A Transcriptome Community-and-Module Approach of the Human Mesoconnectome
Abstract
1. Introduction
2. Materials and Methods
2.1. Database and Graph Estimation
2.2. Gene Community Analysis (Gene-Wise)
2.3. Multilayer Analysis (Module-Wise)
3. Results and Discussion
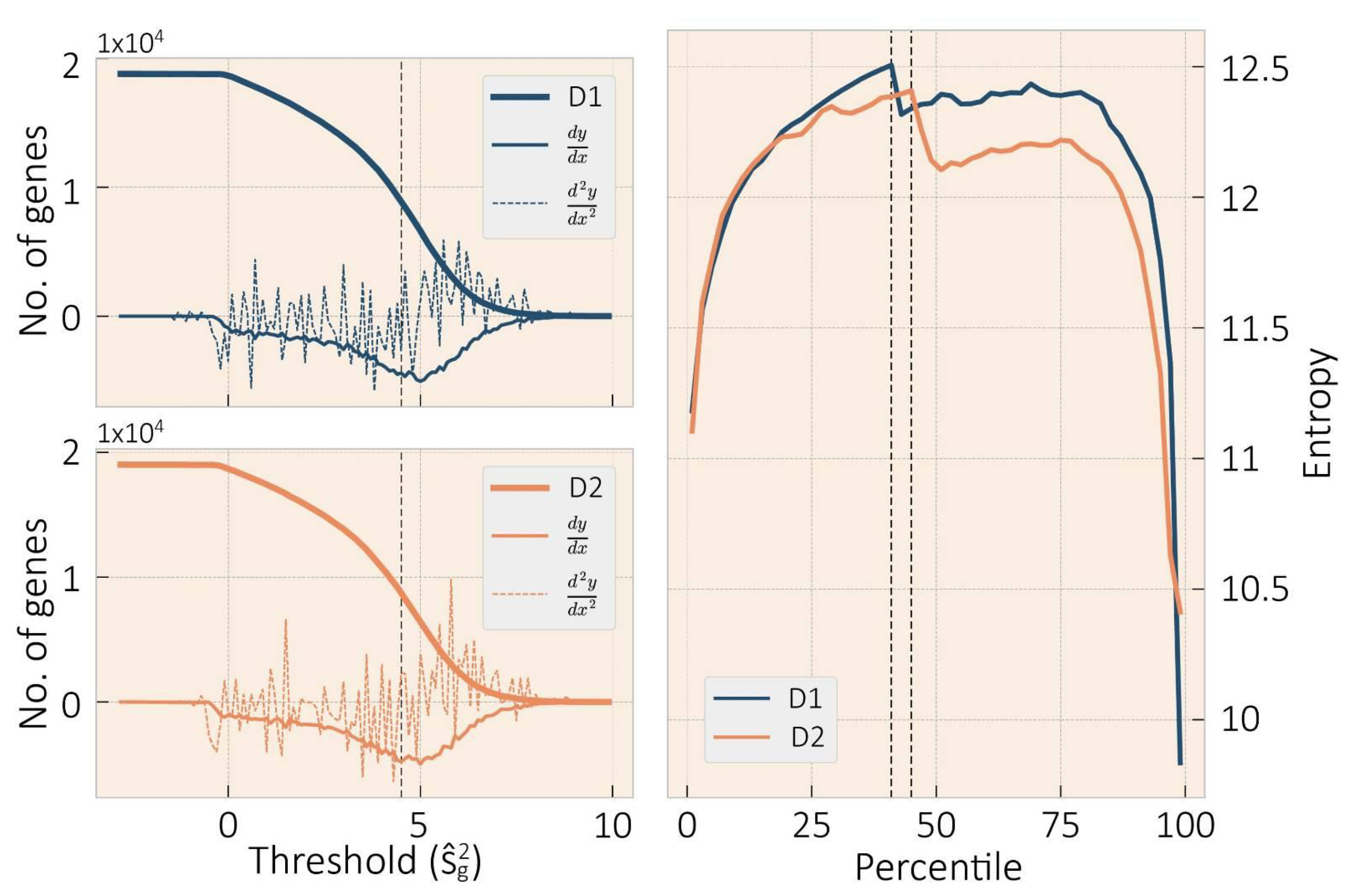
3.1. Data Preprocessing
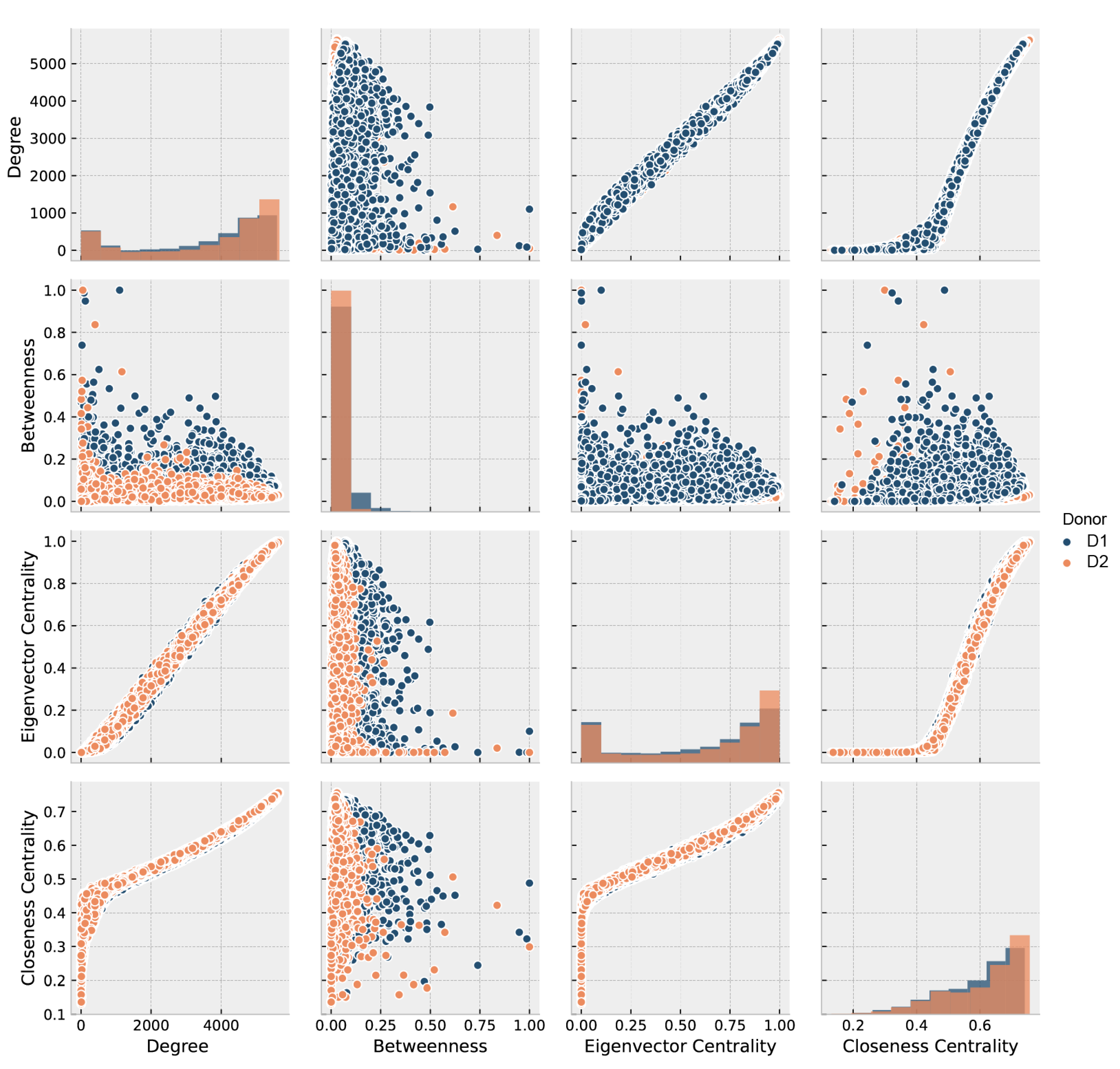
3.2. Gene-Wise Analysis
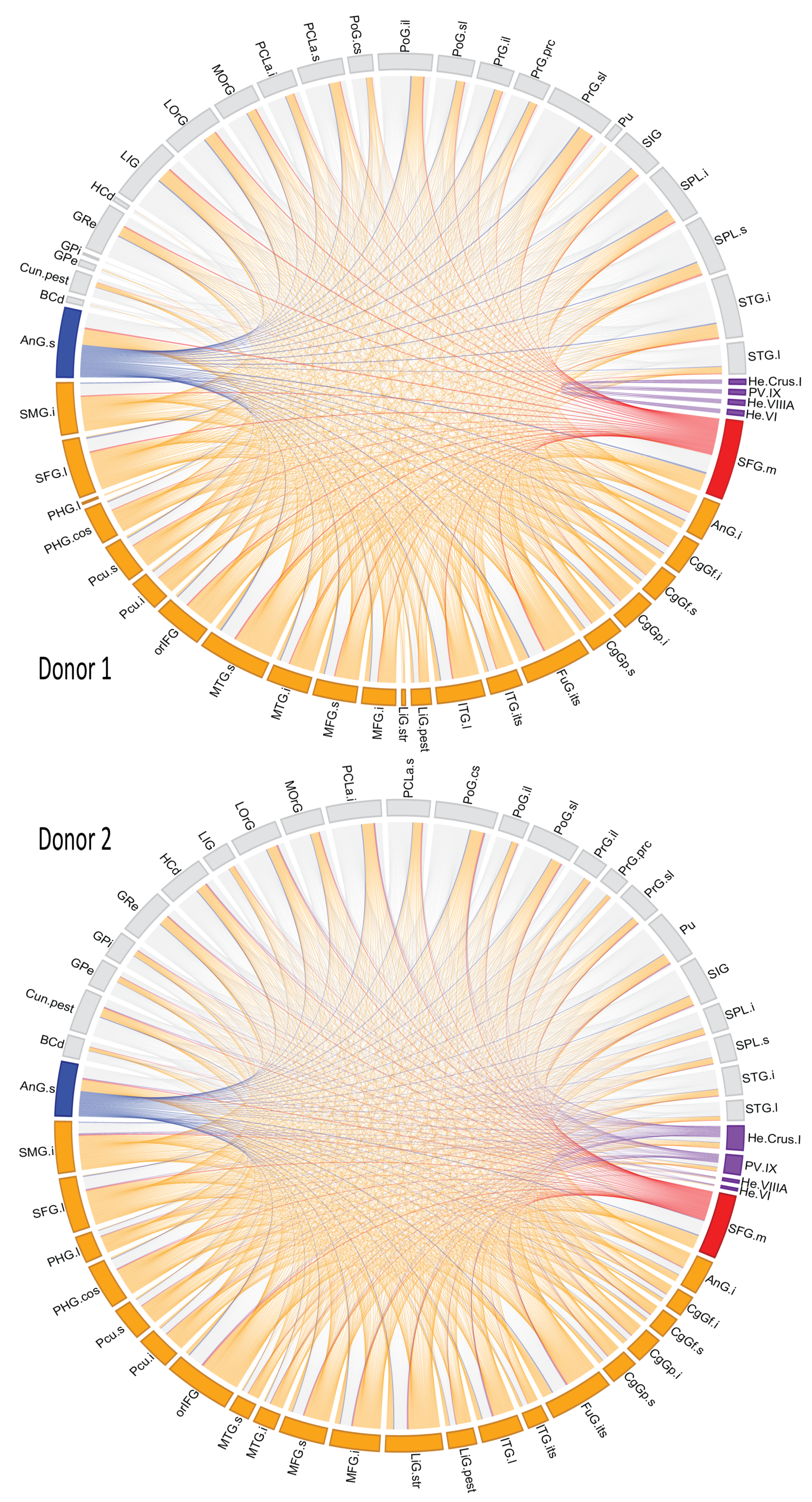
3.3. Module-Wise Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bassett, D.S.; Sporns, O. Network neuroscience. Nat. Neurosci. 2017, 20, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Betzel, R.F.; Medaglia, J.D.; Papadopoulos, L.; Baum, G.L.; Gur, R.; Gur, R.; Roalf, D.; Satterthwaite, T.D.; Bassett, D.S. The modular organization of human anatomical brain networks: Accounting for the cost of wiring. Netw. Neurosci. 2017, 1, 42–68. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O.; Betzel, R.F. Modular Brain Networks. Annu. Rev. Psychol. 2016, 67, 613–640. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.Q.; Halgren, E. A Whole-Cortex Probabilistic Diffusion Tractography Connectome. Eneuro 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Wirsich, J.; Amico, E.; Giraud, A.L.; Goñi, J.; Sadaghiani, S. Multi-timescale hybrid components of the functional brain connectome: A bimodal EEG-fMRI decomposition. Netw. Neurosci. 2020, 4, 658–677. [Google Scholar] [CrossRef] [PubMed]
- Wodeyar, A.; Cassidy, J.M.; Cramer, S.C.; Srinivasan, R. Damage to the structural connectome reflected in resting-state fMRI functional connectivity. Netw. Neurosci. 2020, 4, 1197–1218. [Google Scholar] [CrossRef] [PubMed]
- Coquelet, N.; Tiège, X.D.; Destoky, F.; Roshchupkina, L.; Bourguignon, M.; Goldman, S.; Peigneux, P.; Wens, V. Comparing MEG and high-density EEG for intrinsic functional connectivity mapping. NeuroImage 2020, 210, 116556. [Google Scholar] [CrossRef]
- Kringelbach, M.L.; Cruzat, J.; Cabral, J.; Knudsen, G.M.; Carhart-Harris, R.; Whybrow, P.C.; Logothetis, N.K.; Deco, G. Dynamic coupling of whole-brain neuronal and neurotransmitter systems. Proc. Natl. Acad. Sci. USA 2020, 117, 9566–9576. [Google Scholar] [CrossRef] [PubMed]
- Petkoski, S.; Jirsa, V.K. Transmission time delays organize the brain network synchronization. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2019, 377, 20180132. [Google Scholar] [CrossRef] [PubMed]
- Courtiol, J.; Guye, M.; Bartolomei, F.; Petkoski, S.; Jirsa, V.K. Dynamical Mechanisms of Interictal Resting-State Functional Connectivity in Epilepsy. J. Neurosci. 2020, 40, 5572–5588. [Google Scholar] [CrossRef] [PubMed]
- Jirsa, V.; Proix, T.; Perdikis, D.; Woodman, M.; Wang, H.; Gonzalez-Martinez, J.; Bernard, C.; Bénar, C.; Guye, M.; Chauvel, P.; et al. The Virtual Epileptic Patient: Individualized whole-brain models of epilepsy spread. NeuroImage 2017, 145, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Friedmann, D.; Pun, A.; Adams, E.L.; Lui, J.H.; Kebschull, J.M.; Grutzner, S.M.; Castagnola, C.; Tessier-Lavigne, M.; Luo, L. Mapping mesoscale axonal projections in the mouse brain using a 3D convolutional network. Proc. Natl. Acad. Sci. USA 2020, 117, 11068–11075. [Google Scholar] [CrossRef] [PubMed]
- Tasic, B.; Yao, Z.; Graybuck, L.T.; Smith, K.A.; Nguyen, T.N.; Bertagnolli, D.; Goldy, J.; Garren, E.; Economo, M.N.; Viswanathan, S.; et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 2018, 563, 72–78. [Google Scholar] [CrossRef]
- He, Z.; Han, D.; Efimova, O.; Guijarro, P.; Yu, Q.; Oleksiak, A.; Jiang, S.; Anokhin, K.; Velichkovsky, B.; Grünewald, S.; et al. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat. Neurosci. 2017, 20, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Bentley, B.; Branicky, R.; Barnes, C.L.; Chew, Y.L.; Yemini, E.; Bullmore, E.T.; Vértes, P.E.; Schafer, W.R. The Multilayer Connectome of Caenorhabditis elegans. PLoS Comput. Biol. 2016, 12, e1005283. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.Y.G.; Fornito, A.; Fulcher, B.D. Scaling of gene transcriptional gradients with brain size across mouse development. NeuroImage 2021, 224, 117395. [Google Scholar] [CrossRef] [PubMed]
- Huntenburg, J.M.; Yeow, L.Y.; Mandino, F.; Grandjean, J. Gradients of functional connectivity in the mouse cortex reflect neocortical evolution. NeuroImage 2021, 225, 117528. [Google Scholar] [CrossRef] [PubMed]
- Fornito, A.; Arnatkevičiūtė, A.; Fulcher, B.D. Bridging the Gap between Connectome and Transcriptome. Trends Cogn. Sci. 2019, 23, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Nord, A.S.; Pattabiraman, K.; Visel, A.; Rubenstein, J.L. Genomic Perspectives of Transcriptional Regulation in Forebrain Development. Neuron 2015, 85, 27–47. [Google Scholar] [CrossRef]
- Richiardi, J.; Altmann, A.; Milazzo, A.C. Correlated gene expression supports synchronous activity in brain networks. Science 2015, 348, 1241–1244. [Google Scholar] [CrossRef]
- Arnatkevičiūtė, A.; Fulcher, B.D.; Pocock, R.; Fornito, A. Hub connectivity, neuronal diversity, and gene expression in the Caenorhabditis elegans connectome. PLoS Comput. Biol. 2018, 14, e1005989. [Google Scholar] [CrossRef]
- Fulcher, B.D.; Fornito, A. A transcriptional signature of hub connectivity in the mouse connectome. Proc. Natl. Acad. Sci. USA 2016, 113, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Fornito, A.; Arnatkevičiūtė, A.; Fulcher, B.; Oldham, S.; Tiego, J.; Bellgrove, M. Genetic Influences on Brain Network Hubs. Biol. Psychiatry 2020, 87, S86–S87. [Google Scholar] [CrossRef]
- Arnatkevičiūtė, A.; Fulcher, B.D.; Fornito, A. Uncovering the Transcriptional Correlates of Hub Connectivity in Neural Networks. Front. Neural Circuits 2019, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Arnatkevičiūtė, A.; Fulcher, B.D.; Fornito, A. A practical guide to linking brain-wide gene expression and neuroimaging data. NeuroImage 2019, 189, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.T.; Chen, C.; Grant, A.D.; Padi, M. Generating Ensembles of Gene Regulatory Networks to Assess Robustness of Disease Modules. Front. Genet. 2021, 11, 603264. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Mendoza, L.; Monzon-Sandoval, J.; Urrutia, A.O.; Gutierrez, H. Emergence of co-expression in gene regulatory networks. PLoS ONE 2021, 16, e0247671. [Google Scholar] [CrossRef]
- de Anda-Jáuregui, G.; Alcalá-Corona, S.A.; Espinal-Enríquez, J.; Hernández-Lemus, E. Functional and transcriptional connectivity of communities in breast cancer co-expression networks. Appl. Netw. Sci. 2019, 4, 1–13. [Google Scholar] [CrossRef]
- Mohamadlou, H.; Podgorski, G.J.; Flann, N.S. Modular genetic regulatory networks increase organization during pattern formation. Biosystems 2016, 146, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Changeux, J.P.; Goulas, A.; Hilgetag, C.C. A Connectomic Hypothesis for the Hominization of the Brain. Cereb. Cortex 2020, 31, 2425–2449. [Google Scholar] [CrossRef]
- Barabási, D.L.; Barabási, A.L. A Genetic Model of the Connectome. Neuron 2020, 105, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Kovács, I.A.; Barabási, D.L.; Barabási, A.L. Uncovering the genetic blueprint of the C. elegans nervous system. Proc. Natl. Acad. Sci. USA 2020, 117, 33570–33577. [Google Scholar] [CrossRef] [PubMed]
- Betzel, R.F.; Medaglia, J.D.; Bassett, D.S. Diversity of meso-scale architecture in human and non-human connectomes. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Moreau, C.A.; Raznahan, A.; Bellec, P.; Chakravarty, M.; Thompson, P.M.; Jacquemont, S. Dissecting autism and schizophrenia through neuroimaging genomics. Brain 2021. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Jones, B.M.; Traniello, I.M.; Bukhari, S.A.; Halfon, M.S.; Hofmann, H.A.; Huang, S.; Katz, P.S.; Keagy, J.; Lynch, V.J.; et al. Behavior-related gene regulatory networks: A new level of organization in the brain. Proc. Natl. Acad. Sci. USA 2020, 117, 23270–23279. [Google Scholar] [CrossRef] [PubMed]
- Van den Heuvel, M.P.; Scholtens, L.H.; Kahn, R.S. Multiscale Neuroscience of Psychiatric Disorders. Biol. Psychiatry 2019, 86, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Allen Institute for Brain Science. Allen Human Brain Atlas. 2010. Available online: human.brain-map.org (accessed on 20 March 2020).
- Monaco, A.; Amoroso, N.; Bellantuono, L.; Lella, E.; Lombardi, A.; Monda, A.; Tateo, A.; Bellotti, R.; Tangaro, S. Shannon entropy approach reveals relevant genes in Alzheimer’s disease. PLoS ONE 2020, 14, e0226190. [Google Scholar] [CrossRef] [PubMed]
- Monaco, A.; Monda, A.; Amoroso, N.; Bertolino, A.; Blasi, G.; Carlo, P.D.; Papalino, M.; Pergola, G.; Tangaro, S.; Bellotti, R. A complex network approach reveals a pivotal substructure of genes linked to schizophrenia. PLoS ONE 2019, 13, e0190110. [Google Scholar] [CrossRef]
- Newman, M.E.J. Finding community structure in networks using the eigenvectors of matrices. Phys. Rev. E 2006, 74, 036104. [Google Scholar] [CrossRef]
- Fortunato, S. Community detection in graphs. Phys. Rep. 2010, 486, 75–174. [Google Scholar] [CrossRef]
- Brionne, A.; Juanchich, A.; Hennequet-Antier, C. ViSEAGO: A Bioconductor package for clustering biological functions using Gene Ontology and semantic similarity. BioData Min. 2019, 12, 1–13. [Google Scholar] [CrossRef]
- Wang, J.Z.; Du, Z.; Payattakool, R.; Yu, P.S.; Chen, C.F. A new method to measure the semantic similarity of GO terms. Bioinformatics 2007, 23, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Gene Ontology Semantic Similarity Analysis Using GOSemSim. In Stem Cell Transcriptional Networks; Humana: New York, NY, USA, 2020; Chapter 11; pp. 207–215. [Google Scholar]
- Yu, G.; Li, F.; Qin, Y.; Bo, X.; Wu, Y.; Wang, S. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 2010, 26, 976–978. [Google Scholar] [CrossRef]
- Domenico, M.D.; Porter, M.A.; Arenas, A. MuxViz: A tool for multilayer analysis and visualization of networks. J. Complex Netw. 2014, 3, 159–176. [Google Scholar] [CrossRef]
- Brin, S.; Page, L. The anatomy of a large-scale hypertextual Web search engine. Comput. Netw. ISDN Syst. 1998, 30, 107–117. [Google Scholar] [CrossRef]
- Domenico, M.D.; Solé-Ribalta, A.; Omodei, E.; Gómez, S.; Arenas, A. Ranking in interconnected multilayer networks reveals versatile nodes. Nat. Commun. 2015, 6, 1–6. [Google Scholar] [CrossRef]
- Azimi-Tafreshi, N.; Gómez-Gardeñes, J.; Dorogovtsev, S.N. k-core percolation on multiplex networks. Phys. Rev. E 2014, 90, 032816. [Google Scholar] [CrossRef] [PubMed]
- Mondragon, R.J.; Iacovacci, J.; Bianconi, G. Multilink communities of multiplex networks. PLoS ONE 2018, 13, e0193821. [Google Scholar] [CrossRef] [PubMed]
- Mauro, A.A.; Ghalambor, C.K. Trade-offs, Pleiotropy, and Shared Molecular Pathways: A Unified View of Constraints on Adaptation. Integr. Comp. Biol. 2020, 60, 332–347. [Google Scholar] [CrossRef]
- Watanabe, K.; Stringer, S.; Frei, O.; Mirkov, M.U.; de Leeuw, C.; Polderman, T.J.C.; van der Sluis, S.; Andreassen, O.A.; Neale, B.M.; Posthuma, D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019, 51, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Fagny, M.; Austerlitz, F. Polygenic Adaptation: Integrating Population Genetics and Gene Regulatory Networks. Trends Genet. 2021, 37, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Hermisson, J.; Schlötterer, C. Polygenic adaptation: A unifying framework to understand positive selection. Nat. Rev. Genet. 2020, 21, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Broido, A.D.; Clauset, A. Scale-free networks are rare. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Meghanathan, N. Assortativity Analysis of Real-World Network Graphs based on Centrality Metrics. Comput. Inf. Sci. 2016, 9, 7. [Google Scholar] [CrossRef]
- Meghanathan, N. Correlation Coefficient Analysis of Centrality Metrics for Complex Network Graphs. In Advances in Intelligent Systems and Computing; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 11–20. [Google Scholar]
- Zhou, B.; Lu, X.L.; Holme, P. Universal evolution patterns of degree assortativity in social networks. Soc. Netw. 2020, 63, 47–55. [Google Scholar] [CrossRef]
- Jiang, J.; Wen, S.; Yu, S.; Xiang, Y.; Zhou, W.; Hassan, H. The structure of communities in scale-free networks. Concurr. Comput. Pract. Exp. 2016, 29, e4040. [Google Scholar] [CrossRef]
- Murakami, M.; Ishikura, S.; Kominami, D.; Shimokawa, T.; Murata, M. Robustness and efficiency in interconnected networks with changes in network assortativity. Appl. Netw. Sci. 2017, 2, 6. [Google Scholar] [CrossRef]
- Imms, P.; Domínguez, J.F.; Burmester, A.; Seguin, C.; Clemente, A.; Dhollander, T.; Wilson, P.H.; Poudel, G.; Caeyenberghs, K. Navigating the link between processing speed and network communication in the human brain. Brain Struct. Funct. 2021, 226, 1281–1302. [Google Scholar] [CrossRef]
- Murphy, A.C.; Bertolero, M.A.; Papadopoulos, L.; Lydon-Staley, D.M.; Bassett, D.S. Multimodal network dynamics underpinning working memory. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Ju, H.; Bassett, D.S. Dynamic representations in networked neural systems. Nat. Neurosci. 2020, 23, 908–917. [Google Scholar] [CrossRef]
- Shimono, M.; Hatano, N. Efficient communication dynamics on macro-connectome, and the propagation speed. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zarkali, A.; McColgan, P.; Ryten, M.; Reynolds, R.; Leyland, L.A.; Lees, A.J.; Rees, G.; Weil, R.S. Differences in network controllability and regional gene expression underlie hallucinations in Parkinson’s disease. Brain 2020, 143, 3435–3448. [Google Scholar] [CrossRef] [PubMed]
- Gatt, J.M.; Burton, K.L.; Williams, L.M.; Schofield, P.R. Specific and common genes implicated across major mental disorders: A review of meta-analysis studies. J. Psychiatr. Res. 2015, 60, 1–13. [Google Scholar] [CrossRef]
- Thompson, P.M.; Ge, T.; Glahn, D.C.; Jahanshad, N.; Nichols, T.E. Genetics of the connectome. NeuroImage 2013, 80, 475–488. [Google Scholar] [CrossRef]
- Alcalá-Corona, S.A.; Espinal-Enríquez, J.; de Anda-Jáuregui, G.; Hernández-Lemus, E. The Hierarchical Modular Structure of HER2+ Breast Cancer Network. Front. Physiol. 2018, 9, 1423. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Wang, C.; Zhang, X.; Yang, Q.; Shao, R.; Lai, J.; Du, C. Highly interwoven communities of a gene regulatory network unveil topologically important genes for maize seed development. Plant J. 2017, 92, 1143–1156. [Google Scholar] [CrossRef]
- Szedlak, A.; Smith, N.; Liu, L.; Paternostro, G.; Piermarocchi, C. Evolutionary and Topological Properties of Genes and Community Structures in Human Gene Regulatory Networks. PLoS Comput. Biol. 2016, 12, e1005009. [Google Scholar] [CrossRef]
- Firoz, A.; Malik, A.; Singh, S.K.; Jha, V.; Ali, A. Comparative Analysis of Glycogene Expression in Different Mouse Tissues Using RNA-Seq Data. Int. J. Genom. 2014, 2014, 1–18. [Google Scholar] [CrossRef]
- Tunç, B.; Verma, R. Unifying Inference of Meso-Scale Structures in Networks. PLoS ONE 2015, 10, e0143133. [Google Scholar] [CrossRef]
- Kim, D.J.; Min, B.K. Rich-club in the brain’s macrostructure: Insights from graph theoretical analysis. Comput. Struct. Biotechnol. J. 2020, 18, 1761–1773. [Google Scholar] [CrossRef]
- Griffa, A.; den Heuvel, M.P.V. Rich-club circuitry: Function, evolution and vulnerability. Neurocircuitry 2018, 20, 121–131. [Google Scholar]
- Fisher, J.T.; Hopp, F.R.; Weber, R. A Practical Introduction to Network Neuroscience for Communication Researchers. Commun. Methods Meas. 2020, 15, 60–79. [Google Scholar] [CrossRef]
- Sarzynska, M.; Leicht, E.A.; Chowell, G.; Porter, M.A. Null models for community detection in spatially embedded, temporal networks. J. Complex Netw. 2015, 4, 363–406. [Google Scholar] [CrossRef][Green Version]
- Maffezzini, C.; Calvo-Garrido, J.; Wredenberg, A.; Freyer, C. Metabolic regulation of neurodifferentiation in the adult brain. Cell. Mol. Life Sci. 2020, 77, 2483–2496. [Google Scholar] [CrossRef]
- Götz, M.; Nakafuku, M.; Petrik, D. Neurogenesis in the Developing and Adult Brain—Similarities and Key Differences. Cold Spring Harb. Perspect. Biol. 2016, 8, a018853. [Google Scholar] [CrossRef]
- Zhang, J.; Jiao, J. Molecular Biomarkers for Embryonic and Adult Neural Stem Cell and Neurogenesis. BioMed Res. Int. 2015, 2015, 1–14. [Google Scholar] [CrossRef]
- Cabrera-Reyes, E.A.; Vanoye–Carlo, A.; Rodríguez-Dorantes, M.; Vázquez-Martínez, E.R.; Rivero-Segura, N.A.; Collazo-Navarrete, O.; Cerbón, M. Transcriptomic analysis reveals new hippocampal gene networks induced by prolactin. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Lizen, B.; Moens, C.; Mouheiche, J.; Sacré, T.; Ahn, M.T.; Jeannotte, L.; Salti, A.; Gofflot, F. Conditional Loss of Hoxa5 Function Early after Birth Impacts on Expression of Genes with Synaptic Function. Front. Mol. Neurosci. 2017, 10, 369. [Google Scholar] [CrossRef]
- Jové, M.; Mota-Martorell, N.; Torres, P.; Portero-Otin, M.; Ferrer, I.; Pamplona, R. New insights into human prefrontal cortex aging with a lipidomics approach. Expert Rev. Proteom. 2021, 18, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Praharaj, S.K.; Prabhu, R.V.K.; Prabhu, M.M. Lipidomics and cognitive dysfunction—A Narrative review. Turk. J. Biochem. 2020, 45. [Google Scholar] [CrossRef]
- Pfisterer, U.; Khodosevich, K. Neuronal survival in the brain: Neuron type-specific mechanisms. Cell Death Dis. 2017, 8, e2643. [Google Scholar] [CrossRef]
- Cornejo, V.H.; Luarte, A.; Couve, A. Global and local mechanisms sustain axonal proteostasis of transmembrane proteins. Traffic 2017, 18, 255–266. [Google Scholar] [CrossRef]
- Luarte, A.; Cornejo, V.H.; Bertin, F.; Gallardo, J.; Couve, A. The axonal endoplasmic reticulum: One organelle-many functions in development, maintenance, and plasticity. Dev. Neurobiol. 2017, 78, 181–208. [Google Scholar] [CrossRef] [PubMed]
- Maiti, P.; Manna, J.; Ilavazhagan, G.; Rossignol, J.; Dunbar, G.L. Molecular regulation of dendritic spine dynamics and their potential impact on synaptic plasticity and neurological diseases. Neurosci. Biobehav. Rev. 2015, 59, 208–237. [Google Scholar] [CrossRef] [PubMed]
- Tiberi, M.; Chiurchiù, V. Specialized Pro-resolving Lipid Mediators and Glial Cells: Emerging Candidates for Brain Homeostasis and Repair. Front. Cell. Neurosci. 2021, 15, 136. [Google Scholar] [CrossRef]
- Nakanishi, H.; Ni, J.; Nonaka, S.; Hayashi, Y. Microglial circadian clock regulation of microglial structural complexity, dendritic spine density and inflammatory response. Neurochem. Int. 2021, 142, 104905. [Google Scholar] [CrossRef] [PubMed]
- Vasic, V.; Barth, K.; Schmidt, M.H. Neurodegeneration and Neuro-Regeneration—Alzheimer’s Disease and Stem Cell Therapy. Int. J. Mol. Sci. 2019, 20, 4272. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.W.; Helmeste, D.M.; Leonard, B.E. Neurodegeneration, Neuroregeneration, and Neuroprotection in Psychiatric Disorders. In Modern Trends in Pharmacopsychiatry; Karger, A.G.S., Ed.; Karger Publishers: Basel, Switzerland, 2017; pp. 107–123. [Google Scholar]
- Regensburger, M.; Prots, I.; Winner, B. Adult Hippocampal Neurogenesis in Parkinson’s Disease: Impact on Neuronal Survival and Plasticity. Neural Plast. 2014, 2014, 454696. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Bähr, M.; Doeppner, T.R. From Tumor Metastasis towards Cerebral Ischemia—Extracellular Vesicles as a General Concept of Intercellular Communication Processes. Int. J. Mol. Sci. 2019, 20, 5995. [Google Scholar] [CrossRef] [PubMed]
- Plate, K.H. Mechanisms of Angiogenesis in the Brain. J. Neuropathol. Exp. Neurol. 1999, 58, 313–320. [Google Scholar] [CrossRef]
- Liddelow, S.A. Development of the choroid plexus and blood-CSF barrier. Front. Neurosci. 2015, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Lun, M.P.; Monuki, E.S.; Lehtinen, M.K. Development and functions of the choroid plexus–cerebrospinal fluid system. Nat. Rev. Neurosci. 2015, 16, 445–457. [Google Scholar] [CrossRef]
- Aydin, B.; Arga, K.Y. Co-expression Network Analysis Elucidated a Core Module in Association With Prognosis of Non-functioning Non-invasive Human Pituitary Adenoma. Front. Endocrinol. 2019, 10, 361. [Google Scholar] [CrossRef]
- Osat, S.; Radicchi, F.; Papadopoulos, F. k-core structure of real multiplex networks. Phys. Rev. Res. 2020, 2, 023176. [Google Scholar] [CrossRef]
- Malvestio, I.; Cardillo, A.; Masuda, N. Interplay between $$k$$-core and community structure in complex networks. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, L.; Yao, Z.; Yu, S. A Multi-layer Network Topology Visualization Layout Based on Louvain Community Detection. In Proceedings of the 2018 IEEE Third International Conference on Data Science in Cyberspace (DSC), Guangzhou, China, 18–21 June 2018. [Google Scholar]
- Liu, W.; Suzumura, T.; Ji, H.; Hu, G. Finding overlapping communities in multilayer networks. PLoS ONE 2018, 13, e0188747. [Google Scholar] [CrossRef] [PubMed]
- Brandes, U.; Delling, D.; Gaertler, M.; Gorke, R.; Hoefer, M.; Nikoloski, Z.; Wagner, D. On Modularity Clustering. IEEE Trans. Knowl. Data Eng. 2008, 20, 172–188. [Google Scholar] [CrossRef]
- Briggs, R.G.; Khan, A.B.; Chakraborty, A.R.; Abraham, C.J.; Anderson, C.D.; Karas, P.J.; Bonney, P.A.; Palejwala, A.H.; Conner, A.K.; O’Donoghue, D.L.; et al. Anatomy and White Matter Connections of the Superior Frontal Gyrus. Clin. Anat. 2019, 33, 823–832. [Google Scholar] [CrossRef]
- Li, W.; Qin, W.; Liu, H.; Fan, L.; Wang, J.; Jiang, T.; Yu, C. Subregions of the human superior frontal gyrus and their connections. NeuroImage 2013, 78, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, G.F.; Ralph, M.A.L.; Simons, J.S. A Unifying Account of Angular Gyrus Contributions to Episodic and Semantic Cognition. Trends Neurosci. 2021, 44, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lin, Q.; Han, Z.; He, Y.; Bi, Y. Intrinsic functional network architecture of human semantic processing: Modules and hubs. NeuroImage 2016, 132, 542–555. [Google Scholar] [CrossRef]
- Tanaka, S.; Kirino, E. Increased Functional Connectivity of the Angular Gyrus During Imagined Music Performance. Front. Hum. Neurosci. 2019, 13, 92. [Google Scholar] [CrossRef]
- Mossad, S.I.; Vandewouw, M.M.; Smith, M.L.; Taylor, M.J. The preterm social brain: Altered functional networks for Theory of Mind in very preterm children. Brain Commun. 2021, 3, fcaa237. [Google Scholar] [CrossRef] [PubMed]
- Habas, C. Functional Connectivity of the Cognitive Cerebellum. Front. Syst. Neurosci. 2021, 15, 27. [Google Scholar] [CrossRef]
- Seitzman, B.A.; Gratton, C.; Marek, S.; Raut, R.V.; Dosenbach, N.U.; Schlaggar, B.L.; Petersen, S.E.; Greene, D.J. A set of functionally-defined brain regions with improved representation of the subcortex and cerebellum. NeuroImage 2020, 206, 116290. [Google Scholar] [CrossRef] [PubMed]
- Marek, S.; Siegel, J.S.; Gordon, E.M.; Raut, R.V.; Gratton, C.; Newbold, D.J.; Ortega, M.; Laumann, T.O.; Adeyemo, B.; Miller, D.B.; et al. Spatial and Temporal Organization of the Individual Human Cerebellum. Neuron 2018, 100, 977–993.e7. [Google Scholar] [CrossRef]
- Smallwood, J.; Bernhardt, B.C.; Leech, R.; Bzdok, D.; Jefferies, E.; Margulies, D.S. The default mode network in cognition: A topographical perspective. Nat. Rev. Neurosci. 2021, 22, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Raichle, M.E. The Brain’s Default Mode Network. Annu. Rev. Neurosci. 2015, 38, 433–447. [Google Scholar] [CrossRef]
- Szinte, M.; Knapen, T. Visual Organization of the Default Network. Cerebral Cortex 2019, 30, 3518–3527. [Google Scholar] [CrossRef]
- Shen, W.; Tu, Y.; Gollub, R.L.; Ortiz, A.; Napadow, V.; Yu, S.; Wilson, G.; Park, J.; Lang, C.; Jung, M.; et al. Visual network alterations in brain functional connectivity in chronic low back pain: A resting state functional connectivity and machine learning study. Neuroimage Clin. 2019, 22, 101775. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, S.; Yin, S.; Ren, W.; He, R.; Li, J. The fronto-insular cortex causally mediates the default-mode and central-executive networks to contribute to individual cognitive performance in healthy elderly. Hum. Brain Mapp. 2018, 39, 4302–4311. [Google Scholar] [CrossRef]
- Huang, Z.; Tarnal, V.; Vlisides, P.E.; Janke, E.L.; McKinney, A.M.; Picton, P.; Mashour, G.A.; Hudetz, A.G. Anterior insula regulates brain network transitions that gate conscious access. Cell Rep. 2021, 35, 109081. [Google Scholar] [CrossRef] [PubMed]
- Fareri, D.S.; Smith, D.V.; Delgado, M.R. The influence of relationship closeness on default-mode network connectivity during social interactions. Soc. Cogn. Affect. Neurosci. 2020, 15, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Spagna, A.; Hajhajate, D.; Liu, J.; Bartolomeo, P. Visual mental imagery engages the left fusiform gyrus, but not the early visual cortex: A meta-analysis of neuroimaging evidence. Neurosci. Biobehav. Rev. 2021, 122, 201–217. [Google Scholar] [CrossRef]
- Palejwala, A.H.; O’Connor, K.P.; Milton, C.K.; Anderson, C.; Pelargos, P.; Briggs, R.G.; Conner, A.K.; O’Donoghue, D.L.; Glenn, C.A.; Sughrue, M.E. Anatomy and white matter connections of the fusiform gyrus. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Metzger, B.A.; Magnotti, J.F.; Wang, Z.; Nesbitt, E.; Karas, P.J.; Yoshor, D.; Beauchamp, M.S. Responses to Visual Speech in Human Posterior Superior Temporal Gyrus Examined with iEEG Deconvolution. J. Neurosci. 2020, 40, 6938–6948. [Google Scholar] [CrossRef] [PubMed]
- Ozker, M.; Schepers, I.M.; Magnotti, J.F.; Yoshor, D.; Beauchamp, M.S. A Double Dissociation between Anterior and Posterior Superior Temporal Gyrus for Processing Audiovisual Speech Demonstrated by Electrocorticography. J. Cogn. Neurosci. 2017, 29, 1044–1060. [Google Scholar] [CrossRef] [PubMed]
- Perri, R.L.; Berchicci, M.; Bianco, V.; Quinzi, F.; Spinelli, D.; Russo, F.D. Awareness of perception and sensory–motor integration: ERPs from the anterior insula. Brain Struct. Funct. 2018, 223, 3577–3592. [Google Scholar] [CrossRef] [PubMed]
- Bastuji, H.; Frot, M.; Perchet, C.; Hagiwara, K.; Garcia-Larrea, L. Convergence of sensory and limbic noxious input into the anterior insula and the emergence of pain from nociception. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lhomond, O.; Teasdale, N.; Simoneau, M.; Mouchnino, L. Supplementary Motor Area and Superior Parietal Lobule Restore Sensory Facilitation Prior to Stepping When a Decrease of Afferent Inputs Occurs. Front. Neurol. 2019, 9, 1132. [Google Scholar] [CrossRef] [PubMed]
- Zeharia, N.; Hofstetter, S.; Flash, T.; Amedi, A. A Whole-Body Sensory-Motor Gradient is Revealed in the Medial Wall of the Parietal Lobe. J. Neurosci. 2019, 39, 7882–7892. [Google Scholar] [CrossRef] [PubMed]
- Medaglia, J.D.; Harvey, D.Y.; White, N.; Kelkar, A.; Zimmerman, J.; Bassett, D.S.; Hamilton, R.H. Network Controllability in the Inferior Frontal Gyrus Relates to Controlled Language Variability and Susceptibility to TMS. J. Neurosci. 2018, 38, 6399–6410. [Google Scholar] [CrossRef] [PubMed]
- Aminoff, E.M.; Kveraga, K.; Bar, M. The role of the parahippocampal cortex in cognition. Trends Cogn. Sci. 2013, 17, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Takahashi, K.; Liu, F.C. FOXP Genes, Neural Development, Speech and Language Disorders. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2009; pp. 117–129. [Google Scholar]
- Krick, C.M.; Argstatter, H.; Grapp, M.; Plinkert, P.K.; Reith, W. Heidelberg Neuro-Music Therapy Restores Attention-Related Activity in the Angular Gyrus in Chronic Tinnitus Patients. Front. Neurosci. 2017, 11, 418. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ou, Y.; Su, Q.; Pan, P.; Shan, X.; Chen, J.; Liu, F.; Zhang, Z.; Zhao, J.; Guo, W. Enhanced Global-Brain Functional Connectivity in the Left Superior Frontal Gyrus as a Possible Endophenotype for Schizophrenia. Front. Neurosci. 2019, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, R. Minimal Selfhood and the Origins of Consciousness, 1st ed.; BoD–Books on Demand: Hamburg, Germany, 2018. [Google Scholar]





| Donor | Community | # Genes | Donor | Community | # Genes |
|---|---|---|---|---|---|
| D1 | D1-C1 | 1138 | D2 | D2-C1 | 889 |
| D1-C2 | 1356 | D2-C2 | 1127 | ||
| D1-C3 | 1830 | D2-C3 | 862 | ||
| D1-C4 | 1841 | D2-C4 | 872 | ||
| D1-C5 | 753 | D2-C5 | 1893 | ||
| D1-C6 | 941 | D2-C6 | 887 | ||
| D2-C7 | 1141 |
| Layer | Modularity | |
|---|---|---|
| D1 | D2 | |
| 1 | 0.241 | 0.222 |
| 2 | 0.229 | 0.307 |
| 3 | 0.18 | 0.341 |
| 4 | 0.222 | 0.178 |
| 5 | 0.193 | 0.236 |
| 6 | 0.34 | 0.261 |
| 7 | 0.241 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paredes, O.; López, J.B.; Covantes-Osuna, C.; Ocegueda-Hernández, V.; Romo-Vázquez, R.; Morales, J.A. A Transcriptome Community-and-Module Approach of the Human Mesoconnectome. Entropy 2021, 23, 1031. https://doi.org/10.3390/e23081031
Paredes O, López JB, Covantes-Osuna C, Ocegueda-Hernández V, Romo-Vázquez R, Morales JA. A Transcriptome Community-and-Module Approach of the Human Mesoconnectome. Entropy. 2021; 23(8):1031. https://doi.org/10.3390/e23081031
Chicago/Turabian StyleParedes, Omar, Jhonatan B. López, César Covantes-Osuna, Vladimir Ocegueda-Hernández, Rebeca Romo-Vázquez, and J. Alejandro Morales. 2021. "A Transcriptome Community-and-Module Approach of the Human Mesoconnectome" Entropy 23, no. 8: 1031. https://doi.org/10.3390/e23081031
APA StyleParedes, O., López, J. B., Covantes-Osuna, C., Ocegueda-Hernández, V., Romo-Vázquez, R., & Morales, J. A. (2021). A Transcriptome Community-and-Module Approach of the Human Mesoconnectome. Entropy, 23(8), 1031. https://doi.org/10.3390/e23081031






