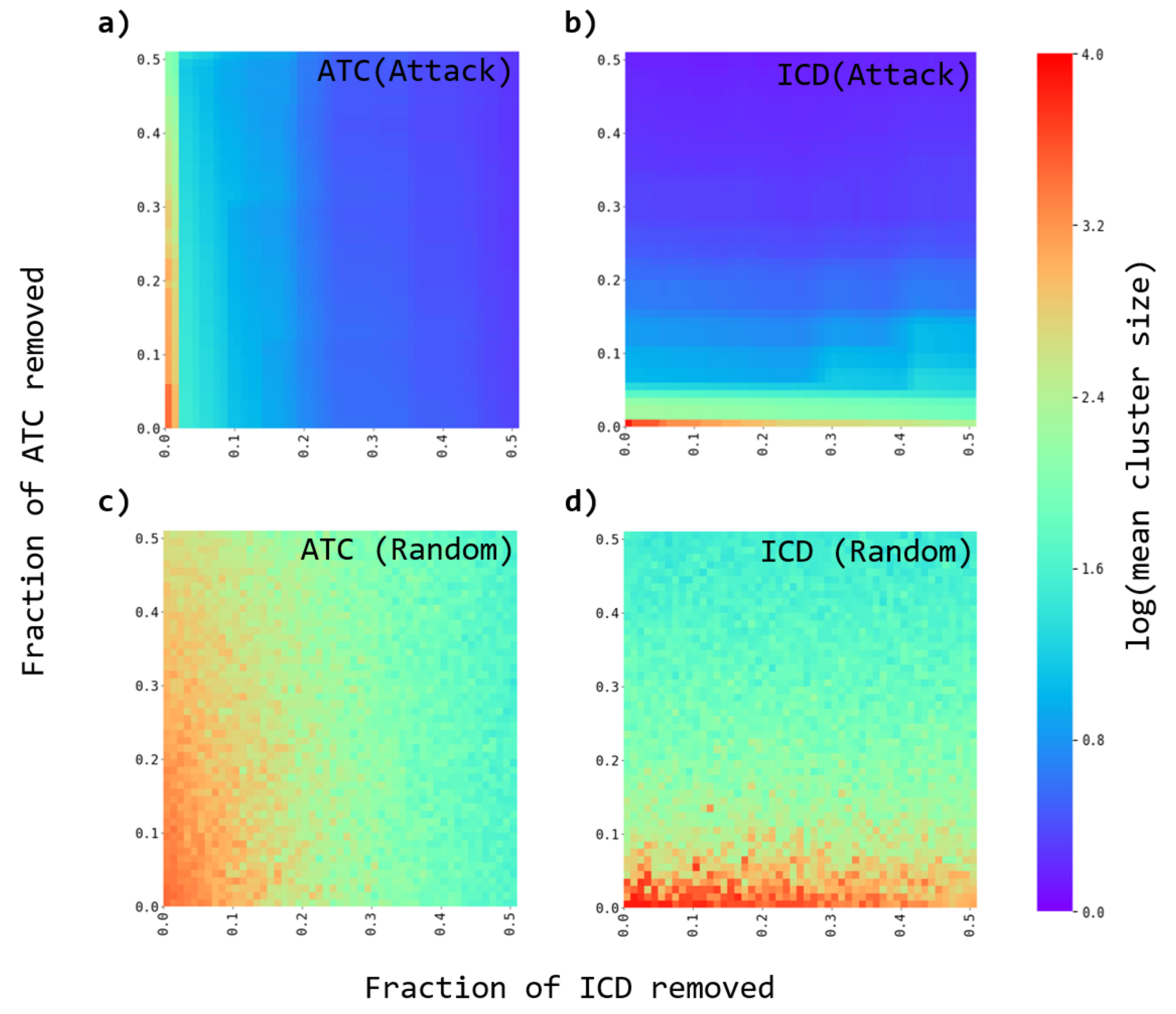
3.3. Robustness of the Networks
- A fraction of either ATC or ICD nodes are removed from the original and the randomized bipartite networks. Two strategies are considered. The nodes to be removed are either chosen as the most connected ones (directed attacks), or at random (random failures).
- The average cluster size is evaluated to evaluate the effect of the node’s removal.
- The process is repeated for several fractions of removed nodes.
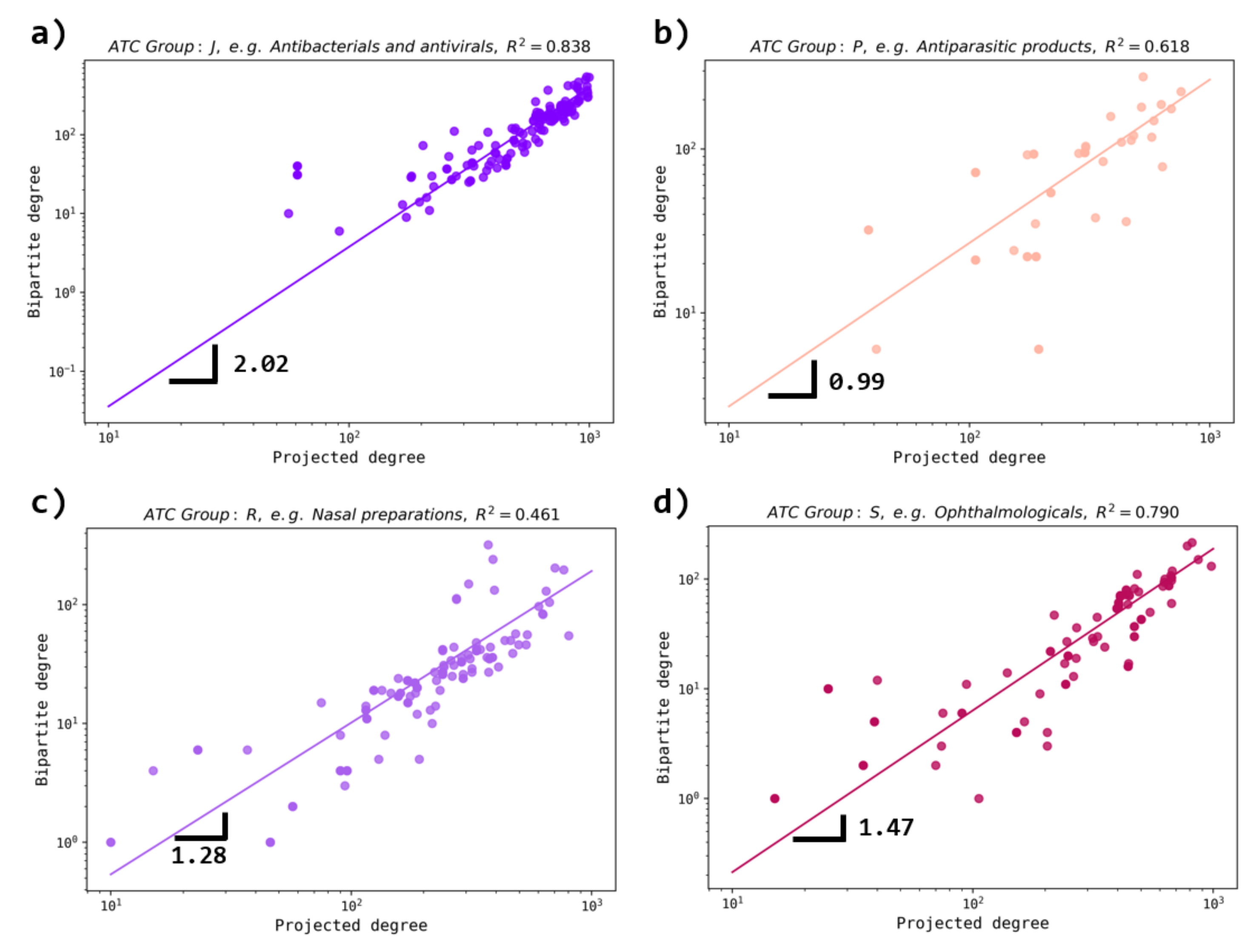
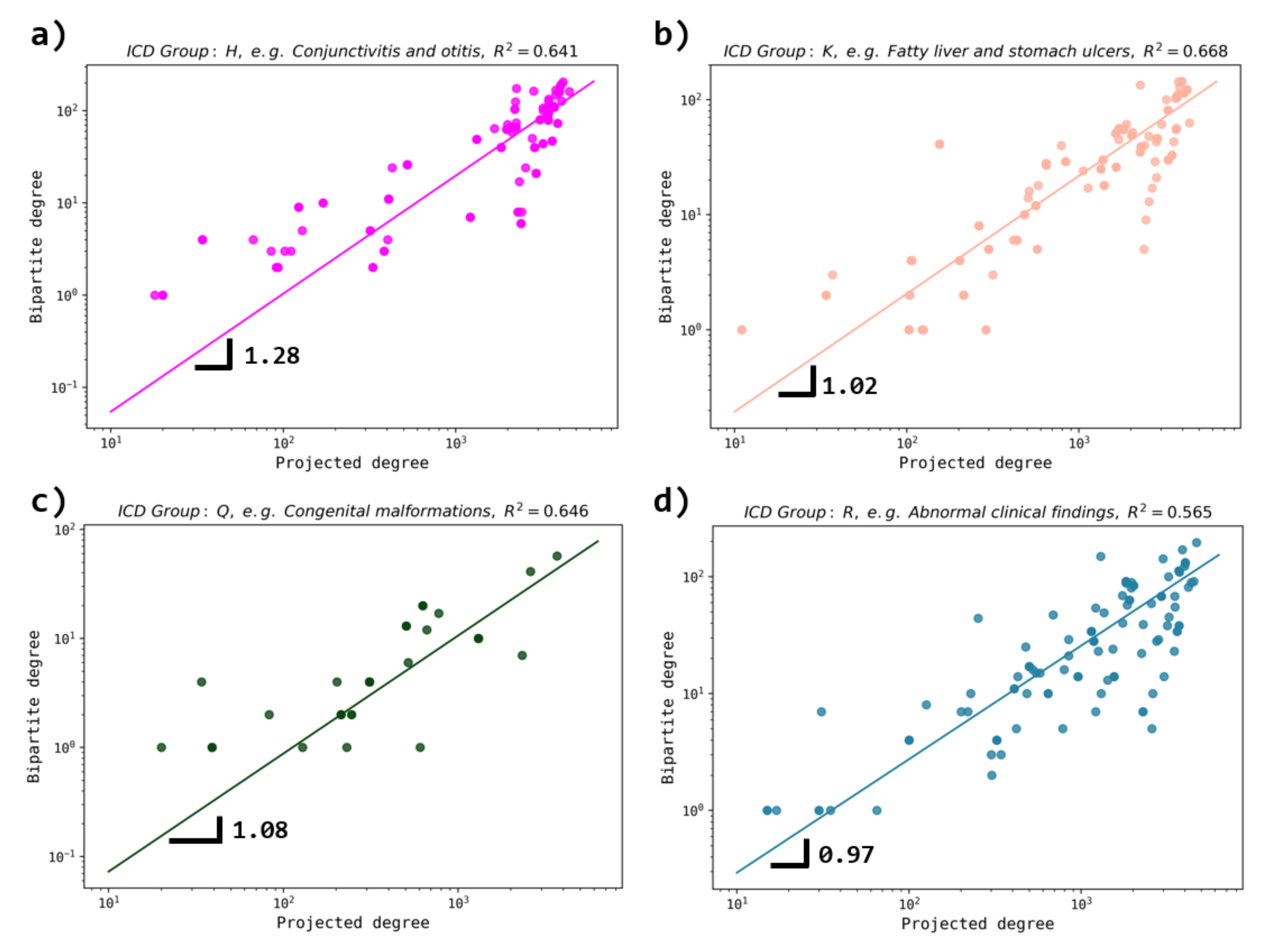
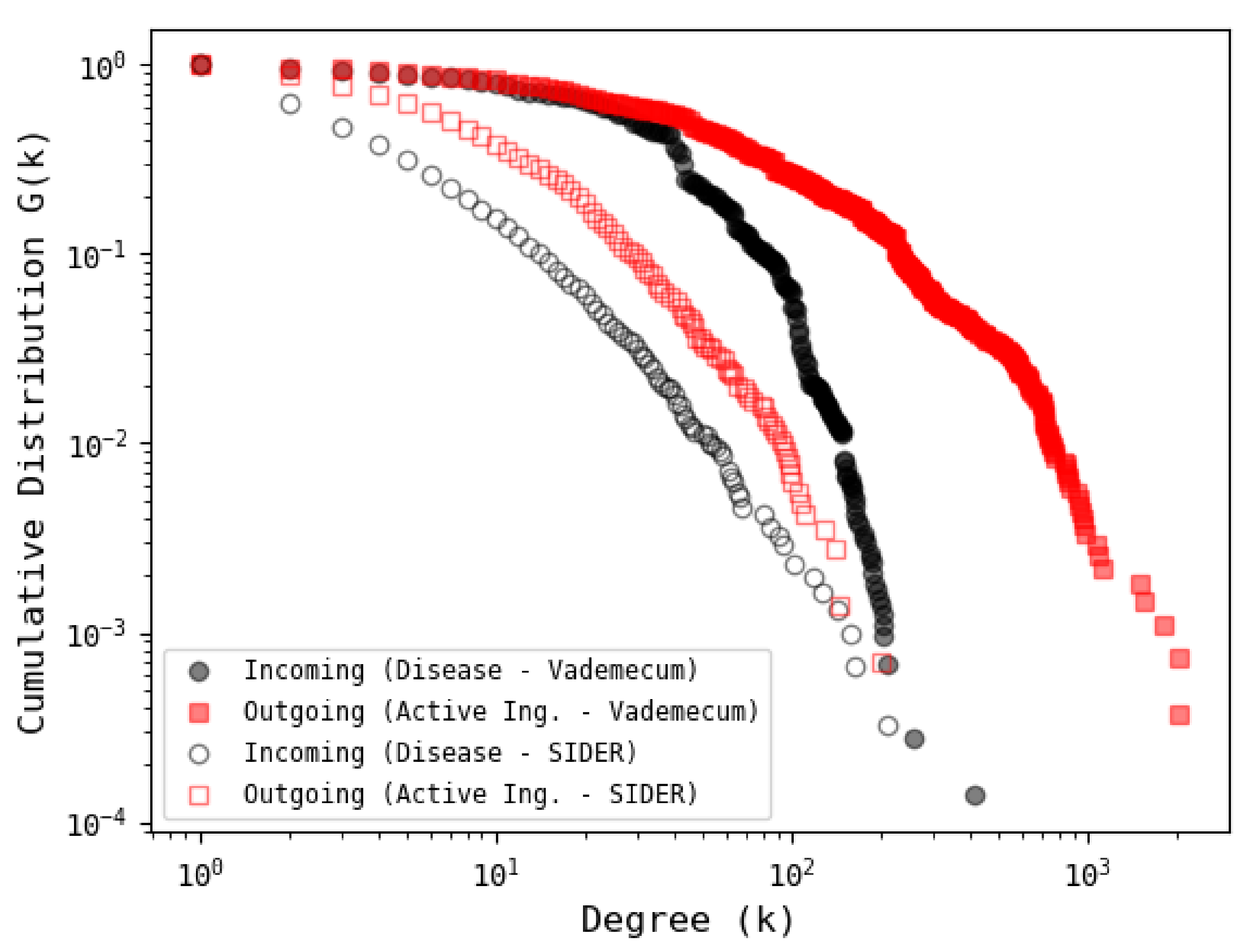
3.4. Comparison with Other Database
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krantz, A. Diversification of the drug discovery process. Nat. Biotechnol. 1998, 16, 1294. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Guney, E.; Menche, J.; Vidal, M.; Barábasi, A.L. Network-based in silico drug efficacy screening. Nat. Commun. 2016, 7, 10331. [Google Scholar] [CrossRef] [Green Version]
- Barabási, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [Green Version]
- Loscalzo, J. Network Medicine; Harvard University Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Pawson, T.; Linding, R. Network medicine. FEBS Lett. 2008, 582, 1266–1270. [Google Scholar] [CrossRef] [Green Version]
- Musa, A.; Ghoraie, L.S.; Zhang, S.D.; Glazko, G.; Yli-Harja, O.; Dehmer, M.; Haibe-Kains, B.; Emmert-Streib, F. A review of connectivity map and computational approaches in pharmacogenomics. Brief. Bioinform. 2017, 19, 506–523. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Li, S. A co-module approach for elucidating drug–disease associations and revealing their molecular basis. Bioinformatics 2012, 28, 955–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Jiang, R.; Zhang, M.Q.; Li, S. Network-based global inference of human disease genes. Mol. Syst. Biol. 2008, 4, 189. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Hu, Q.; Li, S. Systematic Analysis of New Drug Indications by Drug-Gene-Disease Coherent Subnetworks. CPT Pharmacometrics Syst. Pharmacol. 2014, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Ren, J.; Medo, M.; Zhang, Y.C. Bipartite network projection and personal recommendation. Phys. Rev. E 2007, 76, 046115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillaume, J.L.; Latapy, M. Bipartite graphs as models of complex networks. Phys. A Stat. Mech. Appl. 2006, 371, 795–813. [Google Scholar] [CrossRef]
- Ramasco, J.J.; Dorogovtsev, S.N.; Pastor-Satorras, R. Self-organization of collaboration networks. Phys. Rev. E 2004, 70, 036106. [Google Scholar] [CrossRef] [Green Version]
- Newman, M.E. The structure of scientific collaboration networks. Proc. Natl. Acad. Sci. USA 2001, 98, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.K.; Kaski, K.; Fortunato, S. World citation and collaboration networks: Uncovering the role of geography in science. Sci. Rep. 2012, 2, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre-Plans, J.; Piñero, J.; Menche, J.; Sanz, F.; Furlong, L.I.; Schmidt, H.H.; Oliva, B.; Guney, E. Proximal pathway enrichment analysis for targeting comorbid diseases via network endopharmacology. Pharmaceuticals 2018, 11, 61. [Google Scholar] [CrossRef] [Green Version]
- Zitnik, M.; Agrawal, M.; Leskovec, J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics 2018, 34, i457–i466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AY, M.; Goh, K.I.; Cusick, M.E.; Barabasi, A.L.; Vidal, M. Drug–target network. Nat. Biotechnol. 2007, 25, 1119–1127. [Google Scholar]
- Ma’ayan, A.; Jenkins, S.L.; Goldfarb, J.; Iyengar, R. Network analysis of FDA approved drugs and their targets. Mt. Sinai J. Med. 2007, 74, 27–32. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Song, Y.; Guan, J.; Luo, L.; Zhuang, Z. Inferring new indications for approved drugs via random walk on drug-disease heterogenous networks. BMC Bioinform. 2016, 17, 539. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Yue, X.; Huang, F.; Liu, R.; Chen, Y.; Ruan, C. Predicting drug-disease associations and their therapeutic function based on the drug-disease association bipartite network. Methods 2018, 145, 51–59. [Google Scholar] [CrossRef]
- Azuaje, F.J.; Zhang, L.; Devaux, Y.; Wagner, D.R. Drug-target network in myocardial infarction reveals multiple side effects of unrelated drugs. Sci. Rep. 2011, 1, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Li, W.; Liu, G.; Tang, Y. Network-Based Methods for Prediction of Drug-Target Interactions. Front. Pharmacol. 2018, 9, 1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spain, V.V. Vidal Vademecum Spain, Su Fuente de Conocimiento Farmacológico. 2010. Available online: https://www.vademecum.es/ (accessed on 15 June 2021).
- López-Rodríguez, I.; Reyes-Manzano, C.F.; Reyes-Ramírez, I.; Contreras-Uribe, T.J.; Guzmán-Vargas, L. Drugs, Active Ingredients and Diseases Database in Spanish. Augmenting the Resources for Analyses on Drug–Illness Interactions. Data 2021, 6, 3. [Google Scholar] [CrossRef]
- Organization, W.H. World Health Organization, Anatomical Therapeutic Chemical Classification System. 2018. Available online: https://www.whocc.no/ (accessed on 15 June 2021).
- Newman, M. Networks, Oxford University Press: Oxford, UK, 2018.
- Perez-Garcia, L.A.; Mejias-Carpio, I.E.; Delgado-Noguera, L.A.; Manzanarez-Motezuma, J.P.; Escalona-Rodriguez, M.A.; Sordillo, E.M.; Mogollon-Rodriguez, E.A.; Hernandez-Pereira, C.E.; Marquez-Colmenarez, M.C.; Paniz-Mondolfi, A.E. Ivermectin: Repurposing a multipurpose drug for Venezuela’s humanitarian crisis. Int. J. Antimicrob. Agents 2020, 56, 106037. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J. One drug, many uses. Biotechnol. Healthc. 2005, 2, 56. [Google Scholar]
- Fruchterman, T.M.; Reingold, E.M. Graph drawing by force-directed placement. Softw. Pract. Exp. 1991, 21, 1129–1164. [Google Scholar] [CrossRef]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef]
- Campbell, S.; Soman-Faulkner, K. Antiparasitic Drugs. StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Novack, G.D. Repurposing medications. Ocul. Surf. 2021, 19, 336. [Google Scholar] [CrossRef]
- Gautam, C.; Mahajan, S.S.; Sharma, J.; Singh, H.; Singh, J. Repurposing potential of ketamine: Opportunities and challenges. Indian J. Psychol. Med. 2020, 42, 22–29. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Kolmogorov, A.N. Three Approaches to the Quantitative Definition of Information. Probl. Inf. Transm. 1965, 1, 1–7. [Google Scholar] [CrossRef]
- Zenil, H. A Review of Methods for Estimating Algorithmic Complexity: Options, Challenges, and New Directions. Entropy 2020, 22, 612. [Google Scholar] [CrossRef]
- Zenil, H.; Hernández-Orozco, S.; Kiani, N.A.; Soler-Toscano, F.; Rueda-Toicen, A.; Tegnér, J. A decomposition method for global evaluation of shannon entropy and local estimations of algorithmic complexity. Entropy 2018, 20, 605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zenil, H.; Soler-Toscano, F.; Dingle, K.; Louis, A.A. Correlation of automorphism group size and topological properties with program-size complexity evaluations of graphs and complex networks. Phys. A Stat. Mech. Appl. 2014, 404, 341–358. [Google Scholar] [CrossRef] [Green Version]
- Ventresca, M. Using algorithmic complexity to differentiate cognitive states in fmri. In International Conference on Complex Networks and their Applications; Springer: Berlin/Heidelberg, Germany, 2018; pp. 663–674. [Google Scholar]
- Zenil, H.; Kiani, N.A.; Tegnér, J. Symmetry and Correspondence of Algorithmic Complexity over Geometric, Spatial and Topological Representations. Entropy 2018, 20, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, M.; Letunic, I.; Jensen, L.J.; Bork, P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2015, 44, D1075–D1079. [Google Scholar] [CrossRef]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2013, 42, D1091–D1097. [Google Scholar] [CrossRef] [Green Version]
- Ursu, O.; Holmes, J.; Bologa, C.G.; Yang, J.J.; Mathias, S.L.; Stathias, V.; Nguyen, D.T.; Schürer, S.; Oprea, T. DrugCentral 2018: An update. Nucleic Acids Res. 2018, 47, D963–D970. [Google Scholar] [CrossRef]

| Metric | Bipartite | Disease | Active Ingredient |
|---|---|---|---|
| Number of nodes | 9981 | 7252 | 2729 |
| Number of edges | 260,995 | 6,188,810 | 454,164 |
| Mean degree | 52.29 | 1706.78 | 332.84 |
| Density | 0.005 | 0.235 | 0.122 |
| Average shortest path length | - | 1.84 | 1.99 |
| Average clustering | - | 0.724 | 0.735 |
| Assortativity | - | 0.217 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Rodríguez, I.; Reyes-Manzano, C.F.; Guzmán-Vargas, A.; Guzmán-Vargas, L. The Complex Structure of the Pharmacological Drug–Disease Network. Entropy 2021, 23, 1139. https://doi.org/10.3390/e23091139
López-Rodríguez I, Reyes-Manzano CF, Guzmán-Vargas A, Guzmán-Vargas L. The Complex Structure of the Pharmacological Drug–Disease Network. Entropy. 2021; 23(9):1139. https://doi.org/10.3390/e23091139
Chicago/Turabian StyleLópez-Rodríguez, Irene, Cesár F. Reyes-Manzano, Ariel Guzmán-Vargas, and Lev Guzmán-Vargas. 2021. "The Complex Structure of the Pharmacological Drug–Disease Network" Entropy 23, no. 9: 1139. https://doi.org/10.3390/e23091139
APA StyleLópez-Rodríguez, I., Reyes-Manzano, C. F., Guzmán-Vargas, A., & Guzmán-Vargas, L. (2021). The Complex Structure of the Pharmacological Drug–Disease Network. Entropy, 23(9), 1139. https://doi.org/10.3390/e23091139






