Neural Dynamics under Active Inference: Plausibility and Efficiency of Information Processing
Abstract
1. Introduction
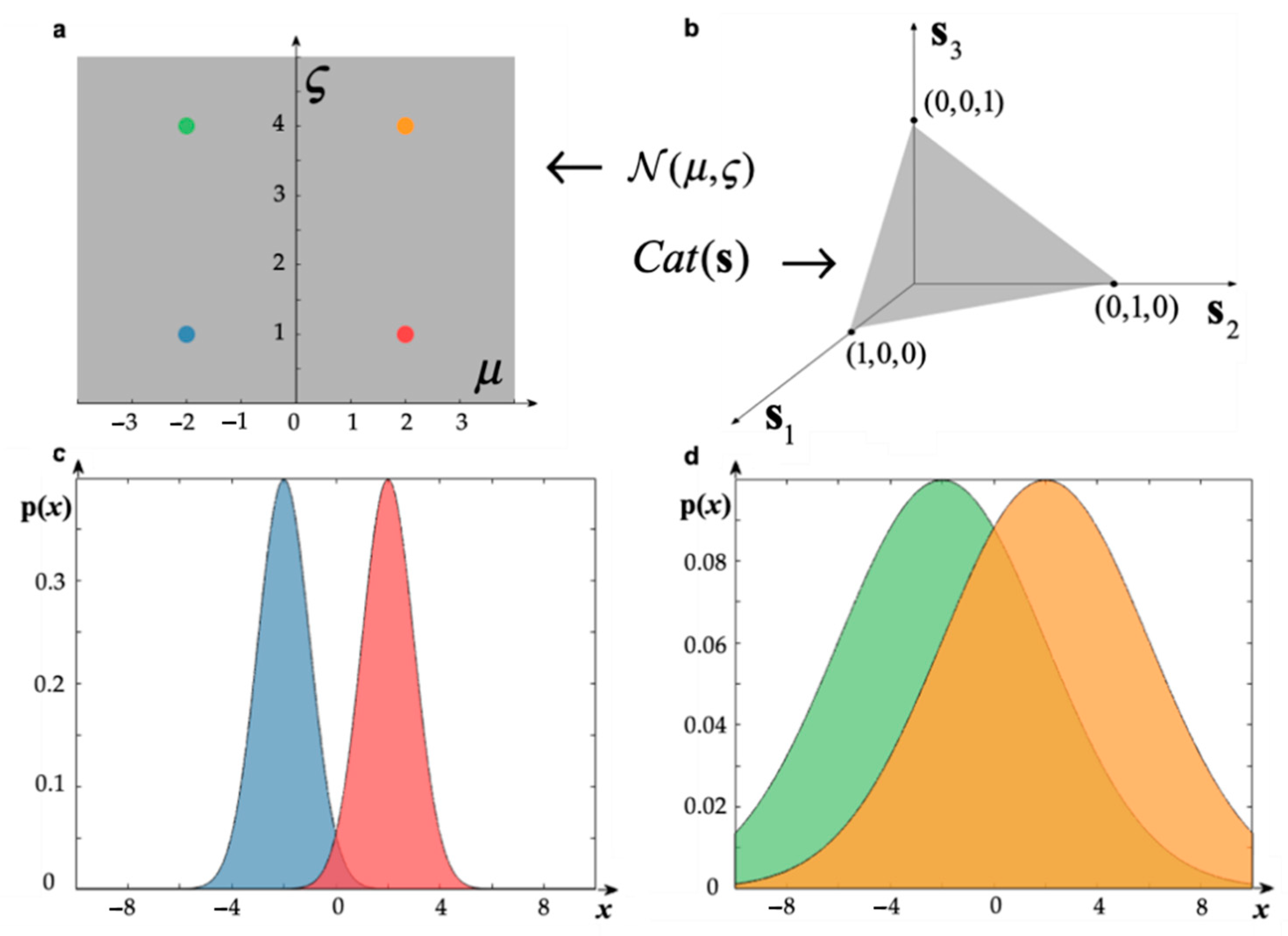
2. The Softmax Activation Function in Neural Population Dynamics
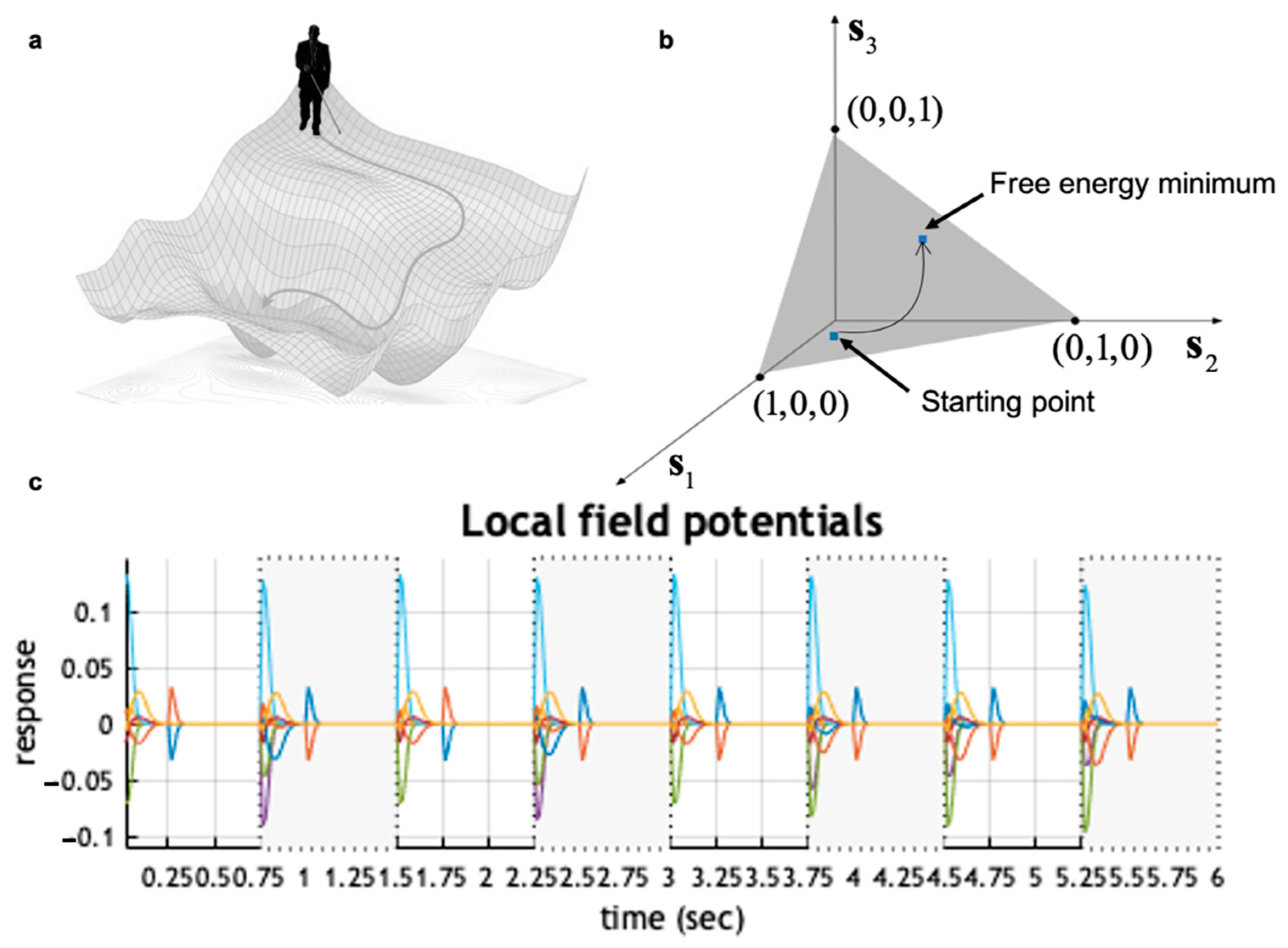
3. Neural Dynamics of Perceptual Inference
4. A Primer on Information Geometry and Natural Gradient Descent
5. Active Inference Approximates Natural Gradient Descent
6. Numerical Simulations
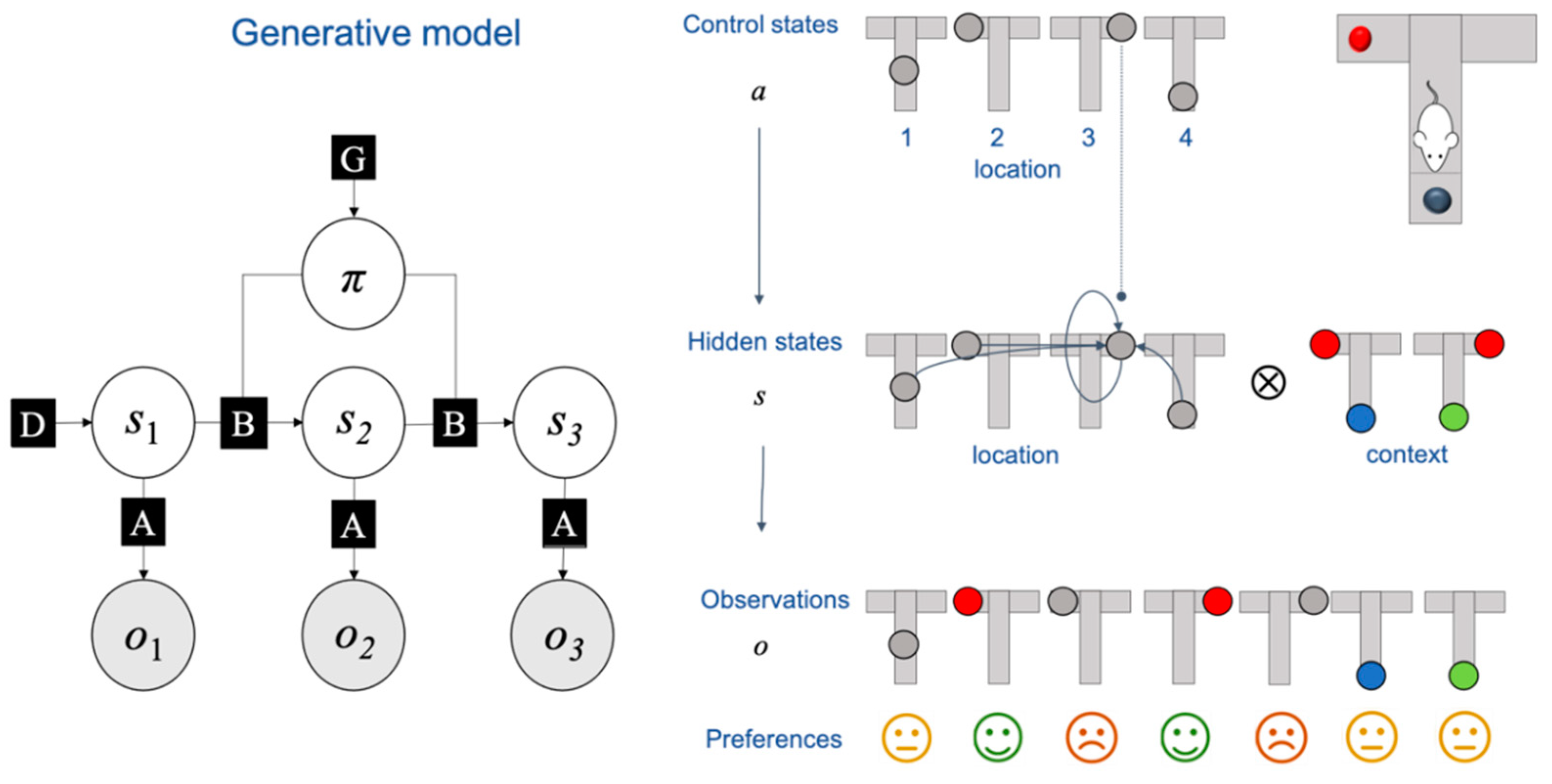
6.1. Methodology
- Observation: The agent receives an observation;
- Perceptual inference: The agent infers hidden states from observations by minimizing free energy;
- Decision-making: The agent executes the action with the lowest expected free energy.
- Learning: The agent learns the generative model by accumulating data on hidden state transitions and on the likelihood mapping between states and outcomes.
6.1.1. Overview of Tasks
6.1.2. Two Schemes
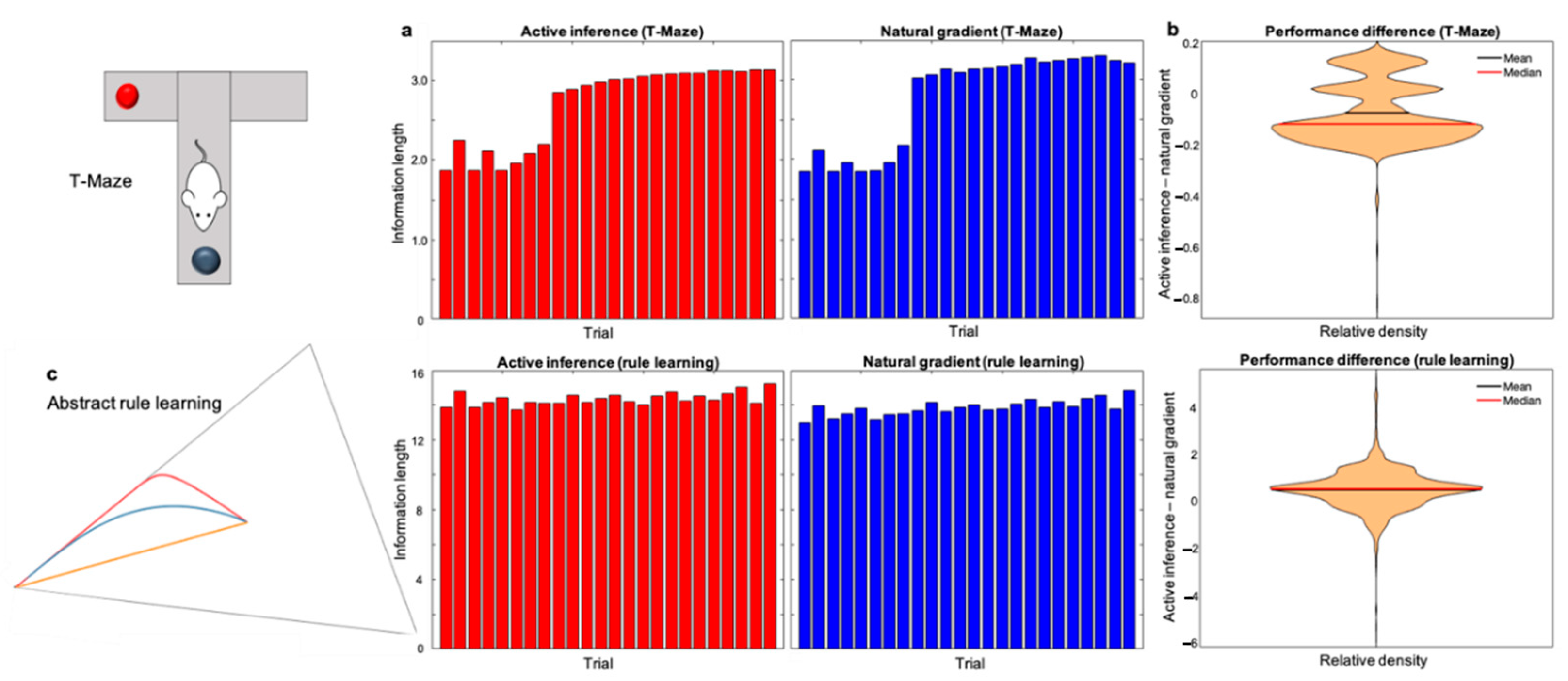
6.1.3. Measuring Information Length
6.2. Results
7. Discussion
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Software Note
Appendix A. Convexity of the Free Energy
- is convex in the interval , which implies that is convex.
- is a linear function, hence it is convex.
- only has positive components, hence is a positive linear combination of polynomials of degree two, which is convex.
Appendix B. Fisher Information Metric Tensor, Information Length and Information Distance
Appendix C. Fisher Information Metric Tensor on the Simplex
Appendix D. Geodesics on the Simplex
Appendix E. Information Distance on the Simplex
References
- Bogacz, R. A tutorial on the free-energy framework for modelling perception and learning. J. Math. Psychol. 2017, 76, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Friston, K. The free-energy principle: A rough guide to the brain? Trends Cogn. Sci. 2009, 13, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Friston, K.; Kilner, J.; Harrison, L. A free energy principle for the brain. J. Physiol. 2006, 100, 70–87. [Google Scholar] [CrossRef] [PubMed]
- Friston, K.J. Life as we know it. J. R. Soc. Interface 2013, 10, 20130475. [Google Scholar] [CrossRef] [PubMed]
- Friston, K. A Free Energy Principle for a Particular Physics. arXiv:190610184 [q-bio]. 2019. Available online: http://arxiv.org/abs/1906.10184 (accessed on 29 February 2020).
- Parr, T.; Da Costa, L.; Friston, K. Markov blankets, information geometry and stochastic thermodynamics. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2020, 378, 20190159. [Google Scholar] [CrossRef]
- Da Costa, L.; Parr, T.; Sajid, N.; Veselic, S.; Neacsu, V.; Friston, K. Active inference on discrete state-spaces: A synthesis. J. Math. Psychol. 2020, 99, 102447. [Google Scholar] [CrossRef]
- Friston, K. A theory of cortical responses. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 815–836. [Google Scholar] [CrossRef] [PubMed]
- Beal, M.J. Variational Algorithms for Approximate Bayesian Inference. Ph.D. Thesis, University of London, London, UK, May 2003; p. 281. [Google Scholar]
- Jordan, M.I.; Ghahramani, Z.; Jaakkola, T.S.; Saul, L.K. An Introduction to Variational Methods for Graphical Models. In Learning in Graphical Models; Jordan, M.I., Ed.; Springer: Dordrecht, The Netherlands, 1998; pp. 105–161. [Google Scholar] [CrossRef]
- Wainwright, M.J.; Jordan, M.I. Graphical Models, Exponential Families, and Variational Inference. FNT Mach. Learn. 2007, 1, 1–305. [Google Scholar] [CrossRef]
- Buckley, C.L.; Kim, C.S.; McGregor, S.; Seth, A.K. The free energy principle for action and perception: A mathematical review. J. Math. Psychol. 2017, 81, 55–79. [Google Scholar] [CrossRef]
- Friston, K. The free-energy principle: A unified brain theory? Nat. Rev. Neurosci. 2010, 11, 127–138. [Google Scholar] [CrossRef]
- Colombo, M.; Wright, C. First principles in the life sciences: The free-energy principle, organicism, and mechanism. Synthese 2018. [Google Scholar] [CrossRef]
- Kappen, H.J.; Gómez, V.; Opper, M. Optimal control as a graphical model inference problem. Mach. Learn. 2012, 87, 159–182. [Google Scholar] [CrossRef]
- Da Costa, L.; Sajid, N.; Parr, T.; Friston, K.; Smith, R. The Relationship between Dynamic Programming and Active Inference: The Discrete, Finite-Horizon Case. arXiv:200908111 [cs, math, q-bio]. 2020. Available online: http://arxiv.org/abs/2009.08111 (accessed on 31 January 2021).
- Millidge, B.; Tschantz, A.; Seth, A.K.; Buckley, C.L. On the Relationship between Active Inference and Control as Inference. arXiv:200612964 [cs, stat]. 2020. Available online: http://arxiv.org/abs/2006.12964 (accessed on 28 June 2020).
- Watson, J.; Imohiosen, A.; Peters, J. Active Inference or Control as Inference? A Unifying View. arXiv:201000262 [cs, stat]. 2020. Available online: http://arxiv.org/abs/2010.00262 (accessed on 27 January 2021).
- Aitchison, L.; Lengyel, M. With or without you: Predictive coding and Bayesian inference in the brain. Curr. Opin. Neurobiol. 2017, 46, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Knill, D.C.; Pouget, A. The Bayesian brain: The role of uncertainty in neural coding and computation. Trends Neurosci. 2004, 27, 712–719. [Google Scholar] [CrossRef]
- Lake, B.M.; Ullman, T.D.; Tenenbaum, J.B.; Gershman, S.J. Building Machines That Learn and Think Like People. arXiv:160400289 [cs, stat]. 2016. Available online: http://arxiv.org/abs/1604.00289 (accessed on 11 August 2019).
- Rao, R.P.N.; Ballard, D.H. Predictive coding in the visual cortex: A functional interpretation of some extra-classical receptive-field effects. Nat. Neurosci. 1999, 2, 79–87. [Google Scholar] [CrossRef]
- Bastos, A.M.; Usrey, W.M.; Adams, R.A.; Mangun, G.R.; Fries, P.; Friston, K.J. Canonical Microcircuits for Predictive Coding. Neuron 2012, 76, 695–711. [Google Scholar] [CrossRef] [PubMed]
- Friston, K.; Kiebel, S. Predictive coding under the free-energy principle. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 1211–1221. [Google Scholar] [CrossRef]
- Kaplan, R.; Friston, K.J. Planning and navigation as active inference. Biol. Cybern. 2018, 112, 323–343. [Google Scholar] [CrossRef] [PubMed]
- Pezzulo, G. An Active Inference view of cognitive control. Front. Psychol. 2012, 3, 478. [Google Scholar] [CrossRef]
- Schwöbel, S.; Kiebel, S.; Marković, D. Active Inference, Belief Propagation, and the Bethe Approximation. Neural Comput. 2018, 30, 2530–2567. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Tani, J. Goal-Directed Planning for Habituated Agents by Active Inference Using a Variational Recurrent Neural Network. Entropy 2020, 22, 564. [Google Scholar] [CrossRef]
- Çatal, O.; Verbelen, T.; Nauta, J.; Boom, C.D.; Dhoedt, B. Learning Perception and Planning With Deep Active Inference. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 3952–3956. [Google Scholar] [CrossRef][Green Version]
- Çatal, O.; Wauthier, S.; Verbelen, T.; De Boom, C.; Dhoedt, B. Deep Active Inference for Autonomous Robot Navigation. arXiv:200303220. 2020. Available online: http://arxiv.org/abs/2003.03220 (accessed on 22 May 2020).
- Sancaktar, C.; van Gerven, M.; Lanillos, P. End-to-End Pixel-Based Deep Active Inference for Body Perception and Action. arXiv:200105847 [cs, q-bio]. 2020 [cited 18 Sep 2020]. Available online: http://arxiv.org/abs/2001.05847 (accessed on 18 September 2020).
- Tschantz, A.; Seth, A.K.; Buckley, C.L. Learning action-oriented models through active inference. PLoS Comput. Biol. 2020, 16, e1007805. [Google Scholar] [CrossRef] [PubMed]
- Schwartenbeck, P.; Passecker, J.; Hauser, T.U.; Fitzgerald, T.H.; Kronbichler, M.; Friston, K.J. Computational mechanisms of curiosity and goal-directed exploration. eLife 2019, 8, 45. [Google Scholar] [CrossRef]
- Tschantz, A.; Millidge, B.; Seth, A.K.; Buckley, C.L. Reinforcement Learning through Active Inference. ICLR. 2020. Available online: http://arxiv.org/abs/2002.12636 (accessed on 9 April 2021).
- Marković, D.; Goschke, T.; Kiebel, S.J. Meta-control of the exploration-exploitation dilemma emerges from probabilistic inference over a hierarchy of time scales. Cogn. Affect. Behav. Neurosci. 2020. [Google Scholar] [CrossRef]
- Friston, K.; Da Costa, L.; Hafner, D.; Hesp, C.; Parr, T. Sophisticated Inference. Neural Comput. 2021, 33, 713–763. [Google Scholar] [CrossRef]
- Lanillos, P.G. Adaptive Robot Body Learning and Estimation Through Predictive Coding. In Proceedings of the2018 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Madrid, Spain, 1–5 October 2018; pp. 4083–4090. [Google Scholar] [CrossRef]
- Lanillos, P.; Pages, J.; Cheng, G. Robot Self/Other Distinction: Active Inference Meets Neural Networks Learning in a Mirror. arXiv:200405473 [cs]. 2020. Available online: http://arxiv.org/abs/2004.05473 (accessed on 18 September 2020).
- Friston, K.J.; Lin, M.; Frith, C.D.; Pezzulo, G.; Hobson, J.A.; Ondobaka, S. Active Inference, Curiosity and Insight. Neural Comput. 2017, 29, 2633–2683. [Google Scholar] [CrossRef]
- Smith, R.; Schwartenbeck, P.; Parr, T.; Friston, K.J. An Active Inference Approach to Modeling Structure Learning: Concept Learning as an Example Case. Front. Comput. Neurosci. 2020, 14. [Google Scholar] [CrossRef]
- Wauthier, S.T.; Çatal, O.; Verbelen, T.; Dhoedt, B. Sleep: Model Reduction. In Deep Active Inference; Springer: Cham, Switzerland, 2020; p. 13. [Google Scholar]
- Cullen, M.; Davey, B.; Friston, K.J.; Moran, R.J. Active Inference in OpenAI Gym: A Paradigm for Computational Investigations into Psychiatric Illness. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2018, 3, 809–818. [Google Scholar] [CrossRef]
- Fountas, Z.; Sajid, N.; Mediano, P.A.M.; Friston, K. Deep Active Inference Agents Using Monte-Carlo Methods. arXiv:200604176 [cs, q-bio, stat]. 2020. Available online: http://arxiv.org/abs/2006.04176 (accessed on 16 July 2020).
- Ueltzhöffer, K. Deep active inference. Biol. Cybern. 2018, 112, 547–573. [Google Scholar] [CrossRef]
- Marković, D.; Reiter, A.M.F.; Kiebel, S.J. Predicting change: Approximate inference under explicit representation of temporal structure in changing environments. PLoS Comput. Biol. 2019, 15, e1006707. [Google Scholar] [CrossRef] [PubMed]
- Markovic, D.; Stojic, H.; Schwoebel, S.; Kiebel, S.J. An Empirical Evaluation of Active Inference in Multi-Armed Bandits. arXiv:210108699 [cs]. 2021. Available online: http://arxiv.org/abs/2101.08699 (accessed on 26 January 2021).
- Sajid, N.; Ball, P.J.; Friston, K.J. Active Inference: Demystified and Compared. arXiv:190910863 [cs, q-bio]. 2020. Available online: http://arxiv.org/abs/1909.10863 (accessed on 30 April 2020).
- Parr, T.; Friston, K.J. Active inference and the anatomy of oculomotion. Neuropsychologia 2018, 111, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Parr, T. The Computational Neurology of Active Vision. Ph.D. Thesis, University College London, London, UK, 2019. [Google Scholar]
- Mirza, M.B.; Adams, R.A.; Mathys, C.D.; Friston, K.J. Scene Construction, Visual Foraging, and Active Inference. Front. Comput. Neurosci. 2016, 10, 56. [Google Scholar] [CrossRef]
- Parr, T.; Friston, K.J. Uncertainty, epistemics and active inference. J. R. Soc. Interface 2017, 14, 20170376. [Google Scholar] [CrossRef]
- Adams, R.A.; Stephan, K.E.; Brown, H.R.; Frith, C.D.; Friston, K.J. The Computational Anatomy of Psychosis. Front. Psychiatry 2013, 4. [Google Scholar] [CrossRef]
- Smith, R.; Kirlic, N.; Stewart, J.L.; Touthang, J.; Kuplicki, R.; Khalsa, S.S.; Feinstein, J.; Paulus, M.P.; Aupperle, R.L. Greater decision uncertainty characterizes a transdiagnostic patient sample during approach-avoidance conflict: A computational modeling approach. PsyArXiv 2020. [Google Scholar] [CrossRef]
- Smith, R.; Schwartenbeck, P.; Stewart, J.L.; Kuplicki, R.; Ekhtiari, H.; Investigators, T.; Martin, P. Imprecise Action Selection in Substance Use Disorder: Evidence for Active Learning Impairments When Solving the Explore-Exploit Dilemma. PsyArXiv 2020. [Google Scholar] [CrossRef]
- Smith, R.; Kuplicki, R.; Feinstein, J.; Forthman, K.L.; Stewart, J.L.; Paulus, M.P.; Tulsa 1000 Investigators; Khalsa, S.S. A Bayesian computational model reveals a failure to adapt interoceptive precision estimates across depression, anxiety, eating, and substance use disorders. PLoS Comput. Biol. 2020, 16, e1008484. [Google Scholar] [CrossRef]
- Millidge, B. Deep active inference as variational policy gradients. J. Math. Psychol. 2020, 96, 102348. [Google Scholar] [CrossRef]
- Friston, K.J.; Fitzgerald, T.H.B.; Rigoli, F.; Schwartenbeck, P.; Pezzulo, G. Active Inference: A Process Theory. Neural Comput. 2017, 29, 1–49. [Google Scholar] [CrossRef]
- Parr, T.; Friston, K.J. The Anatomy of Inference: Generative Models and Brain Structure. Front. Comput. Neurosci. 2018, 12, 90. [Google Scholar] [CrossRef] [PubMed]
- Schwartenbeck, P.; Fitzgerald, T.H.B.; Mathys, C.; Dolan, R.; Friston, K. The Dopaminergic Midbrain Encodes the Expected Certainty about Desired Outcomes. Cereb. Cortex 2015, 25, 3434–3445. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, T.H.B.; Dolan, R.J.; Friston, K. Dopamine, reward learning, and active inference. Front. Comput. Neurosci. 2015, 9. [Google Scholar] [CrossRef] [PubMed]
- Schwartenbeck, P.; Fitzgerald, T.; Dolan, R.J.; Friston, K.J. Exploration, novelty, surprise, and free energy minimization. Front. Psychol. 2013, 4, 710. [Google Scholar] [CrossRef]
- Schwartenbeck, P.; Fitzgerald, T.H.B.; Mathys, C.; Dolan, R.; Kronbichler, M.; Friston, K. Evidence for surprise minimization over value maximization in choice behavior. Sci. Rep. 2015, 5, 16575. [Google Scholar] [CrossRef]
- Sengupta, B.; Stemmler, M.B.; Friston, K.J. Information and Efficiency in the Nervous System—A Synthesis. PLoS Comput. Biol. 2013, 9, e1003157. [Google Scholar] [CrossRef]
- Levy, W.B.; Baxter, R.A. Energy Efficient Neural Codes. Neural Comput. 1996, 8, 531–543. [Google Scholar] [CrossRef]
- Dan, Y.; Atick, J.J.; Reid, R.C. Efficient Coding of Natural Scenes in the Lateral Geniculate Nucleus: Experimental Test of a Computational Theory. J. Neurosci. 1996, 16, 3351–3362. [Google Scholar] [CrossRef]
- Lewicki, M.S. Efficient coding of natural sounds. Nat. Neurosci. 2002, 5, 356–363. [Google Scholar] [CrossRef]
- Chen, B.L.; Hall, D.H.; Chklovskii, D.B. Wiring optimization can relate neuronal structure and function. Proc. Natl. Acad. Sci. USA 2006, 103, 4723–4728. [Google Scholar] [CrossRef]
- Raj, A.; Chen, Y.-H. The Wiring Economy Principle: Connectivity Determines Anatomy in the Human Brain. PLoS ONE 2011, 6, e14832. [Google Scholar] [CrossRef][Green Version]
- Barlow, H. Redundancy reduction revisited. Comput. Neural Syst. 2001, 12, 241–253. [Google Scholar] [CrossRef]
- Barlow, H.B. Possible Principles Underlying the Transformations of Sensory Messages; The MIT Press: 1961. Available online: https://www.universitypressscholarship.com/view/10.7551/mitpress/9780262518420.001.0001/upso-9780262518420-chapter-13 (accessed on 9 April 2021).
- Binder, M.D.; Hirokawa, N.; Windhorst, U. (Eds.) Efficient Coding Hypothesis. Encyclopedia of Neuroscience; Springer: Berlin/Heidelberg, Germany, 2009; p. 1037. [Google Scholar] [CrossRef]
- Denève, S.; Machens, C.K. Efficient codes and balanced networks. Nat. Neurosci. 2016, 19, 375–382. [Google Scholar] [CrossRef]
- Chelaru, M.I.; Dragoi, V. Efficient coding in heterogeneous neuronal populations. Proc. Natl. Acad. Sci. USA 2008, 105, 16344–16349. [Google Scholar] [CrossRef] [PubMed]
- Kostal, L.; Lánský, P.; Rospars, J.-P. Efficient Olfactory Coding in the Pheromone Receptor Neuron of a Moth. PLoS Comput. Biol. 2008, 4, e1000053. [Google Scholar] [CrossRef]
- Olshausen, B.A.; Field, D.J. Natural image statistics and efficient coding. Net. Comput. Neural Syst. 1996, 7, 333–339. [Google Scholar] [CrossRef]
- Olshausen, B.A.; O’Connor, K.N. A new window on sound. Nat. Neurosci. 2002, 5, 292–294. [Google Scholar] [CrossRef]
- Simoncelli, E.P.; Olshausen, B.A. Natural Image Statistics and Neural Representation. Annu. Rev. Neurosci. 2001, 24, 1193–1216. [Google Scholar] [CrossRef]
- Karklin, Y.; Simoncelli, E.P. Efficient coding of natural images with a population of noisy Linear-Nonlinear neurons. In Advances in Neural Information Processing Systems 24; Shawe-Taylor, J., Zemel, R.S., Bartlett, P.L., Pereira, F., Weinberger, K.Q., Eds.; Curran Associates, Inc.: New York, NY, USA, 2011; pp. 999–1007. [Google Scholar]
- Olshausen, B.A.; Field, D.J. Emergence of simple-cell receptive field properties by learning a sparse code for natural images. Nat. Cell Biol. 1996, 381, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Olshausen, B.A.; Field, D.J. Vision and the Coding of Natural Images: The human brain may hold the secrets to the best image-compression algorithms. Am. Sci. 2000, 88, 238–245. [Google Scholar] [CrossRef]
- Bennett, C.H. Notes on Landauer’s principle, reversible computation, and Maxwell’s Demon. Stud. Hist. Philos. Sci. Part B: Stud. Hist. Philos. Mod. Phys. 2003, 34, 501–510. [Google Scholar] [CrossRef]
- Landauer, R. Irreversibility and Heat Generation in the Computing Process. IBM J. Res. Dev. 1961, 5, 183–191. [Google Scholar] [CrossRef]
- Ito, S. Stochastic Thermodynamic Interpretation of Information Geometry. Phys. Rev. Lett. 2018, 121, 030605. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E. Measuring Thermodynamic Length. Phys. Rev. Lett. 2007, 99, 100602. [Google Scholar] [CrossRef]
- Amari, S.-I. Natural Gradient Works Efficiently in Learning. Neural Comput. 1998, 10, 251–276. [Google Scholar] [CrossRef]
- Wilson, H.R.; Cowan, J.D. A mathematical theory of the functional dynamics of cortical and thalamic nervous tissue. Biol. Cybern. 1973, 13, 55–80. [Google Scholar] [CrossRef]
- Fricker, D.; Verheugen, J.A.H.; Miles, R. Cell-attached measurements of the firing threshold of rat hippocampal neurones. J. Physiol. 1999, 517, 791–804. [Google Scholar] [CrossRef]
- Marreiros, A.C.; Daunizeau, J.; Kiebel, S.J.; Friston, K.J. Population dynamics: Variance and the sigmoid activation function. NeuroImage 2008, 42, 147–157. [Google Scholar] [CrossRef]
- Marreiros, A.C.; Kiebel, S.J.; Daunizeau, J.; Harrison, L.M.; Friston, K.J. Population dynamics under the Laplace assumption. NeuroImage 2009, 44, 701–714. [Google Scholar] [CrossRef]
- Friston, K.J.; Harrison, L.; Penny, W. Dynamic causal modelling. NeuroImage 2003, 19, 1273–1302. [Google Scholar] [CrossRef]
- Deco, G.; Jirsa, V.K.; Robinson, P.A.; Breakspear, M.; Friston, K. The Dynamic Brain: From Spiking Neurons to Neural Masses and Cortical Fields. PLoS Comput. Biol. 2008, 4, e1000092. [Google Scholar] [CrossRef]
- Moran, R.J.; Pinotsis, D.A.; Friston, K.J. Neural masses and fields in dynamic causal modeling. Front. Comput. Neurosci. 2013, 7, 57. [Google Scholar] [CrossRef]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006. [Google Scholar]
- Stone, J.V. Artificial Intelligence Engines: A Tutorial Introduction to the Mathematics of Deep Learning; Sebtel Press: Warszawa, Poland, 2019. [Google Scholar]
- Engel, J. Polytomous logistic regression. Stat. Neerlandica 1988, 42, 233–252. [Google Scholar] [CrossRef]
- Huang, F.-L.; Hsieh, C.-J.; Chang, K.-W.; Lin, C.-J. Iterative scaling and coordinate descent methods for maximum entropy. ACL-IJCNLP 2009 Conf. Short Pap. 2009. [Google Scholar] [CrossRef]
- Rodríguez, G. Lecture Notes on Generalized Linear Models. Available online: https://data.princeton.edu/wws509/notes/ (accessed on 9 April 2021).
- Åström, K. Optimal control of Markov processes with incomplete state information. J. Math. Anal. Appl. 1965, 10, 174–205. [Google Scholar] [CrossRef]
- Kullback, S.; Leibler, R.A. On Information and Sufficiency. Ann Math Statist. 1951, 22, 79–86. [Google Scholar] [CrossRef]
- Joyce, J.M. Kullback-Leibler Divergence. International Encyclopedia of Statistical Science; Lovric, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 720–722. [Google Scholar] [CrossRef]
- Rezende, D.J. Short Notes on Divergence Measures. 2018. Available online: https://danilorezende.com/wp-content/uploads/2018/07/divergences.pdf (accessed on 9 April 2021).
- Stachenfeld, K.L.; Botvinick, M.M.; Gershman, S.J. The hippocampus as a predictive map. Nat. Neurosci. 2017, 20, 1643–1653. [Google Scholar] [CrossRef]
- Hafting, T.; Fyhn, M.; Molden, S.; Moser, M.-B.; Moser, E.I. Microstructure of a spatial map in the entorhinal cortex. Nat. Cell Biol. 2005, 436, 801–806. [Google Scholar] [CrossRef]
- Chen, L.L.; Lin, L.-H.; Green, E.J.; Barnes, C.A.; McNaughton, L.A. Head-direction cells in the rat posterior cortex. Exp. Brain Res. 1994, 16, 8–23. [Google Scholar] [CrossRef]
- Taube, J.; Muller, R.; Ranck, J. Head-direction cells recorded from the postsubiculum in freely moving rats. I. Description and quantitative analysis. J. Neurosci. 1990, 10, 420–435. [Google Scholar] [CrossRef]
- Stein, R.B.; Weber, D.J.; Aoyagi, Y.; Prochazka, A.; Wagenaar, J.B.M.; Shoham, S.; Normann, R.A. Coding of position by simultaneously recorded sensory neurones in the cat dorsal root ganglion. J. Physiol. 2004, 560, 883–896. [Google Scholar] [CrossRef]
- Wagenaar, J.B.; Ventura, V.; Weber, D.J. State-space decoding of primary afferent neuron firing rates. J. Neural Eng. 2011, 8, 016002. [Google Scholar] [CrossRef]
- Weber, D.; Stein, R.; Everaert, D.; Prochazka, A. Decoding Sensory Feedback From Firing Rates of Afferent Ensembles Recorded in Cat Dorsal Root Ganglia in Normal Locomotion. IEEE Trans. Neural Syst. Rehabil. Eng. 2006, 14, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Hubel, D.H.; Wiesel, T.N. Receptive fields of single neurones in the cat’s striate cortex. J. Physiol. 1959, 148, 574–591. [Google Scholar] [CrossRef]
- Amari, S. Information Geometry and Its Applications; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Ay, N.; Jost, J.; Lê, H.V.; Schwachhöfer, L. Information Geometry; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Nielsen, F. An Elementary Introduction to Information Geometry. arXiv:180808271 [cs, math, stat]. 2018. Available online: http://arxiv.org/abs/1808.08271 (accessed on 11 August 2019).
- Cencov, N.N. Statistical Decision Rules and Optimal Inference; American Mathematical Society: Providence, RI, USA, 1982. [Google Scholar]
- Liang, T.; Poggio, T.; Rakhlin, A.; Stokes, J. Fisher-Rao Metric, Geometry, and Complexity of Neural Networks. arXiv:171101530 [cs, stat]. 2017. Available online: http://arxiv.org/abs/1711.01530 (accessed on 11 August 2019).
- Cover, T.M.; Thomas, J.A. Elements of Information Theory; Wiley: New Jersey, NJ, USA, 2006. [Google Scholar]
- Amari, S.; Douglas, S.C. Why natural gradient? In Proceedings of the 1998 IEEE International Conference on Acoustics, Speech and Signal Processing, ICASSP ’98 (Cat No98CH36181), Seattle, WA, USA, 12–15 May 1998; pp. 1213–1216. [Google Scholar] [CrossRef]
- Bernacchia, A.; Lengyel, M.; Hennequin, G. Exact natural gradient in deep linear networks and its application to the nonlinear case. Adv. Neural Inf. Process. Syst. 2018, 31, 5941–5950. [Google Scholar]
- Zonghai, S.; Buhai, S. The Projection Adaptive Natural Gradient Online Algorithm for SVM. Available online: https://www.infona.pl//resource/bwmeta1.element.ieee-art-000005573523 (accessed on 9 April 2021).
- Zhang, L.; Cichocki, A.; Amari, S. Natural gradient algorithm for blind separation of overdetermined mixture with additive noise. IEEE Signal Process. Lett. 1999, 6, 293–295. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, H.; Zhong, F. Natural gradient-projection algorithm for distribution control. Optim. Control. Appl. Methods 2008, 30, 495–504. [Google Scholar] [CrossRef]
- Duan, T.; Anand, A.; Ding, D.Y.; Thai, K.K.; Basu, S.; Ng, A.; Schuler, A. NGBoost: Natural Gradient Boosting for Probabilistic Prediction. Available online: http://proceedings.mlr.press/v119/duan20a.html (accessed on 9 April 2021).
- Loeliger, H.-A. An Introduction to factor graphs. IEEE Signal Process. Mag. 2004, 21, 28–41. [Google Scholar] [CrossRef]
- Mirza, M.B.; Adams, R.A.; Mathys, C.; Friston, K.J. Human visual exploration reduces uncertainty about the sensed world. PLoS ONE 2018, 13, e0190429. [Google Scholar] [CrossRef]
- Parr, T.; Markovic, D.; Kiebel, S.J.; Friston, K.J. Neuronal message passing using Mean-field, Bethe, and Marginal approximations. Sci. Rep. 2019, 9, 1889. [Google Scholar] [CrossRef]
- Yedidia, J.S.; Freeman, W.T.; Weiss, Y. Constructing Free-Energy Approximations and Generalized Belief Propagation Algorithms. IEEE Trans. Inf. Theory 2005, 51, 2282–2312. [Google Scholar] [CrossRef]
- Dauwels, J. On Variational Message Passing on Factor Graphs. In Proceedings of the 2007 IEEE International Symposium on Information Theory, Nice, France, 24–29 June 2007; pp. 2546–2550. [Google Scholar] [CrossRef]
- Winn, J.; Bishop, C.M. Variational Message Passing. J. Mach. Learn. Res. 2005, 34, 661–694. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da Costa, L.; Parr, T.; Sengupta, B.; Friston, K. Neural Dynamics under Active Inference: Plausibility and Efficiency of Information Processing. Entropy 2021, 23, 454. https://doi.org/10.3390/e23040454
Da Costa L, Parr T, Sengupta B, Friston K. Neural Dynamics under Active Inference: Plausibility and Efficiency of Information Processing. Entropy. 2021; 23(4):454. https://doi.org/10.3390/e23040454
Chicago/Turabian StyleDa Costa, Lancelot, Thomas Parr, Biswa Sengupta, and Karl Friston. 2021. "Neural Dynamics under Active Inference: Plausibility and Efficiency of Information Processing" Entropy 23, no. 4: 454. https://doi.org/10.3390/e23040454
APA StyleDa Costa, L., Parr, T., Sengupta, B., & Friston, K. (2021). Neural Dynamics under Active Inference: Plausibility and Efficiency of Information Processing. Entropy, 23(4), 454. https://doi.org/10.3390/e23040454







